Abstract
Proteases are implicated in several aspects of the physiology of microorganisms, as well as in host-pathogen interactions. Aminopeptidases are also emerging as novel drug targets in infectious agents. In this study, we have characterized an aminopeptidase from the spirochete Borrelia burgdorferi, the causative agent of Lyme disease. The aminopeptidolytic activity was identified in cell extracts from B. burgdorferi by using the substrate leucine-7-amido-4-methylcoumarin. A protein displaying this activity was purified from B. burgdorferi by a two-step chromatographic procedure, yielding a ∼300-kDa homo-oligomeric enzyme formed by monomers of ∼50 kDa. Gel enzymography experiments showed that enzymatic activity depends on the oligomeric structure of the protease but does not involve interchain disulfide bonds. The enzyme was identified by peptide mass fingerprinting as the putative aminopeptidase II of B. burgdorferi, encoded by the gene BB0069. It shares significant identity to members of the M29/T family of metallopeptidase, is sensitive to bestatin, has a neutral pH optimum, and displays maximal activity at 60°C. Its activity is 1.75-fold higher at the temperature of the mammalian host than at that of the insect host of the pathogen. The activity of this thermophilic aminopeptidase of B. burgdorferi (TAPBb) depends on Zn2+, and temperatures over 70°C promoted its inactivation through a transition from the hexameric state to the monomeric state. Since B. burgdorferi is deficient in pathways for amino acid synthesis, TAPBb could play a role in supplying required amino acids. Alternatively, the enzyme could be involved in peptide and/or protein processing.
Lyme disease is a multisystemic illness resulting from infection with the spirochete Borrelia burgdorferi and related organisms (51). It is most commonly spread to humans by the bite of infected, nymphal-stage hard ticks of the genus Ixodes. Lyme disease has been reported in 20 countries in temperate areas of North America, Europe, and Asia; in the United States, reported human cases outnumber those of all other arthropod-borne diseases combined. It represents a significant health problem because of its potentially severe cardiovascular, neurological, and arthritic complications, difficulty in diagnosis and treatment, and high prevalence in some geographic regions. B. burgdorferi is an invasive spirochete, as evidenced by its migration from the initial lesion, called erythema migrans, to distant tissues. Overall, the pathogenesis of B. burgdorferi appears to be dependent on its ability to colonize, grow, and persist in human tissue for months to years and to elicit tissue damage due to immunopathological reactions. The corresponding factors that may account for these activities include mechanisms for acquisition of required nutrients, motility through mammalian host tissues, shared epitopes with human proteins, and ability to evade the immune system (45, 23, 13).
The type strain B. burgdorferi B31 has an unusual genome containing a linear chromosome of 910,725 bp and 22 linear and circular plasmids (16). The small genome size is associated with an apparent absence of genes for synthesis of amino acids, fatty acids, enzyme cofactors, and nucleotides. Thus, B. burgdorferi is dependent upon the mammalian or tick host for the provision of amino acids and other required components. An ABC oligopeptide transporter system (opp) has been well characterized (32, 54, 55), and genes predicted to encode another oligopeptide ABC transporter and a glycine/betaine transporter have been identified (16) (http://www-biology.ucsd.edu/∼msaier/transport/). Otherwise, the mechanisms of amino acid acquisition by B. burgdorferi are poorly understood.
Proteases fulfill important roles in both the basic physiology and pathogenesis of many microbial pathogens, including the ability to provide required amino acids, to process proteins and peptides, to facilitate local infection and dissemination through host tissue by degrading extracellular proteins, and to generate factors that are involved in the entry of a parasite into the host cells (4, 9, 29, 47). These activities may also be important among pathogenic spirochetes. The genomes of B. burgdorferi, Treponema pallidum (the causative agent of syphilis), Treponema denticola (involved in periodontal disease), and Leptospira interrogans (a cause of leptospirosis) contain many genes homologous to those of virulence-related proteases of other pathogens (16, 17, 43, 50). The prolyl-phenylalanine-specific serine protease dentilisin is involved in the pathogenicity of T. denticola (44). A high-molecular-mass protease displaying collagenolytic activity was detected in in vitro-cultured B. burgdorferi by Grab et al. (22); additionally, Heroldova et al. (24) demonstrated the presence of leucine arylamidase activity in all 13 B. burgdorferi sensu lato strains examined, using the API ZYM system. However, these studies were complicated by the presence of serum in the in vitro cultures, which may contribute proteinase activities; also, other groups were not able to detect protease activity in Borrelia cultured in vitro (11). Other factors that may contribute to the dissemination and pathogenesis of B. burgdorferi in the mammalian host involve the activation or induction of host proteolytic activities. A urokinase-type plasminogen activator binds to the surface of the spirochete, resulting in plasmin generation (10). In addition, host matrix metalloproteinases are induced during B. burgdorferi infection and may contribute to tissue pathology (19, 26, 28, 56).
Aminopeptidases (EC 3.4.11) remove N-terminal amino acids from proteins and peptides, are found in animals, plants, and microorganisms, and have several different cellular functions, such as protein maturation and remodeling, hydrolysis of regulatory peptides, antigen processing, and modulation of gene expression. The combined enzymatic activities of both endopeptidases and aminopeptidases mediate hydrolysis of proteins and peptides into free amino acids. Aminopeptidases are considered novel and important pathogen targets for drugs, as evidenced by the relevance of their activities to the life cycle of Plasmodium (18, 25, 38) and the African trypanosome (30). In addition, vaccination of sheep with leucyl aminopeptidase from Fasciola has induced protection against fascioliasis (40).
In this study, we examined the proteolytic activities present in B. burgdorferi B31, utilizing organisms cultured in serum-free medium to obviate the involvement of serum components. We report the identification and characterization of an aminopeptidolytic activity displayed by a thermophilic metalloaminopeptidase of B. burgdorferi (TAPBb). Its enzymatic and biochemical features lead us to consider TAPBb a member of the thermophilic (T)/M29 family of metallopeptidases. TAPBb shows restricted enzymatic activity that depends on its oligomeric structure. We postulate that the TAPBb may play a role in bacterial nutrient supply and/or protein processing.
MATERIALS AND METHODS
Bacteria and preparation of cell extract.
Borrelia burgdorferi B31 clone 5A3 (39) was cultured at 34°C to mid-log phase in a modified, serum-free form of BSK-II medium (41). Cell extracts were prepared from 100-ml bacterial cultures. The cells were collected by centrifugation (6,000 × g for 10 min at 4°C), washed three times in phosphate-buffered saline, lyophilized, and stored at −80°C. The pellet (7.4 × 1010 cells) was resuspended in 7 ml of water containing 1% Triton X-100 and 0.17% lysozyme; the bacteria were immediately disrupted by sonication with six cycles of 30 s each in an ice bath. After removal of the insoluble material by centrifugation (16,000 or 100,000 × g for 30 min at 4°C), the supernatant, referred to hereafter as enzyme extract, was used in the assays or stored at −80°C. Protein concentrations in samples were determined as described by Bradford (5).
Assay of proteolytic activity.
