Abstract
Background:
Fluorescence labeled DNA probes and in situ hybridization methods had shorter turn round time for results revolutionized their clinical application. Signals obtained from these probes are highly specific, yet they can produce fusion signals not necessarily representing fusion of actual genes due to other genes included in the probe design. In this study we evaluated discordance between cytogenetic, FISH and RNAseq results in 3 different patients with hematologic malignancies and illustrated the need to perform next generation sequencing (NGS) or RNASeq to accurately interpret FISH results.
Methods:
Bone marrow or peripheral blood karyotypes and FISH were performed to detect recurring translocations associated with hematologic malignancies in clinical samples routinely referred to our clinical cytogenetics laboratory. When required, NGS was performed on DNA and RNA libraries to detect somatic alterations and gene fusions in some of these specimens. Discordance in results between these methods is further evaluated.
Results:
For a patient with plasma cell leukemia standard FGFR3 / IGH dual fusion FISH assay detected fusion that was interpreted as FGFR3-positive leukemia, whereas NGS/RNASeq detected NSD2::IGH. For a pediatric acute lymphoblastic leukemia patient, a genetic diagnosis of PDGFRB-positive ALL was rendered because the PDGFRB break-apart probe detected clonal rearrangement, whereas NGS detected MEF2D::CSF1R. A MYC-positive B-prolymphocytic leukemia was rendered for another patient with a cytogenetically identified t(8;14) and MYC::IGH by FISH, whereas NGS detected a novel PVT1::RCOR1 not previously reported.
Conclusions:
These are 3 cases in a series of several other concordant results, nevertheless, elucidate limitations when interpreting FISH results in clinical applications, particularly when other genes are included in probe design. In addition, when the observed FISH signals are atypical, this study illustrates the necessity to perform complementary laboratory assays, such as NGS and/or RNASeq, to accurately identify fusion genes in tumorigenic translocations.
Keywords: Hematologic malignancies, inaccurate FISH positive signals, next generation sequencing, RNASeq analysis
Introduction
Somatic genomic rearrangements perturb cellular processes such as tumor suppression, tissue specific differentiation, kinase signaling, or epigenetic regulation, and thus promote tumor development and progression.1-4 Clonal genetic abnormalities are identified in over 90% of hematologic malignancies, and recurrent gene fusions underlie specific morphologic entities; 5 these have been implicated in disease pathogenesis, and personalized treatments are clinically promising.6,7 Standard techniques such as karyotyping and/or fluorescence in situ hybridization (FISH) using gene specific DNA probes are used to detect these genetic abnormalities. Advanced technologies such as next generation sequencing (NGS) covering a panel of disease specific genes and RNASeq to detect gene fusions is used in some cases. FISH probes cover 200kb to 1Mb of genomic length and are highly specific and sensitive. Analysis using FISH probes is time efficient and cost effective, and they can be used on a variety of specimen types.8,9 Application of FISH probes has helped revolutionize the clinical approach to both neoplastic conditions and constitutional syndromes. FISH probe analysis is particularly useful in detecting abnormalities of targeted regions when a karyotype is normal or is not obtained. Such analysis is also useful in detecting genetic evolution of initial clone and development of sub-clonal populations at diagnosis or at disease relapse in tumors. However, FISH probes lack the resolution necessary to accurately identify specific genes disrupted by DNA breaks and may yield inaccurate results due to the proximity of genes included in probe design. 10 False negative FISH results have been reported in a few cases of hematological tumors.11-13 Here, we present 3 examples of inaccurately interpreted FISH results in hematological malignancies and show that complementary laboratory assays such as NGS, including fusion detection by RNASeq are useful in the accurate identification of genes involved in recurring translocations in tumors; these may have therapeutic and/or prognostic implications.
Materials and Methods
Morphology/Immunohistochemistry
Wright-Giemsa-stained peripheral blood (PB) and bone marrow (BM) aspirates and hematoxylin/eosin-stained trephine biopsy and aspirate clots were processed using standard hematopathology procedures and reviewed by hematopathologists. Immunohistochemical analysis was performed on sections from tissue blocks using a MYC specific antibody (mouse monoclonal antibody, clone Y69, Ventana Medical Systems, Tuscan, AZ).
Flowcytometry
Peripheral blood, and BM aspirate samples were routinely processed and immunophenotyped using 10-color FASCanto flow cytometer (Becton Dickinson, San Jose, CA) and analyzed using Cytopaint Classic Software (Leukocyte, Pleasanton, CA) as previously described. 14
Chromosome analysis, FISH, and next generation sequencing
Peripheral blood or BM were cultured without mitogenic stimulation for 24 hours to 96 hours, exposed to colcemid and hypotonic solution, and were harvested using standard protocols for chromosome analysis of lymphoid malignancies. G-banded metaphase spreads on glass slides were analyzed, karyotyped and the abnormalities were described using ISCN 2020. 15 FISH studies were performed on the cultured cells; both interphase cells and sequentially hybridized de-banded metaphases were evaluated to confirm the involvement of different genes mapped to the bands of chromosomal translocation breaks. Commercially available probes for FGFR3, MYC, CCND1, IGH, MAF, Cep4/Cep10, ABL1/BCR, KMT2A, MYC/IGH, MYC break-apart (Abbott Molecular, Abbott Park, IL), MAFB, ETV6/RUNX1, ABL1, ABL2, PDGFRB (Oxford Gene Technology, Cambridge, UK), and PVT1 (Empire Genomics, Depew, NY) were used in this study.
Next generation sequencing was performed on DNA and RNA prepared from sections of the paraffin embedded hematopathology material with paired control specimens from the saliva. Sequencing libraries were generated using Kapa HyperPrep kits (Roche Diagnostics, Cape Town, South Africa). Genes included in the NGS panel in the genomic DNA and in cDNA derived from RNA are captured by IDT xGen baits (ITDNA, Coralville, IA), and run on an Illumina NextSeq 550. The enriched library contained all exons of 1505 cancer genes included in our customized panel (can be found at https://www.utsouthwestern.edu/education/medical-school/departments/pathology/services/once-upon-a-time/assets/comprehensive-pan-cancer-next-generation-sequencin-solid-tumor-panel-aberration-list.pdf). Analysis was performed using custom germline, somatic and mRNA bioinformatic pipeline. 16 Fusions are called based on the Star-Fusion algorithm, 17 and reviewed in IGV (Broad Institute, Cambridge, MA).
