Abstract
Listeria monocytogenes is a facultative intracellular bacterial pathogen that causes serious disease in immunocompromised individuals, pregnant women, and neonates. Bacterial virulence is mediated by the expression of specific gene products that facilitate entry into host cells and enable bacterial replication; the majority of these gene products are regulated by a transcriptional activator known as PrfA. L. monocytogenes strains containing prfA E77K or prfA G155S mutations exhibit increased expression of virulence genes in broth culture and are hypervirulent in mice. To define the scope of the influences of the prfA E77K and prfA G155S mutations on L. monocytogenes pathogenesis, multiple aspects of bacterial invasion and intracellular growth were examined. Enhanced bacterial invasion of host epithelial cells was dependent on the expression of a number of surface proteins previously associated with invasion, including InlA, InlB, and ActA. In addition to these surface proteins, increased production of the hly-encoded secreted hemolysin listeriolysin O (LLO) was also found to significantly enhance bacterial invasion into epithelial cell lines for both prfA mutant strains. Although prfA E77K and prfA G155S strains were similar in their invasive phenotypes, the infection of epithelial cells with prfA E77K strains resulted in host cell plasma membrane damage, whereas prfA G155S strains did not alter plasma membrane integrity. Bacterial infection of human epithelial cells, in which the production of LLO is not required for bacterial entry into the cytosol, indicated that prfA E77K cytotoxic effects were mediated via LLO. Both prfA E77K and prfA G155S strains were more efficient than wild-type bacteria in gaining access to the host cell cytosol and in initiating the polymerization of host cell actin, and both were capable of mediating LLO-independent lysis of host cell vacuoles in cell lines for which L. monocytogenes vacuole disruption normally requires LLO activity. These experiments illuminate the diverse facets of L. monocytogenes pathogenesis that are significantly enhanced by the constitutive activation of PrfA via prfA mutations and underscore the critical role of this protein in promoting L. monocytogenes virulence.
Listeria monocytogenes is a gram-positive intracellular bacterial pathogen responsible for serious infections in immunocompromised individuals, pregnant women, and neonates (17, 26, 48). The bacterium is capable of invading and replicating within the cytosol of a wide variety of mammalian cell types, including macrophages, fibroblasts, endothelial cells, and epithelial cells (62). Invasion of different cell types is mediated by a number of bacterial surface proteins, including ActA, InlA, and InlB, which bind to specific host cell receptors and initiate signaling cascades to promote bacterial entry (12, 13, 29, 57). After invasion, the bacteria escape from membrane-bound vacuoles into the cell cytosol, where replication occurs followed by spread of the bacteria to infect adjacent cells (44, 60). Vacuole escape is mediated by listeriolysin O (LLO) and two phospholipases, a phosphatidylinositol-specific phospholipase C (PI-PLC) and a broad specificity phospholipase, PlcB (10, 32, 38, 49, 55). Not all of the factors that mediate vacuole lysis are required for vacuole escape in all cell types. For example, while LLO is required for the escape of bacteria from the vacuoles or phagosomes of mouse cell lines and primary macrophages, PlcB is sufficient for mediating bacterial escape in human epithelial cell lines (23, 27, 32, 38, 49). Bacteria that are unable to reach the cytosol of infected host cells do not replicate and are severely attenuated in animal models of infection.
The majority of L. monocytogenes virulence genes that have been identified to date are regulated by a transcriptional activator known as PrfA, a member of the Crp/Fnr family of regulatory proteins (33). PrfA, like Crp and Fnr, is thought to require the binding of a small molecule cofactor or some form of posttranslational modification for full activity (51, 63). PrfA recognizes and binds to a 14-bp palindromic DNA sequence located in the −40 region of target promoters (21). The environmental signal that leads to activation of the PrfA protein and full expression of L. monocytogenes virulence genes has not yet been established, although a number of conditions have been described that significantly alter virulence gene expression (for example, pH, temperature, and available carbon sources) (3, 4, 6, 11, 30, 33, 42, 46). PrfA activation appears to occur within the cytosol of infected host cells, since maximum expression of a number of PrfA-regulated genes is induced within this environment (8, 43, 53).
Previous studies designed to identify gene products that contribute to the induction of intracellular bacterial gene expression led to the identification of two L. monocytogenes mutant strains that contained amino acid substitutions within PrfA that appeared to lock the protein into its activated state (54). Broth-grown cultures of bacteria containing either prfA E77K or prfA G155S mutations exhibited high-level expression of gene products that normally are induced within the cytosol of infected host cells. prfA E77K and prfA G155S mutants were more invasive for epithelial cell lines and were fully virulent in a mouse model of infection, with the prfA G155S mutant exhibiting enhanced virulence (54). We therefore sought to examine the effects of the prfA E77K and prfA G155S mutations on multiple facets of L. monocytogenes pathogenesis to define which aspects of infection were enhanced by the mutational activation of PrfA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in the present study are listed in Table 1. L. monocytogenes 10403S (serotype 1/2a) is resistant to streptomycin, has a 50% lethal dose for mice of 2 × 104 (20), and was the parent strain used for the construction of all mutant strains. L. monocytogenes strains were grown on brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) agar plates or in BHI broth and were stored at −70°C in BHI broth containing 20% glycerol. Escherichia coli strains were grown on Luria-Bertani (LB) agar plates or in LB broth (Invitrogen, Carlsbad, Calif.). Antibiotics were used as indicated at the following concentrations: carbenicillin at 50 μg/ml and chloramphenicol at 10 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Straina | Relevant description | Source or reference |
|---|---|---|
| NF-L476 | L. monocytogenes wild-type strain 10403S with actA-gus-plcB transcriptional fusion | 53 |
| NF-L924 | NF-L476 with prfA E77K | 54 |
| NF-L943 | NF-L476 with prfA G155S | 54 |
| DP-L1942 | actA in-frame deletion | 7 |
| DP-L2161 | hly in-frame deletion | 31 |
| NF-L972 | DP-L1942 with prfA E77K | This work |
| NF-L974 | DP-L1942 with prfA G155S | This work |
| NF-L975 | DP-L2161 with prfA E77K | This work |
| NF-L976 | DP-L2161 with prfA G155S | This work |
| DP-L4405 | inlA in-frame deletion | 2 |
| DP-L4404 | inlA and inlB in-frame deletions | 2 |
| NF-L1012 | DP-L4405 with prfA E77K | This work |
| NF-L1017 | DP-L4404 with prfA G155S | This work |
| DP-L1935 | plcB in-frame deletion | 55 |
| NF-L1051 | plcB in-frame deletion with prfA E77K | This work |
| NF-L1052 | plcB in-frame deletion with prfA G155S | This work |
All strains are derived from L. monocytogenes 10403S.
Construction of L. monocytogenes mutant strain derivatives.