B. burgdorferi protease activity was assayed by using a series of peptidyl fluorogenic substrates (purchased from Sigma Chemical Co.): N-glutaryl-Gly-Gly-Phe-4-metoxy-β-naphthylamide, N-succi-nyl-Gly-Pro-7-amido-4-methylcoumarin, L-Leu-7-amido-4-methylcoumarin(L-AMC), N-carbobenzoxy (CBZ)-Leu-4-metoxy-β-naphthylamide, L-Pro-7-amido-4-meth-ylcoumarin, N-succinyl-Gly-Pro-Leu-Gly-Pro-7-amido-4-methylcoumarin,N-succinyl-Leu-Tyr-7-amido-4-methylcoumarin, N-CBZ-Gly-Gly-Arg-7-amido-4-methylcoumarin, Gly-Phe-4-metoxy-β-naphthylamide, N-CBZ-Phe-Arg-7-amido-4-methylcoumarin, Gly-Pro-7-amido-4-methylcoumarin, N-α-CBZ-L-Arg-7-amido-4-methylcoumarin, N-α-CBZ-Arg-Arg-4-metoxy-β-naphthylamide,L-Ala-L-Ala-L-Phe-Ala-7-amido-4-methylcoumarin, N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin, N-succinyl-Ile-Ala-7-amido-4-methylcoumarin, and Gly-Arg-4-metoxy-β-naphthylamide. Enzyme activity was determined by measuring the fluorescence of either 7-amido-4-methylcoumarin (AMC) or 4-metoxy-β-naphthylamide (MNA) released upon hydrolysis of the substrates as described previously (47). Briefly, assays were performed by incubating 1 μl of enzyme extract (3.2 μg of protein) or 30 ng of purified TAPBb, as specified, for 15 min at the desired temperature in 100 μl of 25 mM Tris-HCl (pH 7.5) (reaction buffer), containing 20 μM fluorogenic substrate. The fluorescence of free AMC or MNA released by enzymatic reaction was recorded in a HITACHI F-2000 spectrofluorimeter as described previously (48). Fluorescence intensities were converted into micromoles of either AMC or MNA by measuring the fluorescence of standard solutions of these fluorogenic groups under the same experimental conditions. Enzymatic activity is expressed in milliunits per milligram, where 1 U represents 1 mmol of released fluorochrome · min−1. In-gel enzymatic activity of either enzyme extract (10 μg) or purified TAPBb (0.5 μg) on L-AMC was measured on a sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE) at 25°C as described previously (48).
Purification and electrophoretic analysis of the aminopeptidase.
B. burgdorferi protease, which hydrolyzes L-AMC, was purified from freshly prepared enzyme extract by fast protein liquid chromatography. The enzyme extract (1 ml; 3.2 mg of protein) was buffered with 25 mM Tris-HCl, pH 7.5, and applied to a DEAE-Sepharose CL-6B (Sigma) column (5 by 1 cm) previously equilibrated with the same buffer. The column was washed, and the proteins were eluted with a linear gradient performed in the same buffer from 0.3 to 0.65 M NaCl for 30 min and then with 1.0 M NaCl for 10 min at 0.5-ml/min flow rate. Fractions (2 ml) were collected on ice, and an aliquot of each fraction was assayed with L-AMC. The enzymatically active fractions, which were eluted between 0.46 and 0.52 M NaCl, were pooled and concentrated to 100 μl with a Centricon-100 concentrator (Amicon, Beverly, Calif.) at 4°C. The solution was then subjected to gel permeation chromatography on a Superose-6 HR 10/30 column (Pharmacia) isocratically perfused with 25 mM Tris-HCl, 150 mM NaCl (pH 7.5) at a flow rate of 0.3 ml/min for 80 min. Each 300-μl fraction was immediately stored on ice until the enzyme activity assay. The active fractions were pooled, concentrated as described above, and stored at −20°C. The column was calibrated with bovine serum albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). The purity and the molecular mass of the purified enzyme were analyzed by SDS-8% PAGE (31) and Coomassie blue staining. The oligomeric structure of the aminopeptidase and the presence of interchain disulfide bonds were evaluated by electrophoresis of either the enzyme extract (20 μg) or purified enzyme (1.0 μg) in either the presence or absence of β-mercaptoethanol (5% [vol/vol]) and with and without prior heating to 100°C for 5 min.
B. burgdorferi protease identification by peptide mass fingerprinting.
The purified protein (1.0 μg) was digested with trypsin (Promega, Madison, Wis.) for peptide mass fingerprinting according to the protocol established by Schevchenko et al. (49). The digested sample was applied to a Perkin-Elmer Sciex API 300 electrospray triple-quadrupole mass spectrometer at a flow rate of 0.02 ml/h. Experimentally determined peptide masses were subjected to a protein identity search against the SwissProt database via the program Profound (http://prowl.rockefeller.edu/profound_bin/WebProFound.exe).
Determination of optimal pH and temperature for activity and thermostability of TAPBb.
The optimal pH for TAPBb activity was assayed as described above in 25 mM Bis-Tris-25 mM Tris-HCl-25 mM borate buffer adjusted to the desired pH. To determine the optimal temperature for TAPBb activity, the reactions were performed at 20, 25, 30, 40, 50, 60, 70, or 100°C in the reaction buffer. The enzyme thermostability was evaluated by the incubation of the purified protein at the same temperatures for either 10 or 240 min in the reaction buffer, following by an L-AMC hydrolysis assay at 60°C. Molecular organization of TAPBb preincubated at different temperatures was evaluated by SDS-8% PAGE under nonreducing conditions as described previously (31) and gel permeation chromatography as described above.
Assay of protease inhibition.
The inhibition pattern of the enzyme was assayed in reaction buffer using L-AMC as a substrate. Different concentrations of EDTA, 1,10-phenanthroline, bestatin, L-trans-epoxysuccinylleucylamido-(4-guanidino) butane (E-64), phenylmethylsulfonyl fluoride (PMSF), N-α-tosyl-lysine chloromethyl ketone, leupeptin, pepstatin A, and phosphoramidon were incubated with 30 ng of purified TAPBb in 100 μl of reaction buffer for 15 min at 25°C before the substrate was added. The enzymatic activity was monitored as described above. All inhibitors were from Sigma.
TAPBb kinetics and cation dependence.
The purified TAPBb (30 ng) was incubated with different concentrations (1 to 100 μM) of L-AMC, and the enzyme reaction was carried out as described above. Kinetic parameters were determined by hyperbolic regression as described previously (12). All assays were done in triplicate and repeated at least three times. The Kcat was calculated by the equation Kcat = Vmax/[E]o, where [E]o represents active enzyme concentration. The effects of divalent cations on enzymatic activity were assessed by incubating purified TAPBb in reaction buffer containing either 10 mM EDTA or 250 μM 1,10-phenanthroline for 15 min at 25°C. After extensive dialysis against reaction buffer, L-AMC (final concentration, 20 μM) and CaCl2, MgCl2, CuCl2, ZnCl2, MnCl2, or CoCl2 (final concentration, 0.4 mM) were added to the reaction system, followed by 15 min of incubation. Substrate hydrolysis was measured as described above. Controls consisted of enzymatic reactions carried out either in the absence of divalent cations or without EDTA treatment.