Results
Clinical characteristics and genetics
Patient 1 was a 62-year man with history of COVID-19, and vitamin B12 deficiency, who was diagnosed with IgG kappa multiple myeloma after he experienced several months of progressively worsening back pain. MRI of spine showed diffuse abnormal marrow signal, L4 compression fracture, osseous masses at T1 causing cord compression, right acetabulum, and right pubic bone. Laboratory results were significant for macrocytic anemia (MCV 102), with a hemoglobin of 11 g/dL and elevated total protein of 9.6 raising concern for multiple myeloma.
Baseline myeloma workup included the following: SPEP/IFE: IgG monoclonal protein at 3.6 g/dL, Kappa FLC 1237 mg/L, Lamba LLC 2.4 mg/L, K/L free light chain ratio 515, B2M: 5.09 mg/L. BM biopsy showed 80% to 90% involvement by CD138 + kappa restricted plasma cells with diffuse sheets of large/atypical plasma cells and PB involvement with greater than 2 × 109/L absolute plasma cells; a hematopathology diagnosis of plasma cell leukemia (PCL) was rendered.
He received radiation therapy to the right acetabulum (20 Gy) L4 (20 Gy) C6-T1 (20 Gy) and dexamethasone for immediate disease control. He was started on lenalidomide, bortezomib, and dexamethasone (RVD). After 3 cycles of induction with RVD, he was referred to us for stem cell transplantation. At this visit, he was noted to have leukocytosis, light chains up to 1000 mg/L, and LDH 1741 consistent with aggressive relapse with free light chain escape. A repeat BM biopsy at this point showed persistent kappa restricted plasma cells with 60% bone marrow involvement. Due to refractive disease, chemotherapy continued with filgrastim, doxorubicin, CISplatin, pegfilgrastin, bendamustine, and carfilzomib, but the patient expired.
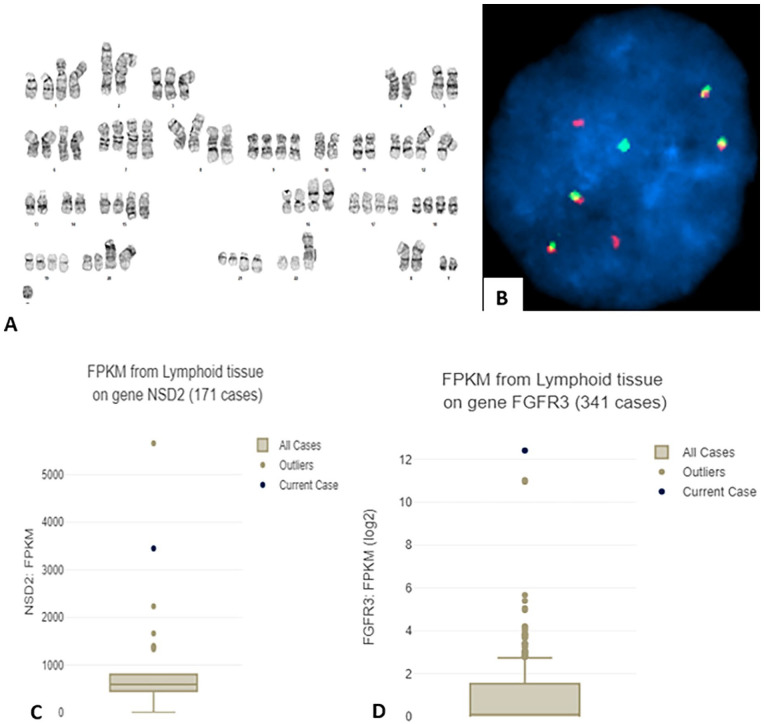
Chromosome analysis of the BM aspirate showed a highly complex karyotype with 2 related clones: 64~77<3n>,X, add(X)(p22.1),Y,+Y,-1,add(1)(p13)x2,-2,der(2)(11qter–>11q12::2p13–>2q21::1q21–>1q23::1q21–>1q23::2q21–>2qter)x2,-4,-5,+6,add(6)(q21)x2,+7,add(7)(q32)x2,+8,der(8)t(8;14)(p21;q11.2)x2,der(8)t(1;8)(p13;q24.1)x2,+9,-10, -11,+12,der(12)t(1;12)(q21;p11.2)x2,-13,-14,+15,add(15)(q15)x2,+16,der(16)t(1;16)(p22;p13.3)x2,+17,+19,+20,der(20)t(13;20)(q12;p13)x2,+21,add(21)(q22)x2,+22,add(22)(p12),+3~4mar[8]/73~78,<3n>,idem,-der(2)x2,+der(2)(11qter–>11q12::2p13–>2q21::1q21–>1q23::1q21–>1q23: :2q21–>2q31::?)x2,+add(5)(q11.2)x2,-add(8)x2,+del(8)(p23p11.2)x2,+mar[6]/46,XY[6] (Figure 1A). FISH analysis with probes for FGFR3, MYC, CCND1, IGH, MAF and MAFB detected FGFR3::IGH (55.5% cells) (Figure 1B). Therefore, a cytogenetic diagnosis of FGFR3 rearranged [t(4;14)] PCL was rendered with poor prognosis. Contrary to the interpretation of FISH results, RNASeq analysis revealed NSD2::IGH and overexpression of both NSD2 and FGFR3 (Figure 1C and D). This is further confirmed by PCR amplification of the fusion junction by primers derived from IGH (GGACTTGGAGGAATGATTCCATGCCAAAGCTTTGCAAGGCTCGCAGTGACCAGGCGCCCGACATG) and from NSD2 (ATTCCAGCTAAGAAAGAGTCTTGTCCAAACACTGGAAGAGACAAAGACCACCTGTTGAAATACAACGT) (Supplemental Figure 1, lane B1). Hybridization to abnormal metaphases with MYC break-apart probe showed deletion of 3′-side of the probe on the der(8) chromosome (image not shown). This signal pattern suggests deletion or loss of 3′-sdie of the probe; however, the possibility that MYC is rearranged or altered cannot be ruled out. Other FISH abnormalities found include del(13) or RB1 loss (37% cells), del(17p) or TP53 loss (34.5% cells), gain for 1q or (extra signals for P18 in 58% cells), and gain of 9 and 15 (33% cells). Other findings in NGS analysis include a deletion in BIRC3 (c.1324+873_173del) and copy number variations of trisomy 7, 12p+, 16p−, 17p−, consistent with the karyotypic abnormalities.