The introduction of the prfA E77K or prfA G155S mutations into L. monocytogenes strains containing in-frame deletions of actA, hly, or inlA or a dual deletion of inlA and inlB was carried out by using allelic exchange as previously described (10, 18). Briefly, pKSV7-derivative plasmid vectors (56) containing the prfA E77K (pNF768) or the prfA G155S (pNF771) mutation (54), along with flanking sequence to facilitate homologous recombination into the correct location within the L. monocytogenes chromosome, were introduced by electroporation into strains DP-L1942 (ΔactA) (7), DP-L2161 (Δhly) (31), DP-L4405 (ΔinlA) (2), and DP-L4404 (ΔinlA ΔinlB) (2). In-frame deletions within plcB in L. monocytogenes prfA E77K (NF-L924) and prfA G155S (NF-L948) strains were constructed by using plasmid pDP1888 (55), which contains an in-frame deletion of plcB in conjunction with DNA flanking sequences for allelic exchange via homologous recombination. For all strains single-copy gene replacement was carried out by allelic exchange as described previously (10, 18). Introduction of ΔplcB into NF-L1051 (prfA E77K actA-gus ΔplcB) and NF-L1052 (prfA G155S actA-gus ΔplcB) was verified by confirmation of loss of lecithinase activity on egg yolk agar. All mutations were verified by DNA sequence analysis of PCR products derived from L. monocytogenes genomic DNA.
Detection of LLO and PlcB activity.
Stationary-phase bacteria were diluted 1:10 into BHI medium and grown at 37°C for 5 h with shaking. The supernatant fluid was assayed for hemolytic activity as previously described (9). plcB-dependent phospholipase production was visualized by using an egg yolk overlay agar plate assay. Chicken egg yolk was added in a 1:1 (vol/vol) ratio to phosphate-buffered saline (PBS) and vortexed to generate a suspension. Then, 5 ml of egg yolk suspension was added to 100 ml of molten LB medium (45°C to 48°C) and 3 ml of egg yolk-agar suspension was overlaid onto an LB agar plate. After solidification of the medium, bacterial strains were gently streaked onto the surface of the plate and incubated at 37°C for 48 h. Phospholipase activity was detected as a zone of opacity surrounding bacterial streaks.
L. monocytogenes intracellular growth in Henle 407 and PtK2 epithelial cell lines.
The cell lines used in these studies were the human-derived epithelial cell line Henle 407 and the Potoroo tridactylis kidney epithelial cell line PtK2 maintained as previously described (55, 58). L. monocytogenes infection of Henle 407 or PtK2 cells grown as monolayers on acid-washed coverslips was carried out as previously described (54). Briefly, L. monocytogenes cultures were grown at 37°C overnight and washed with PBS prior to infection. After 60 min of infection, cell monolayers were washed three times with 37°C PBS, followed by the addition of 37°C Dulbecco modified Eagle medium (Gibco-BRL, Rockville, Md.) plus 10% fetal bovine serum. Gentamicin was then added at a final concentration of 10 μg/ml to kill any remaining extracellular bacteria. Coverslips were removed at the indicated time points postinfection, cell monolayers on each coverslip were lysed by vortexing for 10 s in 5 ml of sterile water, and dilutions of the cell lysate were plated on LB plates. Bacterial CFU were determined after overnight incubation at 37°C on LB plates for each strain in triplicate.
L. monocytogenes association with host cell actin.
L. monocytogenes strains were grown to stationary phase at 37°C overnight in BHI broth. Bacteria were washed with PBS prior to infection of Henle 407 and PtK2 cell monolayers as described above. A multiplicity of infection (MOI) of ca. 300:1 was used for the infection of host cells with strain DP-L2161 (Δhly) and an MOI of ca. 30:1 for NF-L1051 and NF-L1052. The difference in MOIs used for DP-L2161 versus NF-L1051 and NF-L1052 strains was used to enhance the detection of any DP-L2161 bacteria that might have gained access to the host cytosol (NF-L1051 and NF-L1052 strains are more efficient than DP-L2161 at mediating cytosol entry and actin polymerization; hence, these strains were easily detected associated with actin at 7 h postinfection.) Coverslips containing infected cell monolayers were removed at 7 h postinfection and processed for fluorescence microscopy as follows: cell monolayers were fixed by placing a drop of 3.7% formaldehyde in PBS on the coverslip, followed by incubation at room temperature for 10 min. Coverslips were washed by dipping into sterile PBS, and then host cells were permeabilized with 100 μl of PBS plus 0.1% Triton X-100 for 10 min, followed by an additional PBS wash. An aliquot (100 μl) of NBD-phallacidin (Molecular Probes, Eugene, Oreg.) diluted 1:20 into PBS was placed on each coverslip for 20 min at room temperature, and coverslips were then washed by dipping into PBS. A 100-μl aliquot of rabbit polyclonal anti-Listeria antibody (Difco) diluted 1:320 in PBS was then added to each coverslip, followed by a 30-min incubation at room temperature. Tetramethylrhodamine goat anti-rabbit immunoglobulin G (H + I) conujugate (Molecular Probes) was used as a secondary antibody to visualize bacteria. After incubation with the secondary antibody, the coverslips were rinsed in PBS, mounted in Permafluor mounting medium (Immunon, Pittsburgh, Pa.) and allowed to cure in the dark overnight. Bacteria were counted in association with at least 50 randomly chosen infected host cells in at least 10 different fields. Infected cells were assigned into one of two categories: those with five or fewer bacteria associated per host cell and those with >5 bacteria associated per host cell. The two categories were established to facilitate a working distinction between cells in which bacterial replication was likely to have occurred by 7 h postinfection (cells with >5 bacteria associated per cell) and cells in which bacterial replication was less likely to have occurred (cells with ≤5 bacteria associated per cell). Bacteria were considered to be positive for actin association if they were surrounded by actin (actin halos, an early step in polymerization) or were associated with actin tails.
Detection of Henle 407 cell membrane perturbation after infection with L. monocytogenes.
Henle 407 human epithelial cells were grown as cell monolayers on acid-washed coverslips for 2 days prior to infection. L. monocytogenes bacterial cultures were grown to stationary phase in BHI broth culture at 37°C overnight, washed with PBS, and then used to infect cell monolayers at the indicated MOI (Table 2). At 1 h postinfection, coverslips were washed three times with PBS, and epithelial cell membrane integrity was assessed by using the LIVE/DEAD Viability/Cytotoxicity Kit for animal cells (Molecular Probes) according to the protocol recommended by the manufacturer. Briefly, coverslips were incubated at room temperature for 20 to 30 min with 100 μl of a solution generated by the addition of 2 μl of ethidium homodimer and 0.5 μl of calcein AM to 1 ml of PBS. After incubation, coverslips were inverted onto glass slides, the edges were sealed with nail polish, and cells were observed for fluorescence within 1 h with a Nikon Microphot FX microscope equipped with a Photometrics Sensys camera and META-MORPH software. At least 50 cells were examined in 20 independent fields, and each experiment was repeated three times.