RESULTS
B. burgdorferi enzyme extract hydrolyzes the protease substrate Leu-AMC.
In preliminary studies, it was determined that enzyme extracts from B. burgdorferi grown in Barbour-Stoenner-Kelly medium with rabbit serum produce two major bands of proteolytic activity in SDS-PAGE gelatin, whereas that from bacterium grown in the absence of serum did not (data not shown). This finding is consistent with the acquisition of plasmin and other host proteases by B. burgdorferi, as reported previously by other groups (10, 11, 26). To detect proteolytic activity in B. burgdorferi itself, we prepared an enzyme extract from organisms grown in a serum-free medium (41) and incubated it with different synthetic fluorogenic substrates. Among all substrates tested (see Materials and Methods), only L-AMC was hydrolyzed by enzyme extract. The calculated specific enzymatic activity was 1,126 ± 108 mU/mg for enzyme extract obtained upon centrifugation at either 16,000 or 100,000 × g. This indicates that the enzyme is not associated to membranes. Similar specific enzymatic activity was obtained with enzyme extract prepared with B. burgdorferi grown in the presence of serum (data not shown). This proteolytic activity was also assessed by an in-gel procedure that allowed us to estimate the molecular mass of the protease. For this analysis, enzyme extract proteins were separated by SDS-PAGE at 4°C, followed by gel washing for SDS removal. Upon incubation of the gel with L-AMC at pH 7.5, a single fluorescent band corresponding to proteolytic activity was revealed above the 200-kDa molecular weight marker (Fig. 1, lane 1). A protein band comigrating with the enzymatic activity was observed after staining of the same gel (Fig. 1, lane 2).
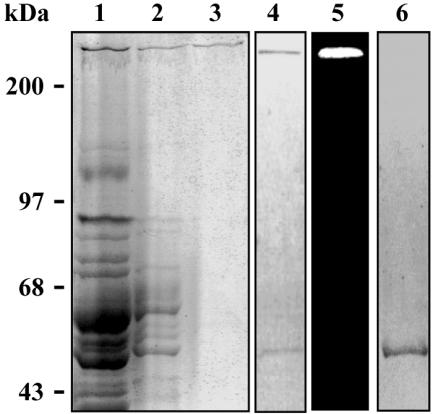
FIG. 1.
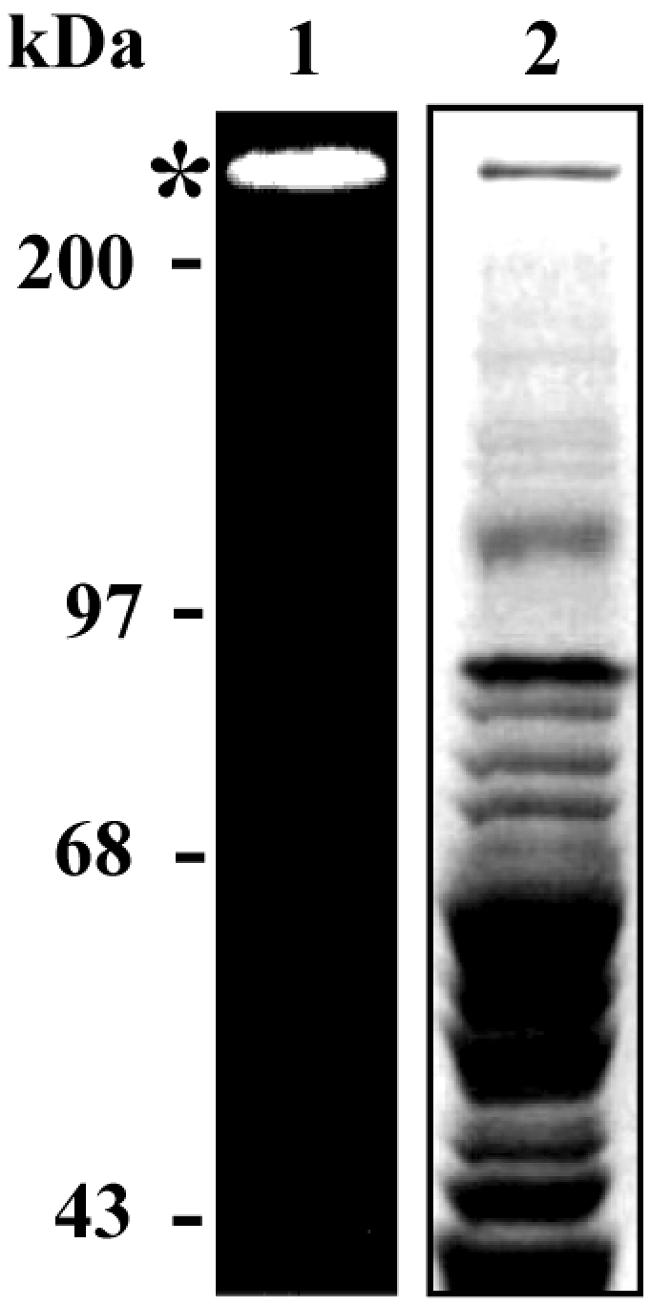
In-gel leucyl aminopeptidase activity of B. burgdorferi protein extract. Enzyme extract, 20 μg, was subjected to SDS-8% PAGE, without previously boiling the sample, at 4°C. The gel was washed and incubated with 20 μM Leu-AMC in reaction buffer at 25°C. The fluorescence of enzymatically released AMC was visualized through UV gel illumination (lane 1, *) prior to gel staining (lane 2).
Purification of the protease and demonstration of its oligomeric form.
The protease displaying activity on L-AMC was purified from B. burgdorferi by subjecting the enzyme extract to a combination of ion exchange and gel permeation chromatography. Table 1 shows a summary of the purification procedure. The protease activity was eluted from a DEAE-Sepharose CL-6B column at 0.46 to 0.52 M concentrations of NaCl and from a Superose-6 HR 10/30 column as a single peak of activity (data not shown), which indicates that under the condition of this experiment, only one enzyme of B. burgdorferi hydrolyzes L-AMC. The last chromatographic step was also used to obtain an estimate of the molecular mass of the enzyme. The enzymatic activity on L-AMC eluted with an apparent molecular mass of 300 kDa (average for four experiments). The same result was obtained when a freshly prepared enzyme extract was applied directly to the column, showing that the ion exchange chromatography did not modify the molecular mass of the protease. The lack of hydrolysis of additional peptidase substrates suggests that the purified enzyme displays narrow activity. The enzyme hydrolyzed L-AMC but not its N-terminal blocked version (N-CBZ-L-AMC). Furthermore, no enzymatic activity was observed on other N-terminal free substrates, such as Pro-AMC, Gly-Phe-MNA, Gly-Pro-AMC, and Gly-Arg-MNA. These data indicate that the enzyme is an aminopeptidase that preferentially removes leucine from N termini of peptides and/or proteins.
TABLE 1.
Purification of aminopeptidase from B. burgdorferi
| Procedure | Sp acta | Total activityb | % Yield | Purification factor |
|---|---|---|---|---|
| Enzyme extract | 6.0 × 10−2 | 20.5 × 10−2 | 100 | 1 |
| DEAE-Sepharose | 15.0 × 10−2 | 18.0 × 10−2 | 88 | 3 |
| Superose 6 | 102.9 × 10−2 | 15.4 × 10−2 | 73 | 32 |
Specific activity: nanomoles of AMC released per minute per milligram of protein.
Total activity: specific activity × total protein.