Figure 1.
(A) G-banded karyotype of leukemia cells in BM; see text for complete description of the karyotype. (B) FISH with FGFR3 (red) and IGH (green) probes showing FGFR3::IGH (4 copies). (C and D) Box and whisker plots showing RNA expression of NSD2 (C) and of FGFR3 (D) using fragments per kilobase millions (FPKM) compared to other clinical cases of the same tissue type according to the corresponding Oncotree root (JCO Clin Cancer Inform 2021;5:221-230).
Patient 2 was a 15-year girl presented to the emergency department with fever and a 2-month history of decreased appetite. A complete blood count showed marked leukocytosis (WBC 648K/µL). A PB smear showed variably sized blasts with round to irregular nuclei, dispersed chromatin, and scant cytoplasm. Flow cytometry revealed a 94% population of cells that were CD10 (bright+), CD11b (partial dim +), CD19 (dim and variably+), CD20 (partial dim +), CD22 (variably +), CD25 (predominantly +), CD33 (partial +), CD34(+), CD36 (partial +), CD38 (partial +), CD45 (partial +), CD79a (+), surface immunoglobulin light chain (−), HLA-DR (+), myeloperoxidase (−), TdT(+2). A diagnosis of pre-B-lymphoblastic leukemia/lymphoma (B-ALL) was rendered. Cerebrospinal fluid was positive for central nervous system involvement at the time of diagnosis.
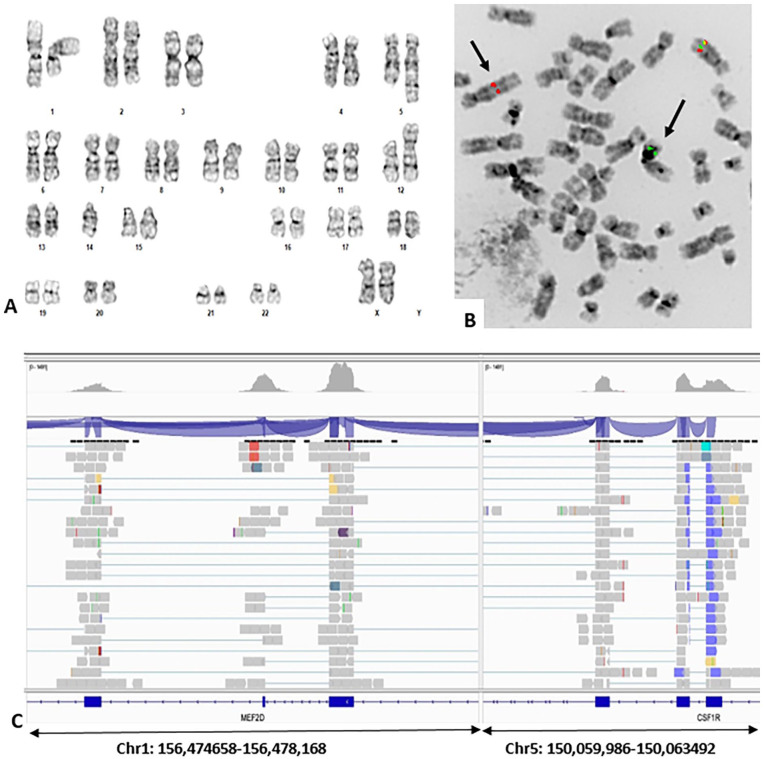
Chromosome analysis of PB showed an abnormal karyotype with 2 related clones - 46,XX,t (1;5)(q23;q33)[9]/45, idem, idic(12;14)(p11.2p11.2)[5]/46, XX[6] (Figure 2A). A high-risk ALL FISH panel including probes Cep4/Cep10, ABL1/BCR, ETV6/RUNX1, ABL1, ABL2, PDGFRB, KMT2A was performed per COG protocol. This panel detected rearrangement in PDGFRB in 69.5% of cells (Figure 2B), and loss of one copy of ETV6 in 19% of cells. These findings correlated with the karyotype findings and suggested a sub-clonal origin of the idic(12;14) clone. A cytogenetic interpretation of PDGFRB rearranged pre-B-ALL with clonal evolution was favored; however, NGS analysis detected a MEF2D::CSF1R (Figure 2C) but not PDGFRB rearrangement, as well as a deletion in IK2ZF1 (chr7:50020408-52450405). Thus, there is a risk of misinterpreting PDGFRB FISH results in the absence of NGS results. Fusion between MEF2D and CSF1R was further confirmed by PCR amplification of the fusion junction using primers from MEF2D (TCAAGTGATGCATTAACCCCTTTCCTGCCTGGGAAGTGATGACTCGCAGGTCGGGCTTGCGGCTGGGGGCTCCAGCTGGGTGCTGTGGGTAGGTGGGGGTGGAGA) and from CSF1R (GGTCGATGAAAGTATAACTGTTGCCCTCATAGCTCTCGATGATCTTCCAGCGGACCTGGTACTTGGGCT) (Supplemental Figure 1, lane D1). At the time of this writing, the patient is under a AALL1131 consolidation Ph- like B-ALL- Dasatinib protocol and was in remission as of the latest follow-up (August 2023).
Figure 2.
(A) G-banded karyotype of PB specimen: 45,XX,t(1;5)(q23;q33),idic(12;14)(p11.2;p11.2). (B) PDGFRB break-apart FISH on a de-banded metaphase of the karyotype in (A) showing rearrangement (black arrows) between der(1) and der(5). (C) MEF2D::CSF1R detected in NGS analysis with genomic coordinates as indicated.