TABLE 2.
Assessment of host cell plasma membrane integrity in infected Henle 407 epithelial cellsa
| Strain genotype | MOI | Permeability to ethidium homodimerb |
|---|---|---|
| Wild type | 938:1 | +++ |
| 462:1 | + | |
| 231:1 | − | |
| Δhly | 1,375:1 | − |
| 938:1 | − | |
| ΔplcB | 938:1 | +++ |
| 462:1 | + | |
| 231:1 | − | |
| prfA E77K | 137:1 | +++ |
| 93:1 | + | |
| 46:1 | − | |
| prfA G155S | 938:1 | +++ |
| 462:1 | + | |
| 231:1 | − | |
| Δhly prfA E77K | 1,375:1 | − |
| 938:1 | − | |
| ΔplcB prfA E77K | 137:1 | +++ |
| 93:1 | + |
Monolayers of Henle 407 cells were infected with bacteria for 1 h. Host cell plasma membrane permeability to ethidium homodimer was visually assessed by fluorescence microscopy. At least 50 total cells were examined in more than 10 independent fields. The results are presented from three independent experiments.
+++, >95% host cells stained positive for ethidium homodimer; +, <25% of host cells stained positive; −, no host cells stain positive.
RESULTS
PrfA-regulated surface proteins contribute to L. monocytogenes prfA E77K and prfA G155S hyper-invasion.
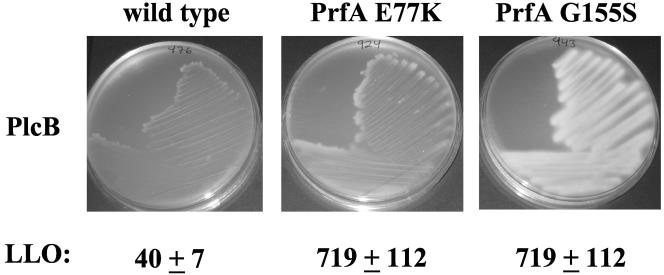
The expression of multiple PrfA-dependent gene products has been shown to be elevated in strains containing prfA E77K or prfA G155S mutations (54). LLO-associated hemolytic activity was increased ∼17-fold for both mutant strains, and increased secretion of PlcB-associated phospholipase was readily detectable (Fig. 1). Interestingly, although the prfA G155S strain secreted levels of hemolytic activity comparable to those of prfA E77K strains, much greater levels of PlcB-associated phospholipase activity were produced. The expression of actA, inlA, and inlB has been shown to be significantly increased in prfA E77K and prfA G155S mutants (54), and each of these surface proteins has been shown to contribute to the invasion of L. monocytogenes into specific cell types (5, 22, 41, 57). To assess the relative contributions of ActA, InlA, and InlB to the increased invasive capacity observed for the L. monocytogenes prfA mutants in epithelial cell lines, in-frame deletions of actA and inlA and a double inlA inlB deletion mutation were introduced into prfA E77K and prfA G155S strains, and bacteria were monitored for their ability to invade and replicate within two cell lines routinely used for L. monocytogenes cellular infections, Henle 407 human intestinal epithelial cells and PtK2 Potoroo tridactylis kidney epithelial cells (14, 27, 34, 35, 38, 52, 65).
FIG. 1.
Increased production of LLO and PlcB by L. monocytogenes prfA E77K and prfA G155S strains. PlcB-associated phospholipase activity was assessed for wild type, prfA E77K, and prfA G155S strains after 48 h of culture growth on LB plates overlaid with 5% egg yolk. Phospholipase activity is detectable as zones of opacity surrounding bacterial streaks. LLO-associated hemolytic activity was determined from bacterial culture supernatants and is expressed as the reciprocal of the dilution at which 50% lysis of sheep erythrocytes was observed.
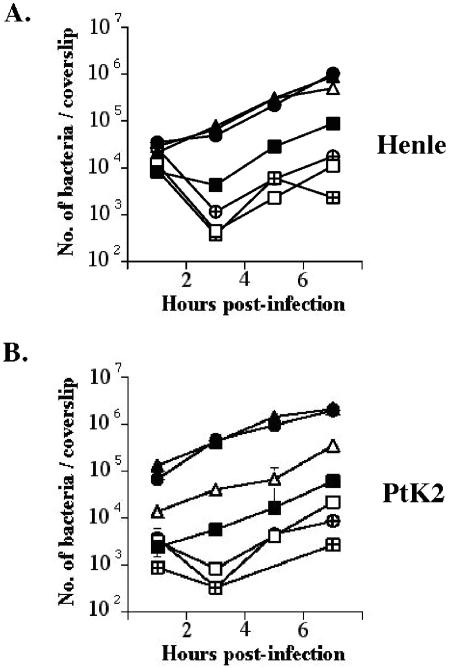
Differences in the invasive capacity of the mutant strains were measured at 3 h postinfection; bacterial CFU recovered at this time point primarily represent bacterial invasion versus intracellular replication since the intracellular doubling times of the L. monocytogenes mutant strains were equivalent to wild type (ca. 80 min). The loss of the inlA gene product or of both inlA and inlB reduced the invasive capacity of L. monocytogenes strains containing wild-type prfA for human epithelial cells by approximately 10-fold (Fig. 2A). ΔinlA strains were also reduced in their invasive capacity for PtK2 cells (Fig. 2B), and the ΔinlA ΔinlB double mutant was slightly less invasive than the ΔinlA single mutant. In contrast, the loss of the inlA gene product in the prfA G155S mutant strain resulted in no significant decrease in bacterial invasion of Henle 407 cells but exhibited an approximately 10-fold decrease in invasion of PtK2 cells (Fig. 2). The ΔinlA ΔinlB prfA E77K strain was dramatically less invasive than the prfA E77K parent strain in both cell types (Fig. 2). These results strongly suggest that the increased expression of the inlB gene product was a major contributor to the increased invasive capacity of the prfA E77K mutant strain for Henle 407 cells and that both InlA and InlB contributed to bacterial invasion of PtK2 cells.
FIG. 2.
Intracellular growth of L. monocytogenes ΔinlA and ΔinlA ΔinlB strains in Henle 407 cells (A) and PtK2 cells (B). L. monocytogenes strains were grown to stationary phase in BHI broth culture and used to infect monolayers of epithelial cells grown on glass coverslips at an MOI of ca. 30:1. At 1 h postinfection, monolayers were washed with PBS, gentamicin was added to kill any extracellular bacteria and, at the indicated time points cell monolayers were lysed and the number of bacteria per coverslip was determined by plating on solid media. The results are expressed as the mean number of bacterial CFU ± the standard error for three coverslips per time point. The findings for one of three experiments with similar results is shown. Symbols: ▪, NF-L476 (wild type); □, DP-L4405 (ΔinlA); ⊞, DP-L4404 (ΔinlA ΔinlB); •, NF-L924 (prfA E77K); ▴, NF-L943 (prfA G155S); ⊕, NF-L1012 (ΔinlA ΔinlB prfA E77K); ▵, NF-L1017 (ΔinlA prfA G155S).