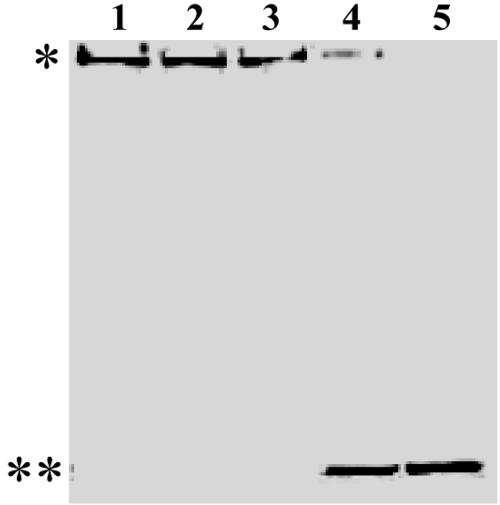
The electrophoretic profile of each step of the purification procedure is shown in Fig. 2. Although the protease was obtained with high purity (Fig. 2, lane 3), in some preparations an additional protein band of about 50 kDa was seen when electrophoresis was carried out under reducing conditions without previous boiling of the sample (Fig. 2, lane 4). However, unlike the higher-molecular-mass band, the 50-kDa band did not hydrolyze the substrate L-AMC, as evidenced by in-gel enzymatic activity experiment (Fig. 2, lane 5). To investigate whether the two bands corresponded to different oligomeric states of the same protein and whether the temperature could interfere with that pattern, the purified protease was boiled for 5 min before SDS-PAGE analysis under reducing conditions (Fig. 2, lane 6). Upon staining of the gel, only the 50-kDa band was revealed. These data suggested that the active 300-kDa protease is an oligomeric protein formed by monomers of about 50 kDa.
FIG. 2.
Electrophoretic analysis of purified aminopeptidase from B. burgdorferi. Samples from crude enzyme extract (10 μg of protein; lane 1), DEAE-Sepharose column fractions (4 μg; lane 2), or Superose-6 column fractions (0.5 μg; lanes 3 to 6) were subjected to SDS-8% PAGE. Electrophoresis took place at 4°C under reducing conditions either with previous boiling of the sample (lane 6) or not (lanes 1 to 5). The gel in lane 4 was previously incubated with Leu-AMC (lane 5) for the in-gel activity assay, as described in the Materials and Methods section. Gels were silver stained.
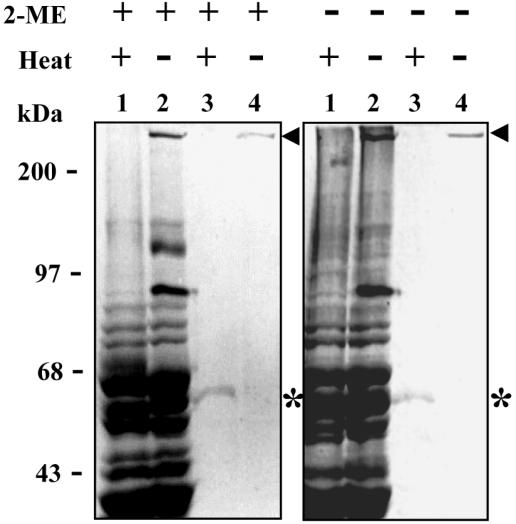
To investigate whether interchain disulfide bonds participated in the arrangement of the oligomer, we subjected either enzyme extract or purified enzyme, previously boiled or not, to SDS-8% PAGE in the presence or absence of β-mercaptoethanol (Fig. 3). The reducing agent did not affect the electrophoretic migration pattern of nonpurified (lanes 1 and 2) and purified (lanes 3 and 4) protease. In contrast, the 300-kDa oligomer was observed in the gels only where the samples have not been previously boiled (lanes 2 and 4), while its 50-kDa monomer was revealed upon sample boiling (lanes 1 and 3). Since high temperature induced the monomerization of the 300-kDa oligomer independently of the presence of β-mercaptoethanol, we conclude that interchain disulfide bonds do not take part in the stabilization of the active protease oligomer.
FIG. 3.
The oligomeric structure of TAPBb does not contain interchain disulfide bonds. Either 20 μg of enzyme extract (lanes 1 and 2) or 2 μg of purified TAPBb (lanes 3 and 4) was subjected to SDS-8% PAGE under reducing (2-ME +) or nonreducing (2-ME −) conditions. Samples in lanes 1 and 3 were heated (+), while those in lanes 2 and 4 were not (−). The gels were stained with Coomassie blue. Arrowheads and asterisks indicate positions of ∼300- and ∼50-kDa bands, respectively.
Molecular identification of the purified protease.
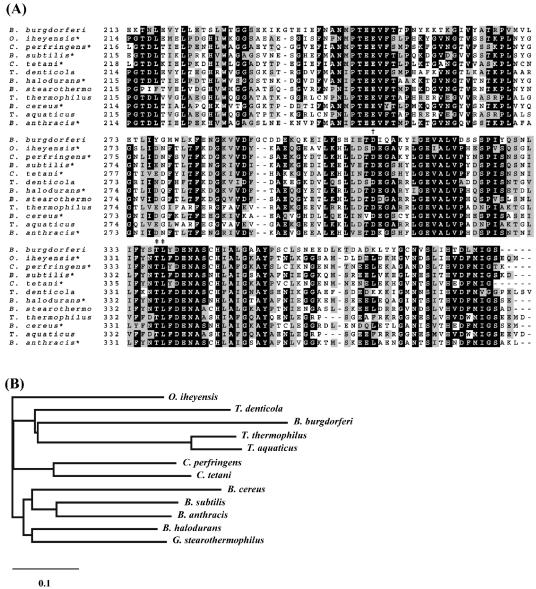
To identify the purified enzyme mediating L-AMC hydrolysis, the enzyme was digested with trypsin and the resulting peptides were submitted to mass spectrometry for peptide mass fingerprinting. Mass values obtained for the detected peptides were compared to those theoretically deduced from sequences deposited in the database. Eight peptides showed very close mass matches to peptides obtained by theoretical digestion of a predicted aminopeptidase of B. burgdorferi (Table 2), encoded by the gene BB0069 (16) (SWISS-PROT/TrEMBL accession no.O51096). This result supports our experimental data showing that the purified enzyme mediates aminopeptidolytic activity. The BB0069 gene comprises an open reading frame that encodes a 412-amino-acid protein with a calculated molecular mass of 46,798 Da and no predicted peptide signal. Based on sequence homology, BB0069 is unrelated to three other predicted aminopeptidases in the B. burgdorferi genome: BB0105 (methionine aminopeptidase); BB0366 (YscI or aminopeptidase I; previously reported by Wallich et al. as APEA_BORBU [SWISS-PROT/TrEMBL]); and BB0627 (PepX; vacuolar X-prolyl dipeptidyl aminopeptidase I). Interestingly, Treponema denticola 35405 contains a homolog of BB0069 (TDE 2337; 34% identity, 57% similarity; Fig. 4A), but no homologs are present in the T. pallidum or L. interrogans genomes. Although BB0069 is within a possible multicistronic operon (BB0069-BB0073), none of the genes in this region are clustered in other organisms or have related predicted functions (data not shown).
TABLE 2.