Patient 3 was a 77-year female who presented with splenomegaly (20 cm) minimal lymphadenopathy, and significant leukocytosis. PB analysis showed absolute lymphocytosis - WBC 74.6/µL, platelets 161, hemoglobin 10.6 g/dL, RBC 4.9, RDW 17.6%, MCV 81.6 fL, and a differential count with 6% segmented neutrophils, 88% lymphocytes, 1% monocytes, 3% eosinophils, 1% basophils, and one nRBC/100 WBC. PB smear showed atypical lymphocytes with round to mildly irregular nuclei, smooth chromatin, prominent nucleoli, and abundant cytoplasm accounting for nearly 80% of lymphocytes. BM also showed atypical lymphocytes with similar morphology. Flow cytometry of a PB specimen identified a clonal B-cell population positive for CD5, CD19, CD20, CD22, CD45, CD52, FMC7, HLA-DR, Ig-lambda, and negative for CD10, CD23, CD200. IHC stains of BM core biopsy was positive for PAX-5, CD20, BCL2, MYC, and negative for LEF-1, Cycling D1, BCL-6, and SOX-11.
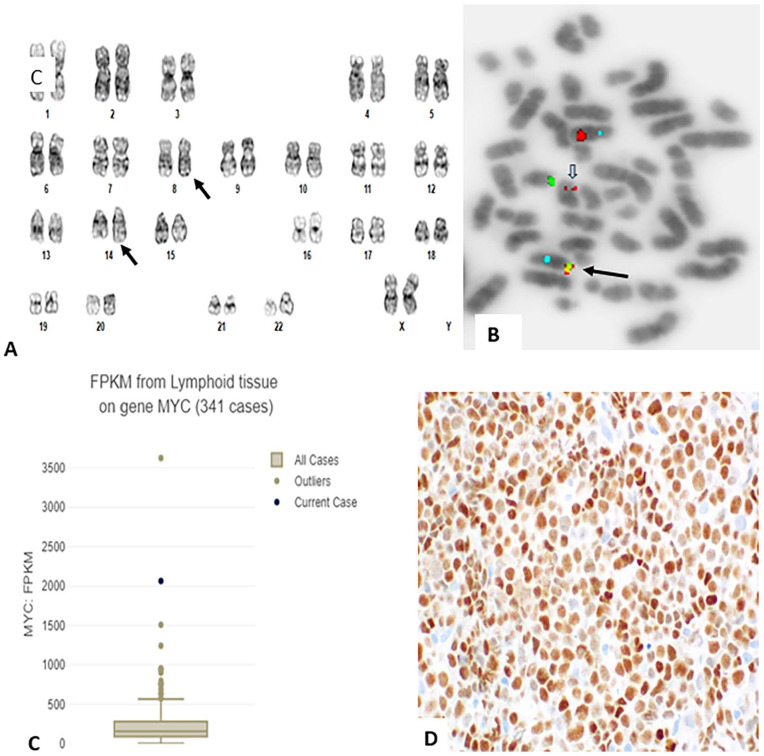
Chromosome analysis of PB showed an abnormal karyotype - 46,XX,t(8;14)(q24;q32)[20] (Figure 3A). Interphase FISH with probes for MYC and IGH showed an atypical pattern with 1 fusion signal in contrast to typical pattern of 2 fusion signals expected for this probe set; sequential de-banded metaphase FISH mapped the fusion signal on the der(8) chromosome (Figure 3B, black arrow). An additional study with MYC break-apart probe confirmed rearrangement in MYC with abnormal signals in 99% of cells (image not shown). From the results of hematopathology, cytogenetics and FISH a diagnosis of B-cell pro-lymphocytic leukemia (B-PLL) with MYC rearrangement was rendered. Contrary to this interpretation NGS analysis detected RCOR1::PVT1 but not MYC::IGH. This is further confirmed by PCR amplification of the fusion junction with primers from RCOR1 (GGTGGCGGTGGCATGAGGGTCGGACCCCAGTACCAGGCGGTGGTGCCCGACTTCGACCCCG) and from PVT1 (AAGGACAGAATAACGGGCTCCCAGATTCACAAGCCCCACCAAGAGGATCACCCCAGGAACGC) (Supplemental Figure 1, lane F1). Thus, in this case FISH results for MYC are also inaccurately interpreted. Detailed analysis of RNASeq data mapped the break in RCOR1 between exons 1 and 2, and the break in PVT1 between exon 7 and 8. Therefore, translocation break at 8q24.1 had occurred on 3′-side of PVT1 and the translocation break at 14q32 had occurred near the 5′-end of RCOR1. This resulted in transposition of RCOR1 along with IGH to 8q24.1. This generated the fusion signal with MYC and IGH probes on the der(8) (Figure 3B, black arrow). In addition, NGS detected sequence variants of possible clinical significance in BCOR (p.Asn1459Ser), CHD2 (p.Gly839Asp), and SF3B1 (p.Gln698_Lys700delinsPro). Other sequence variants of unknown clinical significance also detected included ATM (p.Asp2721Asn), CREBBP (p.Gly2238Arg), SDHA (p.Arg282Gly), and copy number variations on 14q32 (chr14:102681879-102905814) and 17q22 (chr17:57256698-58354393). RNASeq analysis showed significantly increased expression of MYC (Figure 3C), and IHC stain confirmed a high MYC protein expression in about 90% of leukemia cells (Figure 3D).
Figure 3.
(A) G-banded karyotype of PB specimen: 46,XX,t(8;14)(q32;q24.1) (black arrows). (B) FISH with MYC (red), IGH (green) and CEP 8 (aqua) probes showing a fusion signal on der(8)t(8;14) (black arrow), and a residual MYC (red) on der(14)t(8;14) (open arrow). (C) Box and whisker plot showing expression of MYC using fragments per kilobase millions (FPKM) compared to other clinical cases of the same tissue type according to the corresponding Oncotree root (JCO Clin Cancer Inform 2021;5:221-230). (D) MYC protein expression in leukemia cells in BM core biopsy by IHC stain.
Due to advanced age and high-risk disease, the patient was treated with acalabrutinib monotherapy starting in March 2020, and added RCHOP in December 2020. Patient developed refractory disease by April 2021. At this point she received R-GemOx but continued to have persistent disease. Treatment with Lenalidomide + Tafasitamab was initiated in September 2021, but was discontinued in December 2021 due to progressive disease. Salvage therapy initiated in February 2022 with Bendamustine + rituximab + polatuzumab vedotin was suspended after 2 cycles due to poor tolerance. At this time the disease was widespread in abdomen, patient was given palliative radiation therapy, but expired 2 months later.