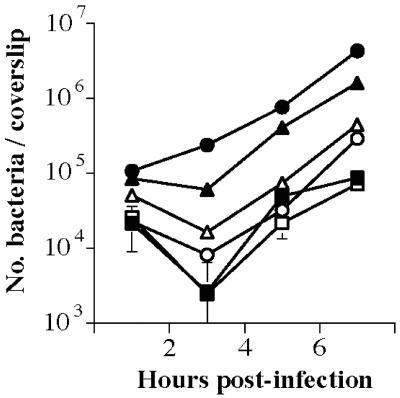
The PrfA-regulated surface protein ActA has also been reported to facilitate L. monocytogenes invasion of host cells through interactions with a heparin sulfate-binding protein (1, 57). Although no significant decrease in invasion was observed for strains lacking actA in the presence of the wild-type prfA allele (Fig. 3), both ΔactA prfA G155S and ΔactA prfA G155S strains were found to have reduced invasive capacity for human epithelial cells but still remained significantly more invasive than wild-type bacteria (Fig. 3). These data support a role for ActA in invasion when the protein is expressed at high levels on the bacterial cell surface, a situation that might occur if cytosolic bacteria were released into the extracellular milieu after host cell lysis.
FIG. 3.
Intracellular growth of L. monocytogenes ΔactA strains in human epithelial cells. L. monocytogenes strains grown to stationary phase in BHI broth culture were used to infect monolayers of Henle 407 epithelial cells grown on coverslips at an MOI of ca. 30:1. At 1 h postinfection, monolayers were washed with PBS, gentamicin was added to kill any extracellular bacteria and, at the indicated time points, the monolayers were lysed and the number of bacteria per coverslip was determined. The results are expressed as the mean number of bacterial CFU ± the standard error for three coverslips per time point. The results for one of three experiments with similar findings are shown. Symbols: ▪, NF-L476 (wild type); □, DP-L1942 (ΔactA); •, NF-L924 (prfA E77K); ○, NF-L972 (ΔactA prfA E77K); ▴, NF-L943 (prfA G155S); ▵, NF-L974 (ΔactA prfA G155S).
LLO increases the invasive capacities of prfA E77K and prfA G155S strains.
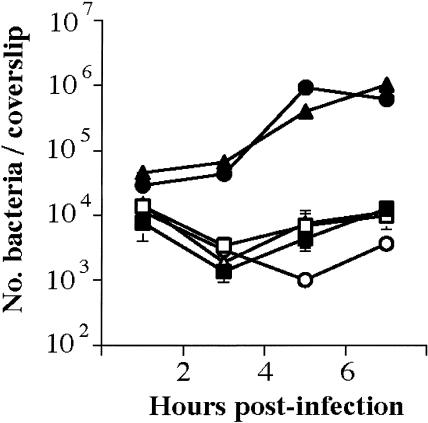
Recent reports have implicated a role for LLO in mediating bacterial invasion of host epithelial cells (16). LLO has been reported to induce the mobilization of extracellular Ca2+ into host cells and to activate downstream Ca2+-dependent signaling pathways that are required for efficient cell invasion (16). To investigate the influences of LLO on the invasion and intracellular growth of prfA mutant strains within Henle 407 human epithelial cells, monolayers were infected with wild-type, prfA E77K, and prfA G155S L. monocytogenes strains and the corresponding hly deletion strains, and bacterial CFU were monitored at specific time points postinfection (Fig. 4) (LLO is not required for bacterial escape from the vacuoles of Henle 407 cells [38]). No significant differences were observed between wild-type and Δhly strains for invasion and/or intracellular replication within Henle 407 cells (Fig. 4). Both prfA E77K and prfA G155S strains were hyperinvasive for host cells; however, invasion was reduced to wild-type levels for both strains in the absence of LLO (Fig. 4). LLO augmentation of invasion was therefore much greater for the mutants than for bacteria expressing wild-type levels of the protein.
FIG. 4.
Intracellular growth of L. monocytogenes Δhly strains in Henle 407 human epithelial cells. L. monocytogenes strains were grown to stationary phase in BHI broth culture and used for the infection of Henle 407 epithelial cells grown on glass coverslips at an MOI of ca. 30:1. At 1 h postinfection, monolayers were washed with PBS, gentamicin was added to kill any extracellular bacteria, and at the indicated time points monolayers were lysed and the number of bacteria per coverslip was determined. The results are expressed as the mean number of bacterial CFU ± the standard error for three coverslips per time point. The results for one of three experiments with similar findings are shown. Symbols: ▪, NF-L476 (wild type); □, DP-L2161 (Δhly); •, NF-L924 (prfA E77K); ○, NF-L975 (Δhly prfA E77K); ▴, NF-L943 (prfA G155S); ▵, NF-L976 (Δhly prfA G155S).