Identification of B. burgdorferi protease by peptide mass fingerprinting
| Tryptic peptide mass (Da)
|
Identified amino acid sequence | |
|---|---|---|
| Experimental dataa (in-gel digestion) | B. burgdorferi aminopeptidase (in silico digestion) | |
| 671.680 | 671.795 | 16GINLQK21 |
| 1,183.352 | 1,183.261 | 288VVDFGCDDEK297 |
| 1,314.408 | 1,314.414 | 159EGSQTLEEFFK169 |
| 1,919.089 | 1,919.249 | 199TLNELNLEKVIFKTEK214 |
| 2,890.105 | 2,890.220 | 236GTEIEFNANMPTEEVFTTP NYKKTCK260 |
| 1,919.152 | 1,919.249 | 199TLNELNLEKVIFKTEK214 |
| 2,187.480 | 2,187.656 | 39ILAKKAYEHAKYVKLNIK57 |
| 1,200.348 | 1,200.444 | 58DIDILKSRLK67 |
Masses are MH+ monoisotopic, with accuracy of 0.1 to 0.2 Da. The enzyme was digested with trypsin, and masses of resulting peptides were determined by electrospray ionization-mass spectrometry and compared to theoretical ones produced by in silico digestion of proteins found in the database (SwissProt).
FIG. 4.
Sequence comparison and relationship of TAPBb to other members of the M29 family of metallopeptidases. (A) Multiple sequence alignment of C-terminal-portion amino acid sequences of aminopeptidases. The putative zinc binding (†) and bestatin ligation residues (‡) are represented. Sequences were obtained from the GenBank/EBI database under the following accession numbers: AAC66461 (Borrelia burgdorferi), BAC14998 (Oceanobacillus iheyensis), TDE2337 (Treponema denticola), P24828 (Geobacillus stearothermophilus), NP_389328 (Bacillus subtilis), NP_562913 (Clostridium perfringens), NP_782566 (Clostridium tetani), NP_243111 (Bacillus halodurans), NP_654259 (Bacillus anthracis), NP_831585 (Bacillus cereus), P42778 (Thermus thermophilus), and P23341 (Thermus aquaticus). (B) Phylogenetic relationship of members of the aminopeptidase M29 family. A phylogram was generated after alignment of the full-length amino acid sequences of the 11 enzymes, using CLUSTAL W software with a PAM250-weight table set with the parameters Ktuple = 1, opening penalty = 3, and gap extension = 5. The scale at the bottom represents the number of amino acid substitutions per site. Asterisks indicate functionally defined enzymes.
Multiple amino acid sequence alignments confirmed that this B. burgdorferi aminopeptidase is a member of the thermophilic metalloaminopeptidase (M29) family of metallopeptidases, also known as the T (thermophilic) family (42). It shared 32 to 36% identity to other members of M29 family, including assigned and unassigned aminopeptidases (Fig. 4A). Sequence alignments also revealed that the C-terminal portion is the most conserved region in this family, reaching 44% identity and 63% similarity between B. burgdorferi and Geobacillus stearothermophilus. Although identity among members of the M29 family and those of the M17, leucyl aminopeptidase family is almost absent (data not shown), sequence alignments suggested conservation of zinc and bestatin binding residues (Fig. 4) (7, 36). In contrast, sequences of members of the M29 family do not comprise the signature NTDAEGRL sequence of the M17 family (7). These data suggest that TAPBb is unrelated to the M17 family, in spite of displaying leucyl aminopeptidase activity. Three-dimensional structures and the active-site residues are not known for any members of the M29 family.
B. burgdorferi aminopeptidase is a neutral, thermophilic, and thermostable enzyme.
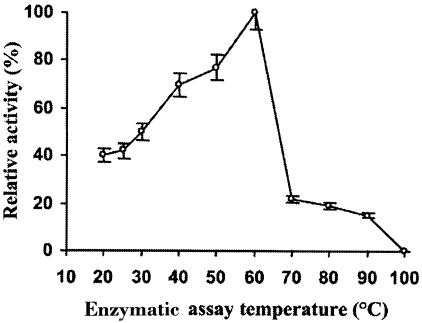
The effect of pH on hydrolysis of L-AMC by the aminopeptidase was determined. It was shown that the enzyme has a strong dependence on neutral pH. Maximal specific activity for the purified enzyme was observed at pH 7.5 (data not shown). At pHs 6.0 and 9.0, the measured enzymatic activities were 20 and 50% of that recorded at pH 7.5. The optimum temperature for aminopeptidase activity on the L-AMC substrate was shown to be 60°C (Fig. 5), and we thus named the enzyme thermophilic aminopeptidase of B. burgdorferi (TAPBb) and named its gene thermophilic aminopeptidase A (tapA). At 20°C the enzymatic activity corresponded to 40% of that measured at 60°C, while at 70°C the enzyme retained 25% of its activity on the substrate. Interestingly, at 40°C the activity of the enzyme was about 1.75 times higher than that measured at 20 to 25°C, the body temperature of the insect. This might suggest that the enzyme activity is regulated depending on the B. burgdorferi host. To determine the thermostability of TAPBb, we assayed substrate hydrolysis by the enzyme at 60°C after its preincubation at different temperatures for either 10 or 240 min (Table 3). The data show that its activity was not significantly modified after a preincubation at 60°C during 240 min. Its preincubation for 10 min at 70°C resulted in significant loss of activity.
FIG. 5.
Optimal temperature for TAPBb activity. Purified aminopeptidase was incubated with Leu-AMC in reaction buffer at different temperatures for 15 min. Enzymatically released AMC was quantitated as described in Materials and Methods. Results are expressed as the percent activity relative to the values obtained at 60°C. Assays were carried out in triplicate, and results represent means for three separate experiments. Standard deviations were less than 8%.
TABLE 3.
TAPBb is thermostablea
| Preincubation temp of enzyme (°C) | Enzymatic activity (% of control)
|
|
|---|---|---|
| 10-min preincubation | 240-min preincubation | |
| 25 | 100 | 98 |
| 37 | 101 | 99 |
| 50 | 105 | 95 |
| 60 | 102 | 87.5 |
| 70 | 25 | 9 |
| 100 | 0 | 0 |
Purified enzyme was preincubated at different temperatures for 10 or 240 min, and then its activity on Leu-AMC was measured at 60°C. Control consisted of enzymatic activity assay without previous incubation of enzyme.
TAPBb enzymatic activity depends on its oligomeric form.
Since preincubation of TAPBb for 10 min at 70°C resulted in significant loss of its activity, we asked whether this enzymatic inactivation was due to monomerization of the oligomer. To address this question, we first incubated TAPBb at different temperatures for 10 min, following by SDS-PAGE analysis. Temperatures up to 60°C did not modify the oligomeric state of the protease, since it migrated as a single high-molecular-mass band in the gel (Fig. 6, lanes 1 to 3). However, upon preincubation at 70°C, the enzyme appeared mostly in its monomeric form, a protein band of 50 kDa (Fig. 6, lane 4). This correlated well with enzymatic activity measured under the same conditions (Table 3). In addition, enzymatic activity was no longer detected upon preincubation at 100°C, most likely due to entire monomerization of the oligomer (Fig. 6, lane 5). Secondly, we evaluated the molecular mass of the active protease, previously subjected to different temperatures, by gel permeation chromatography. Hydrolysis of L-AMC coeluted with the oligomer up to 60°C preincubation, while higher temperatures progressively induced the appearance of the inactive 50-kDa monomer (data not shown). These data indicate that the TAPBb activity depends on its oligomeric form.