Discussion
The use of FISH probes in clinical cytogenetics has become a mainstay due to their ease of use and fast turn-around-time for results. Nevertheless, in situations where the location of chromosomal break is away from the probe foot prints, or where there is a 3-way-translocation, or where there is loss or gain of chromatin at the site of break, analysis with FISH probes can lead to inaccurate results.10,18 Misinterpretation of cytogenetic and/or FISH results can greatly impact personalized treatment. In this study we have shown that standard positive FISH signals can be misinterpreted due to genes that are included in the design of probes, and therefore, complementary laboratory assays such as NGS, RNASeq, and mate-pair sequencing19,20 should be performed for accurate identification of gene fusions involved in driver translocations in tumors.
Plasma cell leukemia, either do-novo or secondary to multiple myeloma (MM) is a rare hematologic malignancy with poor prognosis. The t(4;14)(p16;q32), which is a cryptic change, has been reported in about 8% to 15% of MM/PCL patients; typically, this translocation leads to FGFR3::IGH, and these patients have poor prognosis.21-27 Alternatively, this translocation may also lead to NSD2 (MMSET)::IGH, and these patients have an extremely poor prognosis;28-30 both genes are deregulated by the translocation with different enhancers of the IGH. 30 Since FGFR3 was first identified as the fusion partner with IGH in this translocation, a commercial probe was designed to detect FGFR3::IGH with dual fusion signals. This probe is routinely used for detecting the t(4;14) in newly diagnosed MM or PCL patients, and positive patients are favored a diagnosis of FGFR3-positive MM or PCL. But NSD2 is mapped at 6.25 kb toward the centromeric side of FGFR3, and it is covered in the design of the commercial FGFR3 probe; therefore, FGFR3 / IGH FISH assay cannot discriminate a FGFR3::IGH versus NSD2::IGH. FGFR3 is expressed in only 70% of the t(4;14)+ patients, whereas NSD2 is expressed in all t(4;14)+ patients.31,32 In our patient, cytogenetic and FISH results (FGFR3::IGH) favored a diagnosis of FGFR3 positive [t(4;14)] PCL making the patient eligible for a clinical trial including treatment with the novel pan-tyrosine kinase inhibitor (TKI), dovitinib. However, RNASeq analysis detected NSD2::IGH and overexpression of both NSD2 and FGFR3; therefore, interpretation of FISH results as FGFR3 positive PCL is inaccurate. To the best of our knowledge this is the first documented case in which a t(4;14) activated both NSD2 and FGFR3, simulating a double hit neoplasm. Therefore, it would not be appropriate for this patient to be placed on a clinical trial using dovitinib, and the patient falls into a poorer prognosis category. 33 This case highlights the necessity to use high resolution molecular assays (NGS or RNASeq) to correctly identify the fusion partner in each t(4;14)+ MM or PCL patient. Besides the t(4;14) the karyotype of this patient was complex, had MYC rearrangement and loss of RB1 which are other genetic markers of poor prognosis in MM/PCL.29,34-37
Originating in lymphoid stem cells or progenitor cells, B-ALL is genetically classified into 3 groups: those with sentinel abnormalities (BCR::ABL1, ETV6::RUNX1, TCF3::PBX1, IL3::IGH, KMT2A rearrangement, hypodiploidy, hyperdiploidy), iAMP21, and “B-other” group (Ph-like).5,38 The Ph-like ALL, accounting for 15% to 27% of ALL patients depending on age, is further heterogeneous with a variety of genetic alterations such as ABL class alterations (ABL1, ABL2, PDGFRB, CSF1R), or JAK 2 class (JAK2, CRLF2), or RAS-signaling pathway, or other genes, and have poor outcome.39-43 Identification of the primary or sub-clonal genetic alterations can be used in definitive diagnostic classification, treatment, or risk stratification. Since these patients may respond to tyrosine kinase inhibitors,44,45 screening newly diagnosed Ph-like ALL patients for ABL class genetic changes is standard of care testing for COG enrolled patients. Therefore, FISH with break-apart probes for ABL1, ABL2, PDGFRB is performed on diagnostic specimens. PDGFRB rearrangement is a recurring abnormality in Ph-like ALL.46,47 PDGFRB and CSF1R are closely linked with about 500 bp distance between them. 48 Therefore, FISH probe for PDGFRB cannot distinguish involvement of PDGFRB versus CSF1R in fusions with other genes. Thus, use of commercially available FISH probe for PBGFRB has the risk for misinterpretation of results. First reported in a patient with B-ALL, 49 translocations fusing MEF2D with several partners is another genetic marker of poor outcome in this group of patients; CSF1R is one of the fusion partners and is associated with a worse outcome.48,50-53 Although only a few cases of MEF2D::CSF1R have been reported, these patients stand out and cluster close to Ph-like ALL in gene expression signature than other MEF2D fusion patients. 50 In the absence of results from other complementary assays such as NGS, distinguishing PDGFRB versus CSF1R rearrangement by FISH using PDGFRB probe is not possible. In our patient, standard PDGFRB break-apart FISH results favored inaccurate genetic classification as PDGFRB positive ALL, suggesting that the patient may benefit from treatment with TKIs. 43 Contrary to this result, RNASeq analysis showed CSF1R::MEF2D and therefore, the patient falls into a poorer prognosis and may benefit from treatment with histone deacetylase inhibitors. 50 Thus, these results suggest a necessity for caution in interpreting PDGFRB break-apart FISH results, and for performing fusion screening assays for prognostic classification and for individualized therapeutic management. In addition to MEF2D::CSF1R, this patient also had IK2ZF1 deletion which is another genetic marker for poor prognosis in ALL.53,54
Karyotypes of B-PLL are mostly complex, and t(MYC) is present in 62% of them; these patients frequently carried sequence variants in BCOR, CHD2, and SF3B1, and have an aggressive clinical course.55,56 Therefore, screening for MYC translocation is performed by FISH in diagnostic specimens. Although FISH false negativity for MYC fusion has been reported, 10 false positivity had not been reported yet.