L. monocytogenes prfA E77K damages host cell plasma membranes.
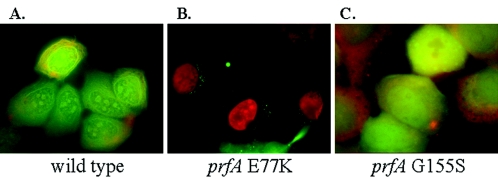
It was observed in several experiments that bacterial CFU recovered from either Henle 407 or PtK2 cells infected with prfA E77K decreased beginning ca. 7 h postinfection (Fig. 4) (K. Mueller, unpublished data). A similar phenomenon has been reported for several other L. monocytogenes mutant strains (15, 25, 37), and in each case the mutant strain was shown to be cytotoxic to the infected host cell, resulting in the loss of host cell membrane integrity and an influx of gentamicin that led to bacterial killing. To determine whether bacterial infection with prfA E77K strains affected host cell membrane integrity, Henle epithelial cells were infected with wild type, prfA E77K, or prfA G155S bacterial strains and monitored for cell viability (esterase activity) or loss of membrane integrity (influx of ethidium homodimer) (Fig. 5). Bacterial infection of Henle 407 cells with either wild-type L. monocytogenes or the prfA G155S mutant resulted in no visible change in cell membrane permeability or cell viability, as indicated by the ability of infected cells to metabolize calcein AM and exclude ethidium homodimer (Fig. 5). When equivalent numbers of prfA E77K bacteria were used to infect Henle cells, evidence of host cell cytotoxicity was observed. Infected host cells were permeable to the ethidium homodimer, as evidenced by nuclear staining at 2 h postinfection (Fig. 5). The L. monocytogenes prfA E77K mutant did not appear, however, to kill all infected host cells at 2 h postinfection since the examination of monolayers at 5 h postinfection revealed that >95% of the infected cells exhibited full plasma membrane integrity, as evidenced by the exclusion of ethidium homodimer (Mueller, unpublished). These observations suggested that cells infected with prfA E77K initially sustained damage to the plasma membrane but that this damage was reparable. At 7 h postinfection, monolayers of prfA E77K-infected cells once again exhibited signs of cytotoxicity, leading to disruption of the monolayer (Mueller, unpublished). Plasma membrane damage was not evident in J774 mouse macrophage-like cells infected with prfA E77K, and the prfA E77K strain formed plaques equivalent in size to those formed by wild-type L. monocytogenes in mouse L2 fibroblast cell monolayers, even in the presence of high concentrations of gentamicin (50 μg/ml) (54; Mueller, unpublished). Host cell membrane perturbation was observed for L. monocytogenes prfA E77K strains after the infection of PtK2 epithelial cells, whereas cells infected with either wild-type or prfA G155S retained the ability to exclude ethidium homodimer (Mueller, unpublished). These data suggest that the membrane perturbations induced by prfA E77K are most readily detected in epithelial cells. Since prfA E77K and prfA G155S strains are similar to one another in their ability to invade epithelial cells (Fig. 2 and 3), the membrane damage associated with prfA E77K cannot be ascribed solely to increased numbers of intracellular bacteria.
FIG. 5.
Assessment of host plasma membrane integrity in cells infected with L. monocytogenes prfA mutants. L. monocytogenes strains were grown to stationary phase in BHI broth culture and used for the infection of monolayers of Henle epithelial cells grown on coverslips at an MOI of ca. 100:1. At 1 h postinfection the cells were washed with PBS, gentamicin was added to kill any extracellular bacteria, and coverslips were stained for cell viability and plasma membrane integrity as described in Materials and Methods. Green fluorescence is indicative of cell esterase activity. The nucleic acids of cells with damaged plasma membranes fluoresces red due to the incorporation of ethidium homodimer.
Two PrfA-dependent gene products have previously been associated with host cell cytotoxicity. Bacteria expressing altered forms of LLO (15, 25, 37) or high levels of PlcB within the cytosol of infected host cells (39) have been shown to elicit plasma membrane damage. To determine whether altered expression of LLO and/or PlcB by the prfA E77K strain was responsible for host cell membrane perturbations, prfA mutant strains containing deletions in hly or plcB were constructed and used for the infection of Henle 407 epithelial cells. Loss of host cell plasma membrane integrity was monitored at 2 h postinfection in response to increasing numbers of bacteria used for infection (Table 2). For wild-type L. monocytogenes, an MOI of 100:1 generally resulted in approximately one intracellular bacterium per epithelial cell (K. Mueller and N. Freitag, unpublished data). Host cell membrane damage was only observed at very high MOI (>900:1) for wild-type L. monocytogenes or for prfA G155S strains (Table 2). No plasma membrane damage was observed when cells were infected with either strain at an MOI of 231:1. Membrane damage was dependent upon the expression of LLO; the MOI could be increased to more than 1,000 CFU per cell in the absence of LLO, with no apparent host cell membrane disruption (Table 2). In contrast, the infection of Henle 407 cells with prfA E77K at an MOI of 137:1 resulted in host cell membrane damage for >95% of the cells within the monolayer. The elimination of plasma membrane perturbations elicited by prfA E77K strains required the use of an MOI as low as 46:1 (Table 2). prfA E77K-associated damage to the host cell plasma membrane was dependent upon the presence of LLO and was not significantly influenced by PlcB. These data indicate that the deregulation of LLO production by PrfA E77K contributes to the transient membrane damage inflicted by L. monocytogenes prfA E77K mutants on infected host cells. Since prfA E77K and prfA G155S produced equivalent levels of LLO activity during growth in broth culture (both in BHI broth and in BHI broth treated with activated charcoal, a condition that induces hly expression) (Fig. 1) (Mueller, unpublished), these data suggest that prfA E77K strains may differ from prfA G155S strains with respect to production of LLO within the cytosol. Alternatively, prfA E77K strains may differentially express a separate factor that works in concert with LLO to promote membrane damage in infected host cells.
prfA E77K and prfA G155S strains are enhanced for escape from host cell vacuoles.
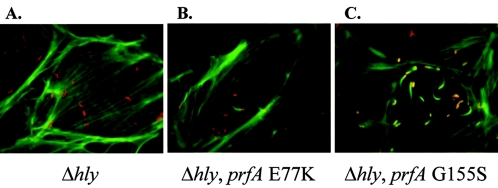
While investigating the effects of LLO production on prfA mutant invasion, bacterial infections of PtK2 cells were monitored in addition to the infections of human epithelial cell lines. Surprisingly, it was observed that in contrast to the infection of PtK2 cells with Δhly strains, in which bacteria remained trapped within host cell vacuoles, Δhly prfA E77K and Δhly prfA G155S mutant strains were capable of vacuolar escape and cytosolic replication in PtK2 cells (Fig. 6). prfA E77K and prfA G155S mutants associated with actin tails within the cytosol of infected PtK2 cells were readily detectable (Fig. 6). To quantitatively assess the ability of the prfA mutants to mediate escape from PtK2 vacuoles in the absence of LLO production, PtK2 cells were infected with L. monocytogenes Δhly, Δhly prfA E77K, and Δhly prfA G155S bacterial strains and then examined for cytosolic localization by monitoring for bacterial association with host cell actin (Table 3).
FIG. 6.
Cytosolic localization of Δhly prfA E77K and Δhly prfA G155S in PtK2 epithelial cells. L. monocytogenes strains were grown to stationary phase in BHI broth culture and used to infect PtK2 epithelial cells grown on glass coverslips at an MOI of 300:1 (DP-L2161 [Δhly]) or 30:1 (NF-L975 [Δhly prfA E77K] and NF-L976 [Δhly prfA G155S]). At 1 h postinfection the monolayers were washed with PBS, and gentamicin was added to kill any extracellular bacteria. At 5 h postinfection (Δhly prfA E77K and Δhly prfA G155S strains) and 7 h postinfection (Δhly strain) monolayers were fixed, permeabilized, and stained for L. monocytogenes with an anti-Listeria antibody and tetramethylrhodamine goat anti-rabbit conjugated secondary antibody to detect bacteria (red) and with NBD-phallacidin to detect F-actin (green). No DP-L2161 were detected associated with host cell actin, despite the increased numbers of bacteria used for infection. In contrast, F-actin comet tails were found associated with Δhly prfA E77K and Δhly prfA G155S, indicating entry of these strains into the host cell cytosol.