FIG. 6.
Electrophoretic profile of TAPBb upon preincubation at different temperatures. Purified protein was incubated at 37 (lane 1), 50 (lane 2), 60 (lane 3), 70 (lane 4), or 100°C (lane 5) for 10 min in the reaction buffer, following by SDS-8% PAGE analysis under nonreducing conditions. The gel was silver stained. Oligomeric (*) and monomeric (**) states of TAPBb are indicated.
TAPBb is a metalloaminopeptidase dependent on zinc.
TAPBb hydrolytic activity towards L-AMC was completely inhibited by 250 μM 1,10-phenanthroline or 100 μM bestatin, while 10 mM EDTA inactivated only 50% of the activity (Table 4). Its activity was not sensitive to E-64, PMSF, N-α-tosyl-lysine chloromethyl ketone, leupeptin, or pepstatin A. The activity of TAPBb previously inhibited by either 10 mM EDTA or 250 μM 1,10-phenanthroline was fully restored with 0.4 mM Zn2+ but not with Ca2+, Mg2+, Mn2+, Cu2+, or Co2+ (Table 4). This indicates that TAPBb is a zinc-dependent metalloaminopeptidase.
TABLE 4.
Inhibition pattern and cation dependence of TAPBba
| Concn and inhibitor | TAPBb activity (% of control) with:
|
|||||
|---|---|---|---|---|---|---|
| No cation | Zn2+ | Mn2+ | Ca2+ | Cu2+ | Co2+ | |
| 250 μM 1,10-phen- anthroline | 0.00 | 98 | 0.00 | 0.20 | 0.70 | 0.20 |
| 10 mM EDTA | 50.0 | 93 | 47 | 52.5 | 49.3 | 53.1 |
| 100 μM bestatin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 2 mM PMSF | 96.4 | NDb | ND | ND | ND | ND |
| 15 μM E-64 | 96.7 | ND | ND | ND | ND | ND |
| 15 μM TLCK | 98.2 | ND | ND | ND | ND | ND |
| 200 μM pepstatin A | 102.5 | ND | ND | ND | ND | ND |
| 200 μM leupeptin | 95.8 | ND | ND | ND | ND | ND |
Purified TAPBb was incubated with inhibitor before the addition of Leu-AMC. Enzyme incubated with 1,10-phenanthroline or EDTA was dialyzed against reaction buffer prior to incubation with the substrate and 0.4 mM CaCl2, MgCl2, CuCl2, ZnCl2, MnCl2, or CoCl2. Reactions took place at 60°C, and results represent means from three independent experiments carried out in triplicate. Standard deviations were less than 10%.
ND, not determined.
TAPBb kinetic parameters.
The Michaelis-Menten constant (Km) and maximal velocity (Vmax) of purified TAPBb were determined by using the hyperbolic regression method with L-AMC as a substrate. The aminopeptidase has a Km value of 45.1 ± 1.9 μM. Its calculated catalytic constant (kcat) and catalytic efficiency (kcat/Km) were 8.03 ± 0.71 S−1 and 17.8 × 10−2 μM−1 · S−1, respectively.
DISCUSSION
In this study, we identified the product of BB0069 as a thermophilic aminopeptidase and named it TAPBb to indicate this activity. TAPBb hydrolyzed the fluorogenic substrate L-AMC in a specific manner, as evidenced by the absence of detectable hydrolysis of any other substrate used. None of the other proteolytic substrates were hydrolyzed by the B. burgdorferi cytosolic extract under the conditions examined. These results suggest that proteolytic activity of B. burgdorferi is highly restrictive, which may explain why this invasive bacterium employs mammalian host proteases with broad specificity for its own benefit (10, 11, 26).
Secreted bacterial aminopeptidases are monomeric, while the intracellular enzymes are either monomeric or multimeric, comprising one, two, four, or six monomers (21). None of the aminopeptidases of the M29 family described to date is hexameric: aminopeptidase T of Thermus aquaticus and Bacillus stearothermophilus are homodimeric enzymes (3, 37), and PepS of Streptococcus thermophilus is monomeric (14). The hexameric quaternary structure of TAPBb is required for activity, but the mechanism of this structural dependence is not currently known. The hexameric structure implies the binding of complementary hydrophobic regions between subunits and reduction of the molecule surface with the medium, thus limiting the amount of water required to stabilize these proteins (20). Disulfide bridges do not take part in the oligomeric assembly of TAPBb, because monomerization did not depend on the presence of a reducing agent. This is in agreement with observations for other members of the M29 family (3, 37) as well as other aminopeptidases, such as leucyl aminopeptidase of bovine lens and aminopeptidase A of Escherichia coli (8, 53). The correlation between the multimeric structure and activity of TAPBb indicates either that the active site is formed at the subunit junctions or that subunit interactions stabilize the active form of the monomers; additional experimentation will be needed to resolve this question.
TAPBb is the first M29 aminopeptidase characterized from any pathogenic microorganism. To date, in addition to bacteria, genes encoding M29 aminopeptidases have been identified in archea (27) and plants (EMBL AJ004922). However, only aminopeptidases from T. aquaticus, Thermus thermophilus, G. stearothermophilus, and S. thermophilus have been conclusively characterized as members of this family (14, 36, 42). Among the features that justify this classification are thermophilicity, high conservation of C termini, and no significant identity with other known aminopeptidases. In addition to considerable sequence identity with members of the M29 family, TAPBb is thermophilic. Its optimal temperature for activity was observed at 60°C, identical to that for the aminopeptidases of G. stearothermophilus and S. thermophilus (3, 14) and similar to that determined for T. aquaticus aminopeptidase (70°C) (35). In contrast to these microorganisms, B. burgdorferi is not thermophilic. However, it is likely that bacteria originated from hyperthermophilic organisms (1, 6). Forterre (15) postulated that the mesophilic prokaryotes existing today must have evolved through a gradual adaptation of thermophilic enzymes to lower temperature optima. Spirochetes have been positioned as one of the most primitive bacterial phyla in the universal evolutionary tree (6). It is of interest that the free-living spirochete Spirochaeta thermophila is thermophilic. Thus, the presence of a thermophilic enzyme in B. burgdorferi may represent an evolutionary holdover from a thermophilic ancestor. The thermophilic properties of TAPBb could alter other properties of the enzyme and hence result in a survival advantage, a subject that requires further investigation.
In members of the M29 family characterized previously, the thermostability and temperature optima of the aminopeptidase are higher in the more thermophilic bacteria. For example, the S. thermophilus aminopeptidase PepS retains 59% of its activity after incubation at 50°C for 20 min (14). Although not produced by a thermophilic bacterium, TAPBb is even more stable than PepS; it maintained 87.5% of its activity after 240 min at 60°C. The hexameric nature of TAPBb may increase its thermostability and thus represent an adaptive advantage relative to monomeric counterparts as PepS. Other highly thermostable enzymes, such as methylthioadenosine phosphorylase of Sulfolobus solfataricus (2) and peptidase B of Salmonella enterica (34), are also hexameric proteins.