Chromosome 8q24.1 domain houses 2 important genes, MYC and PVT1. Spaced at a genomic distance of about 53 kb apart with MYC on the centromeric side and PVT1 on the telomeric side, these genes are involved in important cellular functions such as genomic integrity, proliferation, and programmed cell death.57-60 MYC and PVT1 interact with each other mutually regulating their expression and thus promoting tumorigenesis.61,62 PVT1 over expression lead to increased expression of MYC and it has been correlated with poor patient outcome. 60 PVT1 is promiscuous with several fusion partners, including MYC, and its contribution to tumorigenesis is cancer-type specific. 63 PVT1 rearrangements are infrequent in B-cell neoplasms. In B-cell neoplasms with typical t(8q24) PVT1 fusion with IGL, IGK, SUPT3H, NBEA and WWOX have been reported in isolated cases.33,56,61,64,65 Fusion between PVT1 and MYC has also been reported in tumors with amplified 8q24 region. 66 RCOR1, located on the centromeric side of IGH at 14q32, is a transcriptional corepressor involved in the epigenetic regulation of blood cell development. 67 Oncogenic fusion between RCOR1 and XPC or TRAF3 has been reported in breast cancer. 68 RCOR1 fusions have not been reported in B-cell tumors, but deletions in RCOR1 were associated with poor outcome in a subset of diffuse large B-cell lymphoma. 69 This is the first documentation of PVT1::RCOR1 in tumors.
The case presented here is unique with discordance between chromosome, FISH and NGS findings. The typical t(8;14) identified in karyotype yielded an atypical fusion signal (one fusion signal) with MYC and IGH probes. Atypical fusion between MYC and IGH due to complex rearrangements or single fusion signals due to other types of structural rearrangements of 8q24 have been reported in few cases.10,70,71 In 2 cases the atypical fusion signal originated due to the transposition of entire MYC to the distal side of IGH.10,70 In the other study the atypical fusion was due to cryptic insertion of exon2 and exon 3 of MYC into IGH. 71 However, MYC expression was not evaluated in these cases. Contrary to these previous reports, in the present patient, NGS analysis did not detect a rearrangement in MYC, but instead detected PVT1::RCOR1 and disruption of both genes. RNASeq analysis did not show dramatic overexpression of both fused genes but did show overexpression of MYC; the latter is further confirmed by high positivity for MYC protein by IHC. Structural analysis of the genomic fusion showed translocation of entire IGH to exon 7 of PVT1; this may have disrupted PVT1 and led to transcriptional activation of MYC. This case also illustrates that transposition of entire IGH downstream to MYC can also induce MYC expression, and that the molecular landscape of MYC deregulation in B-cell malignancies may be more diverse and complex and require complementary high resolution molecular assays to detect them. In addition, this case further illustrates the necessity to further evaluate atypical rearrangements detected using MYC FISH probes to accurately identify fusion partners. In addition to this novel fusion and deregulation of MYC, this patient also had sequence variants in BCOR, CHD2, SF3B1, CREBBP which are typically seen in cases with complex to high complex karyotypes, 56 however, the karyotype in this case was not complex.
Recently, Lopes et al 19 reported false positive rearrangement in PDGFRA in a chronic myeloid leukemia patient with a t(4;11)(q12;p15),t(9;22)(q34;q11.2) in the karyotype. FISH with a break-apart probe for PDGFRA detected rearrangement raising the possibility of a novel fusion in the patient. However, mate-pair sequencing located the break at 4q12 in an intronic region between CHIC2 and PDGFRA. In a pediatric tumor, rhabdomyosarcoma, with t(8;13)(p11.2;q14) FISH probe for FGFR1 detected amplification, FISH probe for FOXO1 detected rearrangement and amplification raising the possibility of FOXO1::FGFR1 which has been reported in a previous case. 72 Contrary to FISH findings with FGFR1 probe, NGS analysis detected FOXO1::NSD3, and this was further confirmed by a break-apart probe for NSD3. Since FGFR1 and NSD3 were mapped to 8p11.2, in the absence of NGS results, FISH would have been falsely reported as FGFR1-positive rhabdomyosarcoma. 73
Conclusion
The cases presented here underscore the need to use complementary laboratory methods to fully understand the clinical utility of results from FISH probes designed for detecting recurring fusions in tumors. With increased molecular testing in routine diagnostic work-up, the number of discordant gene alterations between FISH and NGS is expected to be increased. Therefore, it is important to be aware of this finding to avoid misclassification of neoplasms. FISH, NGS and RNASeq technologies complement each other, and NGS and RNASeq may be required to unequivocally detect fusion genes in tumors and for developing patient specific therapeutic and/or management plans.
Supplemental Material
Supplemental material, sj-docx-1-pat-10.1177_2632010X241230262 for RNASeq Analysis for Accurate Identification of Fusion Partners in Tumor Specific Translocations Detected by Standard FISH Probes in Hematologic Malignancies by Prasad Koduru, Weina Chen, Franklin Fuda, Gurbakhash Kaur, Farrukh Awan, Samuel John, Rolando Garcia and Jeffrey Gagan in Clinical Pathology
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: FA has provided consultancy to: Genentech, Astrazeneca, Abbvie, Janssen, Pharmacyclics, Gilead sciences, Kite pharma, Celgene, Karyopharm, MEI Pharma, Verastem, Incyte, Beigene, Johnson and Johnson, Dava Oncology, BMS, Merck, Cardinal Health, ADCT therapeutics, Epizyme, Caribou Biosciences, Cellecter Bisosciences, and received research funding from Pharmacyclics. All other authors report no conflict of interest in performing work included in this manuscript.
Written informed consent was obtained from patients or responsible guardian to use interesting findings from the clinical investigations and/or tests in academic publications after deidentifying the subjects.