TABLE 3.
Vacuole escape and actin association of Δhly prfA E77K and Δhly prfA G155S bacterial strains in Henle 407 and PtK2 epithelial cellsa
| Cell type and bacterial strain genotypea | Mean % cells or bacteria ± SE
|
|||
|---|---|---|---|---|
| Host cells with ≤5 bacteriab | Actin-positive bacteriac | Host cells with >5 bacteriab | Actin-positive bacteriac | |
| P t K2 cells | ||||
| Δhly | 75 ± 9 | 0 ± 0 (263) | 25 ± 9 | 0 ± 0 (402) |
| Δhly prfA E77K | 86 ± 5 | 4 ± 2 (271) | 14 ± 5 | 52 ± 19 (269) |
| Δhly prfA G155S | 66 ± 8 | 3 ± 1 (236) | 34 ± 8 | 47 ± 10 (855) |
| Henle cells | ||||
| Δhly | 87 ± 1 | 1 ± 1 (294) | 13 ± 1 | 51 ± 1 (362) |
| Δhly prfA E77K | 84 ± 6 | 3 ± 0 (259) | 16 ± 6 | 76 ± 4 (984) |
| Δhly prfA G155S | 65 ± 4 | 19 ± 11 (181) | 35 ± 4 | 84 ± 5 (1,333) |
Monolayers of Henle 407 and PtK2 cells were infected with L. monocytogenes grown to stationary phase as described in Materials and Methods. An MOI of 300:1 was used for Δhly strains to facilitate the identification of any cytosolic bacteria; an MOI of 30:1 was used for the mutant prfA constructs. At 7 h postinfection, monolayers were fixed and stained for L. monocytogenes with an anti-Listeria antibody and tetramethylrhodamine goat anti-rabbit antibody to detect bacteria and NBD-phallacidin to detect actin.
At least 50 PtK2 or Henle host cells were counted in at least 10 different microscopic fields. The percentages reflect the mean percentages of host cells associated with either ≤5 bacteria per cell or >5 bacteria per cell ± the standard error. The results are reported from at least three independent experiments.
Bacteria associated with host cells were scored for association with host cell actin. The numbers reflect the percentages of associated bacteria that colocalized with F-actin or were associated with actin tails. The numbers given in parenthesis within the table reflect total numbers of cell-associated bacteria examined. The results are expressed as the means ± the standard errors for three separate experiments.
PtK2 cells were examined at 7 h postinfection, and infected cells were assigned to one of two populations to facilitate comparison: those with ≤5 bacteria per cell and those with >5 bacteria per cell. PtK2 cells with ≤5 bacteria per cells were considered as likely to reflect populations of cells in which bacteria were either inefficient at escape or unable to escape cell vacuoles and thus gain access to the cytosol. Consistent with this assumption, this population was largest for cells infected with the L. monocytogenes Δhly strain, and no Δhly bacteria were observed associated with F-actin (Table 3). A large number of PtK2 cells infected with Δhly prfA E77K or Δhly prfA G155S also contained ≤5 bacteria; however, a small percentage of these cells contained bacteria associated with actin (ca. 4% of cell-associated bacteria) (Table 3). A significant population of Δhly prfA E77K or Δhly prfA G155S strain-infected cells (14 to 34%) contained >5 bacteria and, of these, 50% of the bacteria associated with the cells stained positive for F-actin (Table 3). These results indicate that although the loss of LLO activity reduces the overall efficiency of vacuolar escape for the prfA mutant strains, the presence of the prfA mutations confers upon the bacteria the ability to gain access to the cytosol in a cell line in which bacterial escape is normally strictly dependent upon LLO. LLO-independent escape of the prfA mutant strains was not observed in the mouse macrophage-like cell line J774, and the mutants failed to form plaques indicative of intracellular growth and cell-to-cell spread in the murine L2 fibroblast cell line (Mueller, unpublished), indicating that the vacuoles of these cell types remained resistant to lysis in the absence of LLO production.
The ability of Δhly prfA E77K and Δhly prfA G155S strains to enter the cytosol of PtK2 cells suggested that these strains had an enhanced capacity for vacuole escape that might also be expected contribute to more efficient vacuole membrane lysis in other cell types. To examine this possibility, Henle 407 human epithelial cells were infected with Δhly, Δhly prfA E77K, or Δhly prfA G155S strains, and the bacteria were monitored for association with F-actin. Twenty to thirty percent more Δhly prfA E77K and Δhly prfA G155S bacteria were observed associated with F-actin in comparison to Δhly strains (Table 3). The increase in the numbers of bacteria associated with F-actin for the mutant strains probably reflects not only an increased efficiency in bacterial escape from host cell vacoules (as suggested by bacterial escape in PtK2 cells) but also the immediate presence of ActA on the bacterial cell surface that would alleviate the need for cytosolic induction of ActA expression.
DISCUSSION
The expression of gene products required for L. monocytogenes virulence is regulated by the activation state of PrfA. In the absence of PrfA, the expression of key virulence gene products is effectively eliminated and L. monocytogenes is essentially avirulent (20, 36, 40). The constitutive activation of PrfA via mutation results in the expression of virulence genes under conditions that normally tightly repress expression, and this in turn appears to increase bacterial virulence in animal models of infection (54). The work presented here underscores the diversity and scope of processes underlying L. monocytogenes interactions with host cells that are influenced and enhanced by PrfA activation.
ActA, InlA, and InlB are PrfA-regulated gene products with demonstrated contributions to bacterial invasion of epithelial cells (1, 5, 13, 22, 28, 29, 41, 57). Expression of these bacterial surface proteins is upregulated in prfA E77K and prfA G155S strains, and both prfA mutant strains exhibited increased invasive capacity for epithelial cells (54) (Fig. 2). Both InlA and InlB were found to contribute to the increased invasive capacity of the L. monocytogenes prfA mutant strains, and the important role of these two surface proteins in host cell invasion has been well documented for many cell types (12, 13). Less obvious is the relevance of the contributions made to invasion by ActA. ActA is normally expressed at low to undetectable levels in standard broth culture and prior to bacterial entry into host cells (19, 43, 53). Once L. monocytogenes reaches the cytosol, actA expression increases several hundredfold and becomes one of the most predominant bacterial surface proteins (7, 19, 43, 53). As a result of its expression patterns, ActA appears to contribute little to bacterial invasion of tissue culture cells under normal conditions (Fig. 3). Its contributions are more pronounced when expressed at high levels on the bacterial surface (57), as evidenced by its contributions to invasion for prfA E77K and prfA G155S strains (Fig. 3). It is tempting to speculate that augmentation of bacterial invasion as mediated by ActA may play a role if infected cells containing cytosolic L. monocytogenes are lysed and bacteria are suddenly released into the extracellular milieu. Under these conditions, ActA might help to promote rapid bacterial entry into nearby cells and reduce L. monocytogenes's vulnerability to humoral host defenses.