Bacterial aminopeptidases are subdivided into three main catalytic groups based on their sensitivity to inhibitors: metalloaminopeptidases, whose activity is inhibited by chelating agents, such as EDTA, 1,10-phenanthroline, and bestatin; serine aminopeptidases that are sensitive to PMSF; and cysteine aminopeptidases, those susceptible to a broad range of agents, such as Hg2+, E-64, and iodoacetamide (21). Our data indicate that TAPBb is a metalloaminopeptidase and Zn2+ is its likely cofactor. Although metalloaminopeptidases exhibit a broad range of metal ion dependence, Zn2+ is the most frequently associated cation (42). Members of the M29 family from T. aquaticus and G. stearothermophilus have Co2+ and Mg2+ as cofactors (35, 52). The cofactor for another characterized member of the M29 family, PepS, is unknown; however, it is unlikely to be Zn2+ or Co2+, since low concentrations of these cations inactivate the enzyme (14). Therefore, TAPBb seems to be the first described member of the M29 family to employ Zn2+ to activate the water molecule during catalysis. Among the known metal ligands of metalloproteases, Asp is conserved in TAPBb. It has been proposed that two phylogenetically unrelated subgroups of Zn2+ aminopeptidases, the largest group of metalloproteases, may be distinguished (21). The first comprises aminopeptidases, such as PepN of E. coli, that display the divalent cation binding motif HExxH, and the second subgroup comprises aminopeptidases that show peptide sequence similarities with bovine lens leucine aminopeptidase. TAPBb appears to be unrelated to these subgroups of Zn2+ aminopeptidases, since its peptide sequence shows neither the HExxH motif nor considerable similarities with any other Zn2+-requiring aminopeptidases.
Although functional properties are unknown for any member of the M29 family, aminopeptidases have been implicated in several physiological roles, such as antigen processing, nutritional supply, degradation of proteins and peptides of both endogenous and exogenous origin, protein maturation, bacterial sensitivity to antibiotics, and stability of plasmids (33). In this study, TAPBb comprised the entire leucine aminopeptidase activity in B. burgdorferi extracts. Thus, it most likely plays an important role in protein and peptide processing as well as in leucine recycling. B. burgdorferi lacks genes coding for amino acid biosynthesis enzymes, and leucine transporters have not been identified for this pathogen (16, 46). An analysis of codon usage indicated that Leu content is essentially equivalent in B. burgdorferi, T. pallidum, and E. coli (10.11, 10.34, and 10.6%, respectively). Also, Leu does not appear to be more frequent than expected at the penultimate N-terminal residue position (which would be susceptible to aminopeptidase action upon removal of the N-terminal N-formyl methionine) (data not shown). Therefore, it is unclear why B. burgdorferi would require a Leu-specific aminopeptidase. If a biosynthetic pathway is indeed lacking, leucine must be acquired by the bacterium through hydrolysis of host and endogenous proteins. It thus appears that inactivation of TAPBb by specific inhibitors or through gene disruption would reveal its functional role and consequently its importance in B. burgdorferi physiology and pathogenesis.
Acknowledgments
This work was supported by CNPq and CAPES, Brazil, and by NIH grant AI37277 (S.J.N.).
Editor: J. T. Barbieri
REFERENCES
- 1.Achenbach-Richter, L., R. Gupta, K. O. Stetter, and C. R. Woese. 1987. Were the original eubacteria thermophiles? Syst. Appl. Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 2.Appleby, T. C., I. I. Matheus, M. Porcelli, G. Cacciapuoti, and S. E. Ealick. 2001. Three-dimensional structure of a hyperthermophilic 5′-deoxy-5′-methylthioadenosine phosphorylase from Sulfolobus solfataricus. J. Biol. Chem. 276:39232-39342. [DOI] [PubMed] [Google Scholar]
- 3.Balerna, M., and H. Zuber. 1974. Thermophilic aminopeptidase from Bacillus stearothermophilus. IV. Int. J. Pept. Protein Res. 6:499-514. [DOI] [PubMed] [Google Scholar]
- 4.Bond, M. D., and H. E. Van Wart. 1984. Characterization of the individual collagenases from Clostridium histolyticum. Biochemistry 23:3085-3091. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. R., C. J. Douady, M. J. Italia, W. E. Marshall, and M. J. Stanhope. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281-285. [DOI] [PubMed] [Google Scholar]
- 7.Burley, S. K., P. R. David, and W. N. Lipscomb. 1991. Leucine aminopeptidase, bestatin inhibition and a model for enzyme-catalyzed peptide hydrolysis. Proc. Natl. Acad. Sci. USA 88:6916-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burley, S. K., P. R. David, A. Taylor, and W. N. Lipscomb. 1990. Molecular structure of leucine aminopeptidase at 2.7-A resolution. Proc. Natl. Acad. Sci. USA 87:6878-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, J. L., and J. L. Benach. 2000. The generation of enzymatically active plasmin on the surface of spirochetes. Methods 21:133-141. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, J. L., T. J. Sellati, J. E. Testa, R. R. Kew, M. B. Furie, and J. L. Benach. 1995. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 63:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish-Bowden, A. 1976. Estimation of the dissociation constants of enzyme-substrate complexes from steady-state measurements. Interpretation of pH-independence of Km. Biochem. J. 153:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Embers, M. E., R. Ramamoorthy, and M. T. Philipp. 2004. Survival strategies of Borrelia burgdorferi, the etiologic agent of Lyme disease. Microbes Infect. 6:312-318. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Espla, M. D., and F. Rul. 1999. PepS from Streptococcus thermophilus—a new member of the aminopeptidase T family of thermophilic bacteria. Eur. J. Biochem. 263:502-510. [DOI] [PubMed] [Google Scholar]
- 15.Forterre, P. 1995. Looking for the most “primitive” organism(s) on Earth today, the state of the art. Planet Space Sci. 43:167-177. [DOI] [PubMed] [Google Scholar]
- 16.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, J. C. Venter, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 18.Gavigan, C. S., J. P. Dalton, and A. Bell. 2001. The role of aminopeptidases in haemoglobin degradation in Plasmodium falciparum-infected erythrocytes. Mol. Biochem. Parasitol. 117:37-48. [DOI] [PubMed] [Google Scholar]
- 19.Gebbia, J. A., J. L. Coleman, and J. L. Benach. 2004. Selective induction of matrix metalloproteinases by Borrelia burgdorferi via toll-like receptor 2 in monocytes. J. Infect. Dis. 189:113-119. [DOI] [PubMed] [Google Scholar]
- 20.Godsell, D. S., and A. J. Olson. 1993. Soluble proteins, size, shape and function. Trends Biochem. Sci. 18:65-68. [DOI] [PubMed] [Google Scholar]
- 21.Gonzales, T., and J. Robert-Baudouy. 1996. Bacterial aminopeptidases, properties and functions. FEMS Microbiol. Rev. 18:319-344. [DOI] [PubMed] [Google Scholar]
- 22.Grab, D. J., R. Kennedy, and M. T. Philipp. 1996. Borrelia burgdorferi possesses a collagenolytic activity. FEMS Microbiol. Lett. 144:39-45. [DOI] [PubMed] [Google Scholar]
- 23.Gray, J. S., O. Kahl, R. S. Lane, and G. Stanek. 2002. Lyme borreliosis: biology, epidemiology, and control. CABI Publishing, Wallingford, Oxon, United Kingdom.