Author Contributions: PK analyzed all results and wrote the manuscript, WC and FF performed all hematopathology studies and wrote this part in the manuscript, GK, FA, SJ are clinical care providers and contributed the clinical data on the patients, RG reviewed and wrote FISH studies, JG performed NGS and RNASeq studies and wrote these results in the manuscript.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [DOI] [PubMed] [Google Scholar]
- 2. Ponder BA. Cancer genetics. Nature. 2001;411:336-341. [DOI] [PubMed] [Google Scholar]
- 3. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789-799. [DOI] [PubMed] [Google Scholar]
- 4. Kou F, Wu L, Ren X, Yang L. Chromosome abnormalities: new insights into their clinical significance in cancer. Mol Therap Oncolytics. 2020;17:562-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swerdlow SH, Campo E, Harris NL, et al. (eds). WHO Classification of Tumors of Haematipoietic and Lymphoid Tissues. IARC; 2017. [Google Scholar]
- 6. Krzyszczyk P, Acevedo A, Davidoff EJ, et al. The growing role of precision and personalized medicine for cancer treatment. Technology. 2018;6:79-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nikanjam M, Okamura R, Barkauskas DA, Kurzrock R. Targeting fusions for improved outcomes in oncology treatment. Cancer. 2020;126:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolff DJ, Bagg A, Cooley LD, et al. Guidance for fluorescence in situ hybridization testing in hematologic disorders. J Mol Diagn. 2007;9:134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui M, You L, Ren X, et al. Long non-coding RNA PVT1 and cancer. Biochem Biophys Res Commun. 2016;471:10-13. [DOI] [PubMed] [Google Scholar]
- 10. King RL, McPhail ED, Meyer RG, et al. False-negative rates for MYC fluorescence in situ hybridization probes in B-cell neoplasms. Haematologica. 2019;104: e248-e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Score J, Walz C, Jovanovic JV, et al. Detection and molecular monitoring of FIP1L1-PDGFRA-positive disease by analysis of patient-specific genomic DNA fusion junctions. Leukemia. 2009;23:332-339. [DOI] [PubMed] [Google Scholar]
- 12. Peterson JF, Pitel BA, Smoley SA, et al. Elucidating a false-negative MYC break-apart fluorescence in situ hybridization probe study by next-generation sequencing in a patient with high-grade B-cell lymphoma with IGH/MYC and IGH/BCL2 rearrangements. Mol Case Stud. 2019;5(3):a004077. doi:10.1101/mcs.a004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsson-Arvidsson L, Norberg A, Sjögren H, Johansson B. Frequent false-negative FIP1L1-PDGFRA FISH analyses of bone marrow samples from clonal eosinophilia at diagnosis. Br J Haematol. 2020;188:e76-279. [DOI] [PubMed] [Google Scholar]
- 14. Alsuwaidan A, Koduru P, Fuda F, et al. A combined biomarker of bright CD38 and MYC ⩾55% is highly predictive of double-/triple-hit high-grade B-cell lymphoma. Am J Clin Pathol. 2022;158:338-344. [DOI] [PubMed] [Google Scholar]
- 15. McGowan-Jordan J, Hastings RJ, Moore S. (eds). ISCN - an International System for Human Cytogenetic Nomenclature. Karger; 2020. [DOI] [PubMed] [Google Scholar]
- 16. Raulerson C, Jimenez G, Wakeland B, et al. ANSWER: annotation software for electronic reporting. JCO Clin Cancer Inform. 2022;6:e2100113. [DOI] [PubMed] [Google Scholar]
- 17. Haas BJ, Dobin A, Li B, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scattone A, Catino A, Schirosi L, et al. Discordance between FISH, IHC, and NGS analysis of ALK status in advanced non-small cell lung cancer (NSCLC): a brief report of 7 cases. Transl Oncol. 2019;12:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopes JL, Webley M, Pitel BA, et al. Characterizing false-positive fluorescence in situ hybridization results by mate-pair sequencing in a patient with chronic myeloid leukemia and progression to myeloid blast crisis. Cancer Genet. 2020;243:48-51. [DOI] [PubMed] [Google Scholar]
- 20. Heydt C, Wölwer CB, Velazquez Camacho O, et al. Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med Genomics. 2021;14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chesi M, Nardini E, Brents LA, et al. Frequent translocation t(4;14)(p16.3; q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keats JJ, Reiman T, Belch AR, Pilarski LM. Ten years and counting: so what do we know about t(4;14)(p16;q32) multiple myeloma. Leuk Lymphoma. 2006;47: 2289-2300. [DOI] [PubMed] [Google Scholar]
- 23. Manier S, Salem KZ, Park J, et al. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14:100-113. [DOI] [PubMed] [Google Scholar]
- 24. Winkler JM, Greipp P, Fonseca R. t(4;14)(p16;3;q32) is strongly associated with a shorter survival in myeloma patients. Br J Haematol. 2003;120:170-171. [DOI] [PubMed] [Google Scholar]
- 25. Keats JJ, Reiman T, Maxwell CA, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101: 1520-1529. [DOI] [PubMed] [Google Scholar]
- 26. Cleynen A, Szalat R, Kemal Samur M, et al. Expressed fusion gene landscape and its impact in multiple myeloma. Nat Commun. 2017;8:1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sato S, Kamata W, Okada S, Tamai Y. Clinical and prognostic significance of t(4;14) translocation in multiple myeloma in the era of novel agents. Int J Hematol. 2021;113:207-213. [DOI] [PubMed] [Google Scholar]
- 28. Mirabella F, Wu P, Wardell CP, et al. MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J. 2013;3:e114-e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murase T, Ri M, Narita T, et al. Immunohistochemistry for identification of CCND1, NSD2, and MAF gene rearrangements in plasma cell myeloma. Cancer Sci. 2019;110:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chesi M, Nardini E, Lim RSC, et al. The t(4;14) translocation in myeloma dysregulates both FGFR3and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025-3034. [PubMed] [Google Scholar]
- 31. Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr. A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374-2376. [DOI] [PubMed] [Google Scholar]
- 32. Keats JJ, Maxwell CA, Taylor BJ, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foltz SM, Gao Q, Yoon CJ, et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020;11:2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mikulasova A, Ashby CC, Tytarenko RG, et al. MYC rearrangements in multiple myeloma are complex, can involve more than five different chromosomes and correlate with increased expression of MYC and a distinct downstream gene expression pattern. Blood. 2017;130:65-65. [Google Scholar]