Perhaps most striking was the enhancement of invasion observed in response to high-level expression of the hly-encoded gene product LLO (Fig. 4). A recent report by Dramsi and Cossart (16) has described LLO-enhanced invasion of the human epithelial cell line Hep-2, and has demonstrated that LLO induces the mobilization of extracellular Ca2+ into Hep-2 cells to activate Ca2+-dependent signaling pathways that are required for efficient cell invasion. Our results confirm the contributions of LLO to host cell invasion when the protein is expressed at high levels, since L. monocytogenes prfA mutants were 10-fold more invasive in the presence of LLO expression than in its absence (Fig. 4). It is interesting that LLO appears to mediate an opposite effect on bacterial invasion in macrophage cells. Wadsworth and Goldfine (64) reported that LLO delayed the entry of L. monocytogenes into the mouse macrophage-like cell line J774. It appears therefore that L. monocytogenes in some instances promotes its entry into selected cell types (such as epithelial cells) while delaying its entry into others (such as macrophages). Although macrophages are considered a significant portal of entry for L. monocytogenes into animal hosts (61), activated macrophages have been shown to effectively kill L. monocytogenes (45, 50), thus they would be just the type of cell that L. monocytogenes would do well to avoid.
Strains containing prfA E77K or prfA G155S mutations were enhanced for bacterial escape from host cell vacuoles and actin polymerization (Fig. 6 and Table 3). The enhancement for vacuolar escape was most evident after the infection of PtK2 epithelial cells with the Δhly prfA E77K and Δhly prfA G155S mutant strains. L. monocytogenes that lack the hly-encoded gene product LLO are normally unable to mediate lysis of vacuole membranes in murine cells and within PtK2 cells. The ability of the prfA mutants to reach the cytosol of PtK2 cells in the absence of LLO indicates that the increased expression of other PrfA-dependent gene products, such as perhaps PlcB and PlcA, is capable of mediating lysis of some host cell vacuoles. This enhanced escape ability is not observed for all cell types, as witnessed by the inability of these strains to escape from the vacuoles of mouse J774 macrophage-like cells or mouse L2 fibroblast cells (Mueller, unpublished). These observations imply that the composition of the vacuole membrane differs between PtK2 cells and J774 or L2 cells. It has previously been a puzzle as to why L. monocytogenes can mediate escape from the vacuole in the absence of LLO in certain cell types, such as human epithelial cells, fibroblasts, and dendritic cells (47, 49), but not in other cell types. It is possible that the vacuoles of cells such as J774 macrophages or L2 fibroblasts differ in membrane composition from those present in human cell lines or that the vacuole environment of human cells induces expression of phospholipases that are not induced within the vacuoles of other cell types. The ability of the prfA E77K and prfA G155S mutant strains to mediate LLO-independent vacuole lysis in PtK2 cells indicates that PrfA activation is sufficient for vacuole membrane disruption in PtK2 cells in the absence of LLO. The failure of the prfA mutant strains to mediate vacuole lysis within J774 or L2 cells suggests that these vacuole membranes are sufficiently different in composition to prevent lysis in spite of PrfA activation.
L. monocytogenes strains containing the prfA E77K mutation differed from bacteria containing the prfA G155S in that the former strain produced signs of plasma membrane disruption in infected host cells, whereas the latter strain did not (Fig. 5 and Table 2). prfA E77K-dependent host cell membrane damage was linked to LLO expression, an observation that suggests that the two prfA mutants may differ with respect to the amounts of LLO (or factors that work in concert with LLO) produced by bacteria within host cells. prfA E77K strains secreted amounts of hemolytic activity that were equivalent to prfA G155S strains in broth culture; however, prfA G155S strains exhibited higher levels of expression of actA and plcB (Fig. 1) (54). It is clear that the E77K and G155S substitutions do not result in equivalent alterations of PrfA function and, indeed, the mutations are located in structurally different locations within the protein. The G155S mutation is located near the DNA-binding helix-turn-helix motif within PrfA, whereas E77K is located within a region near the PrfA dimer interface located opposite of the DNA-binding region (59). It is possible that the G155S alters PrfA function by inducing conformational changes in PrfA that expose the DNA-binding region of PrfA and enhance the ability of the protein to bind target promoters, as has been suggested for analogous substitutions within the cyclic AMP receptor protein (24). The E77K mutation may influence a separate aspect of PrfA activation, possibly by enhancing dimer formation. Biochemical and structural analyses of the effects of PrfA mutations on various aspects of PrfA function are currently in progress.
In summary, it is clear that multiple aspects of L. monocytogenes pathogenesis are enhanced by the constitutive activation of PrfA. The prfA E77K and prfA G155S mutations (i) increased bacterial invasion of epithelial cells in a manner dependent upon the expression of several PrfA-regulated proteins (InlA, InlB, ActA, and LLO), (ii) enhanced escape from the vacuoles of epithelial cells and enabled escape from certain vacuoles in the absence of LLO, and (iii) facilitated rapid bacterial association with host cell actin. Each of these activities should serve to promote L. monocytogenes replication and survival within host cells and together may translate into the enhancement of virulence observed for prfA E77K and prf G155S mutant strains in mice (54). The fact that L. monocytogenes strains containing constitutively activated mutant alleles of prfA have distinct phenotypes in vitro and in vivo suggests that in specific environments PrfA is normally inactive or may function at a lower level of activity than that observed for the mutationally activated PrfA proteins. Overexpression of virulence genes by prfA E77K and prfA G155S strains serves as a useful tool for gaining insight into L. monocytogenes pathogenesis and the regulation of virulence genes in vivo. Future work will focus on defining exactly how the E77K and G155S mutations influence PrfA conformational structure, thereby altering the biochemistry of PrfA-catalyzed activities.
Acknowledgments
We thank Hao Shen and Jeff Miller for the gift of the L. monocytogenes inlA and inlA inlB deletion strains and Daniel Portnoy for providing DP-L2161 and DP-L1942. We thank Helene Marquis for providing the plcB deletion plasmid pDP1888 and strain DP-L1935. We also thank members of the Freitag laboratory for helpful discussions.
This study was supported by Public Health Service grant AI41816 (N.E.F.) from the National Institutes of Health and by the M. J. Murdock Charitable Trust.