- 24.Heroldova, M., M. Nemec, Z. Hubalek, and J. Halouzka. 2001. Enzyme activities of Borrelia burgdorferi sensu lato. Folia Microbiol. (Prague) 46:179-182. [DOI] [PubMed] [Google Scholar]
- 25.Howarth, J., and D. G. Lloyd. 2000. Simple 1,2-aminoalcohols as strain-specific antimalarial agents. J. Antimicrob. Chemother. 46:625-628. [DOI] [PubMed] [Google Scholar]
- 26.Hu, L. T., M. A. Eskildsen, C. Masgala, A. C. Steere, E. C. Arner, M. A. Pratta, A. J. Grodzinsky, A. Loening, and G. Perides. 2001. Host metalloproteinases in Lyme arthritis. Arthritis Rheum. 44:1401-1410. [DOI] [PubMed] [Google Scholar]
- 27.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, H. Kikuchi, et al. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 28.Kirchner, A., U. Koedel, V. Fingerle, R. Paul, B. Wilske, and H. W. Pfister. 2000. Upregulation of matrix metalloproteinase-9 in the cerebrospinal fluid of patients with acute Lyme neuroborreliosis. J. Neurol. Neurosurg. Psychiatry 68:368-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemba, M., and D. E. Goldberg. 2002. Biological roles of proteases in parasitic protozoa. Annu. Rev. Biochem. 71:275-305. [DOI] [PubMed] [Google Scholar]
- 30.Knowles, G. 1993. The effects of arphamenine-A, an inhibitor of aminopeptidases, on in-vitro growth of Trypanosoma brucei brucei. J. Antimicrob. Chemother. 32:172-174. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lin, B., S. A. Short, M. Eskildsen, M. S. Klempner, and L. T. Hu. 2001. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp(−) Escherichia coli. Biochim. Biophys. Acta 1499:222-231. [DOI] [PubMed] [Google Scholar]
- 33.Lowther, W. T., and B. W. Matthews. 2002. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem. Rev. 102:4581-4607. [DOI] [PubMed] [Google Scholar]
- 34.Mathew, Z., T. Knox, and C. Miller. 2000. Salmonella enterica serovar Typhimurium peptidase B is a leucyl aminopeptidase with specificity for acidic amino acids. J. Bacteriol. 182:3383-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minagawa, E., S. Kaminogawa, H. Matsuzawa, T. Ohta, and K. Yamauchi. 1988. Isolation and characterization of a thermostable aminopeptidase (aminopeptidase T) from Thermus aquaticus YT-1, an extremely thermophilic bacterium. Agric. Biol. Chem. 52:755-763. [Google Scholar]
- 36.Motoshima, H., E. Minagawa, F. Tsukasaki, and S. Kaminogawa. 1997. Cloning of genes of the aminopeptidase T family from Thermus thermophilus HB8 and Geobacillus stearothermophilus NCIB8924, apparent similarity to the leucyl aminopeptidase family. Biosci. Biotech. Biochem. 61:1710-1717. [DOI] [PubMed] [Google Scholar]
- 37.Motoshima, H., N. Azuma, S. Kaminogawa, M. Ono, E. Minagawa, H. Matsuzawa, T. Ohta, and K. Yamauchi. 1990. Molecular cloning and nucleotide sequence of the aminopeptidase T gene of Thermus aquaticus YT-1 and its high-level expression in Escherichia coli. Agric. Biol. Chem. 54:2385-2392. [PubMed] [Google Scholar]
- 38.Nankya-Kitaka, M. F., G. P. Curley, C. S. Gavigan, A. Bell, and J. P. Dalton. 1998. Plasmodium chabaudi chabaudi and P. falciparum: inhibition of aminopeptidase and parasite growth by bestatin and nitrobestatin. Parasitol. Res. 84:552-558. [DOI] [PubMed] [Google Scholar]
- 39.Norris, S. J., J. K. Howell, S. A. Garza, M. S. Ferdows, and A. G. Barbour. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect. Immun. 63:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piacenza, L., D. Acosta, I. Basmadjian, J. P. Dalton, and C. Carmona. 1999. Vaccination with cathepsin L proteinases and with leucine aminopeptidase induces high levels of protection against fascioliasis in sheep. Infect. Immun. 67:1954-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posey, J., and F. C. Gherardini. 2000. Lack of a role for iron in the Lyme disease pathogen. Science 288:1651-1653. [DOI] [PubMed] [Google Scholar]
- 42.Rawlings, N. D. 1998. Introduction: other families of metallopeptidases, p. 1448-1452. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, London, United Kingdom.
- 43.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 44.Rosen, G., R. Naor, and M. N. Sela. 1999. Multiple forms of the major phenylalanine specific protease in Treponema denticola. J. Periodontal Res. 34:269-276. [DOI] [PubMed] [Google Scholar]
- 45.Saier, M. H., Jr., and J. Garcia-Lara. 2001. The spirochetes: molecular and cellular biology. Horizon Press, Wymondham, Norfolk, United Kingdom.
- 46.Saier, M. H., Jr., and I. T. Paulsen. 2000. Whole genome analyses of transporters in spirochetes: Borrelia burgdorferi and Treponema pallidum. J. Mol. Microbiol. Biotechnol. 2:393-399. [PubMed] [Google Scholar]
- 47.Santana, J. M., P. Grellier, J. Schrevel, and A. R. Teixeira. 1997. A Trypanosoma cruzi-secreted 80 kDa proteinase with specificity for human collagen types I and IV. Biochem. J. 325:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santana, J. M., P. Grellier, M. H. Rodier, J. Schrevel, and A. R. Teixeira. 1992. Purification and characterization of a new 120 kDa alkaline proteinase of Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 187:1466-1473. [DOI] [PubMed] [Google Scholar]
- 49.Schevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-891. [DOI] [PubMed] [Google Scholar]
- 50.Seshadri, R., G. S. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E., Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, and I. T. Paulsen. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA 101:5646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 52.Stoll, E., H. G. Weder, and H. Zuber. 1976. Aminopeptidase II from Bacillus stearothermophilus. Biochim. Biophys. Acta 438:212-220. [PubMed] [Google Scholar]
- 53.Sträter, N., D. J. Sherratt, and S. D. Colloms. 1999. X-ray structure of aminopeptidase A from Escherichia coli and a model for the nucleoprotein complex in Xer site-specific recombination. EMBO J. 18:4513-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, X. G., B. Lin, J. M. Kidder, S. Telford, and L. T. Hu. 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J. Bacteriol. 184:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, X. G., J. M. Kidder, J. P. Scagliotti, M. S. Klempner, R. Noring, and L. T. Hu. 2004. Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (Opp) operon. J. Bacteriol. 186:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, Z., H. Chang, R. P. Trevino, K. Whren, J. Bhawan, and M. S. Klempner. 2003. Selective up-regulation of matrix metalloproteinase-9 expression in human erythema migrans skin lesions of acute lyme disease. J. Infect. Dis. 188:1098-1104. [DOI] [PubMed] [Google Scholar]