Editor: V. J. DiRita
REFERENCES
- 1.Alvarez-Dominguez, C., J. A. Vazquez-Boland, E. Carrasco-Marin, P. Lopez-Mato, and F. Leyva-Cobian. 1997. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 65:78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakardjiev, A. I., B. A. Stacy, S. J. Fisher, and D. A. Portnoy. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect. Immun. 72:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 5.Braun, L., H. Ohayon, and P. Cossart. 1998. The InIB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol. Microbiol. 27:1077-1087. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubert, A., Z. Sokolovic, S.-K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261:323-336. [DOI] [PubMed] [Google Scholar]
- 9.Camilli, A., C. R. Paynton, and D. A. Portnoy. 1989. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc. Natl. Acad. Sci. USA 86:5522-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conte, M. P., G. Petrone, A. M. Di Biase, C. Longhi, M. Penta, A. Tinari, F. Superti, G. Fabozzi, P. Visca, and L. Seganti. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13:23-31. [DOI] [PubMed] [Google Scholar]
- 13.Cossart, P., and P. J. Sansonetti. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242-248. [DOI] [PubMed] [Google Scholar]
- 14.Dabiri, G. A., J. M. Sanger, D. A. Portnoy, and F. S. Southwick. 1990. Listeria monocytogenes moves rapidly through the host cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. USA 87:6068-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decatur, A. L., and D. A. Portnoy. 2000. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290:992-995. [DOI] [PubMed] [Google Scholar]
- 16.Dramsi, S., and P. Cossart. 2003. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 71:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freitag, N. E. 2000. Genetic tools for use with Listeria monocytogenes, p. 488-498. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington D.C.
- 19.Freitag, N. E., and K. E. Jacobs. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect. Immun. 67:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitag, N. E., P. Youngman, and D. A. Portnoy. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of Listeria monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 23.Gaillard, J. L., P. Berche, and P. Sansonetti. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garges, S., and S. Adhya. 1988. Cyclic AMP-induced conformational change of cyclic AMP receptor protein (CRP): intragenic suppressors of cyclic AMP-independent CRP mutations. J. Bacteriol. 170:1417-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glomski, I. J., M. M. Gedde, A. W. Tsang, J. A. Swanson, and D. A. Portnoy. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, M. L., and A. H. Killinger. 1966. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 30:309-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundling, A., M. D. Gonzalez, and D. E. Higgins. 2003. Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J. Bacteriol. 185:6295-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ireton, K., and P. Cossart. 1997. Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes. Annu. Rev. Genet. 31:113-138. [DOI] [PubMed] [Google Scholar]
- 29.Ireton, K., B. Payrastre, and P. Cossart. 1999. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. J. Biol. Chem. 274:17025-17032. [DOI] [PubMed] [Google Scholar]
- 30.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551. [DOI] [PubMed] [Google Scholar]
- 31.Jones, S., and D. A. Portnoy. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 62:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathariou, S., P. Metz, H. Hof, and W. Goebel. 1987. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 34.Lacayo, C. I., and J. A. Theriot. 2004. Listeria monocytogenes actin-based motility varies depending on subcellular location: a kinematic probe for cytoarchitecture. Mol. Biol. Cell 15:2164-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauer, P., J. A. Theriot, J. Skoble, M. D. Welch, and D. A. Portnoy. 2001. Systematic mutational analysis of the amino-terminal domain of the Listeria monocytogenes ActA protein reveals novel functions in actin-based motility. Mol. Microbiol. 42:1163-1177. [DOI] [PubMed] [Google Scholar]
- 36.Leimeister-Wachter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lety, M. A., C. Frehel, P. Berche, and A. Charbit. 2002. Critical role of the N-terminal residues of listeriolysin O in phagosomal escape and virulence of Listeria monocytogenes. Mol. Microbiol. 46:367-379. [DOI] [PubMed] [Google Scholar]
- 38.Marquis, H., V. Doshi, and D. A. Portnoy. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63:4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquis, H., and E. J. Hager. 2000. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 35:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vazquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 41.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of Listeria monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 42.Millenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 43.Moors, M. A., B. Levitt, P. Youngman, and D. A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mounier, J., A. Ryter, M. Coquis-Rondon, and P. J. Sansonetti. 1990. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 58:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myers, J. T., A. W. Tsang, and J. A. Swanson. 2003. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J. Immunol. 171:5447-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, S. F., G. S. A. B. Stewart, and R. G. Kroll. 1992. The use of bacterial luciferase for monitoring the environmental regulation of expression of genes encoding virulence factors in Listeria monocytogenes. J. Gen. Microbiol. 138:2619-2627. [DOI] [PubMed] [Google Scholar]
- 47.Paschen, A., K. E. Dittmar, R. Grenningloh, M. Rohde, D. Schadendorf, E. Domann, T. Chakraborty, and S. Weiss. 2000. Human dendritic cells infected by Listeria monocytogenes: induction of maturation, requirements for phagolysosomal escape and antigen presentation capacity. Eur. J. Immunol. 30:3447-3456. [DOI] [PubMed] [Google Scholar]
- 48.Portnoy, D. A., T. Chakraborty, W. Goebel, and P. Cossart. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Portnoy, D. A., R. D. Schreiber, P. Connelly, and L. G. Tilney. 1989. Gamma interferon limits access of Listeria monocytogenes to the macrophage cytoplasm. J. Exp. Med. 170:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ripio, M.-T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J.-A. Vazquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanger, J. M., B. Mittal, F. S. Southwick, and J. W. Sanger. 1990. Analysis of intracellular motility and actin polymerization induced by Listeria monocytogenes in PtK2 cells. J. Cell Biol. 111:415A. [Google Scholar]
- 53.Shetron-Rama, L. M., H. Marquis, H. G. A. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70:1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48:1537-1551. [DOI] [PubMed] [Google Scholar]
- 55.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 57.Suarez, M., B. Gonzalez-Zorn, Y. Vega, I. Chico-Calero, and J. A. Vazquez-Boland. 2001. A role for ActA in epithelial cell invasion by Listeria monocytogenes. Cell Microbiol. 3:853-864. [DOI] [PubMed] [Google Scholar]
- 58.Theriot, J. A., T. J. Mitchison, L. G. Tilney, and D. A. Portnoy. 1992. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357:257-260. [DOI] [PubMed] [Google Scholar]
- 59.Thirumuruhan, R., K. Rajashankar, A. A. Fedorov, T. Dodatko, M. R. Chance, P. Cossart, and S. C. Almo. 2003. Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes. Protein Data Bank accession code 1OMI (http://www.rscb.org.pdb/).
- 60.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez-Boland, J. A., G. Dominguez-Bernal, B. Gonzalez-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 62.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vega, Y., C. Dickneite, M.-T. Ripio, R. Böckmann, B. Gonzalez-Zorn, S. Novella, G. Gominguez-Bernal, W. Goebel, and W. Vazquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wadsworth, S. J., and H. Goldfine. 2002. Mobilization of protein kinase C in macrophages induced by Listeria monocytogenes affects its internalization and escape from the phagosome. Infect. Immun. 70:4650-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong, K. K., H. G. Bouwer, and N. E. Freitag. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell Microbiol. 6:155-166. [DOI] [PubMed] [Google Scholar]