- 35. Lakshman A, Painuly U, Rajkumar SV, et al. Impact of acquired del(17p) in multiple myeloma. Blood Adv. 2019;3:1930-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abdallah N, Baughn LB, Rajkumar SV, et al. Implications of MYC rearrangements in newly diagnosed multiple myeloma. Clin Can Res. 2020;26:6581-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cardona-Benavides IJ, de Ramón C, Gutiérrez NC. Genetic abnormalities in multiple myeloma: prognostic and therapeutic implications. Cells. 2021;10:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts K, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in ph-like lymphoblastic leukemia. N Engl J Med. 2014;371:1005-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts KG, Gu Z, Payne-Turner D, et al. High frequency and poor outcome of Philadelphia chromosome–like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35:394-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boer JM, Steeghs EM, Marchante JR, et al. Tyrosine kinase fusion genes in pediatric BCR-ABL1-like acute lymphoblastic leukemia. Oncotarget. 2017;8:4618-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cario G, Leoni V, Conter V, et al. Relapses and treatment-related events contributed equally to poor prognosis in children with ABL-class fusion positive B-cell acute lymphoblastic leukemia treated according to AIEOP-BFM protocols. Haematologica. 2020;105:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jain S, Abraham A. BCR-ABL1-like B-acute lymphoblastic leukemia/lymphoma: A comprehensive review. Arch Pathol Lab Med. 2020;144:150-155. [DOI] [PubMed] [Google Scholar]
- 44. Apperley JF, Gardembas M, Melo JV, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002;347:481-487. [DOI] [PubMed] [Google Scholar]
- 45. Giacomo DD, Quintini M, Pierini V, et al. Genomic and clinical findings in myeloid neoplasms with PDGFRB rearrangement. Annl Haematol. 2022;101: 297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwab C, Ryan SL, Chilton L, et al. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): genetic profile and clinical implications. Blood. 2016;127:2214-2218. [DOI] [PubMed] [Google Scholar]
- 47. Heilmann AM, Schrock AB, He J, et al. Novel PDGFRB fusions in childhood B- and T-acute lymphoblastic leukemia. Leukemia. 2017;31:1989-1992. [DOI] [PubMed] [Google Scholar]
- 48. Lilljebjörn H, Ågerstam H, Orsmark-Pietras C, et al. RNA-seq identifies clinically relevant fusion genes in leukemia including a novel MEF2D/CSF1R fusion responsive to imatinib. Leukemia. 2014;28:977-979. [DOI] [PubMed] [Google Scholar]
- 49. Barber KE, Harrison CJ, Broadfield ZJ, et al. Molecular cytogenetic characterization of TCF3 (E2A)/19p13.3 rearrangements in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46:478-486. [DOI] [PubMed] [Google Scholar]
- 50. Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. doi: 10.1038/ncomms13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Y-F, Wang B-Y, Zhang W-N, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lilljebjörn H, Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood. 2017;130:1395-1401. [DOI] [PubMed] [Google Scholar]
- 53. Ohki K, Kiyokawa N, Saito Y, et al. Clinical and molecular characteristics of MEF2D fusion-positive B-cell precursor acute lymphoblastic leukemia in childhood, including a novel translocation resulting in MEF2D-HNRNPH1 gene fusion. Haematologica. 2019;104:128-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dorge P, Meissner B, Zimmermann M, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chong LC, Ben-Neriah S, Slack GW, et al. High-resolution architecture and partner genes of MYC rearrangements in lymphoma with DLBCL morphology. Blood Adv. 2018;2:2755-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chapiro E, Pramil E, Diop M, et al. Genetic characterization of B-cell prolymphocytic leukemia: a prognostic model involving MYC and TP53. Blood. 2019; 134:1821-1831. [DOI] [PubMed] [Google Scholar]
- 57. Shtivelman E, Henglein B, Groitl P, Lipp M, Bishop JM. Identification of a human transcription unit affected by the variant chromosomal translocations 2;8 and 8;22 of Burkitt lymphoma. Proc Natl Acad Sci USA. 1989;86:3257-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Colombo T, Farina L, Macino G, Paci P. PVT1: A rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. doi 10.1155/2015/304208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caforio M, Sorino C, Iacovelli S, et al. Recent advances in searching c-MYC transcriptional cofactors during tumorigenesis. J Exp Clin Cancer Res. 2018; 37:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Onagoruwa OT, Pal G, Ochu C, Ogunwobi OO. Oncogenic role of PVT1 and therapeutic implications. Front Oncol. 2020;10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shtivelman E, Bishop JM. Effects of translocations on transcription from PVT. Mol Cell Biol. 1990;10:1835-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parolia A, Cieślik M, Chinnaiyan AM. Competing for enhancers: PVT1 fine-tunes MYC expression. Cell Res. 2018;28:785-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jin K, Wang S, Zhang Y, et al. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol Life Sci. 2019;76:4275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nagoshi H, Taki T, Hanamura I, et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954-4962. [DOI] [PubMed] [Google Scholar]
- 65. Nakamura Y, Kayano H, Kakegawa E, et al. Identification of SUPT3H as a novel 8q24/MYC partner in blastic plasmacytoid dendritic cell neoplasm with t(6;8)(p21;q24) translocation. Blood Cancer J. 2015;5:e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. L’Abbate A, Macchia G, D’Addabbo P, et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 2014;42:9131-9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yao H, Goldman DC, Fan G, Mandel G, Fleming WH. The corepressor Rcor1 is essential for normal myeloerythroid lineage differentiation. Stem Cells. 2015; 33:3304-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim RN, Moon H-G, Han W, Noh D-Y. Perspective insight into future potential fusion gene transcript biomarker candidates in breast cancer. Int J Mol Sci. 2018;19:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chan FC, Telenius A, Healy S, et al. An RCOR1 loss-associated gene expression signature identifies a prognostically significant DLBCL subgroup. Blood. 2015;125:959-966. [DOI] [PubMed] [Google Scholar]
- 70. Amarillo I, Bui PH, Kantarci S, et al. Atypical rearrangement involving 3′-IGH@ and a breakpoint at least 400 kb upstream of an intact MYC in a CLL patient with an apparently balanced t(8;14)(q24.1;q32) and negative MYC expression. Mol Cytogenet. 2013;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wagener R, Bens S, Toprak UH, et al. Cryptic insertion of MYC exons 2 and 3 into the immunoglobulin heavy chain locus detected by whole genome sequencing in a case of “MYC-negative” Burkitt lymphoma. Haematologica. 2020;105: e202-e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu J, Guzman MA, Pezanowski D, et al. FOXO1–FGFR1 fusion and amplification in a solid variant of alveolar rhabdomyosarcoma. Mod Pathol. 2011;24: 1327-1335. [DOI] [PubMed] [Google Scholar]
- 73. Rakheja D, Park JY, Yang MS, et al. Rhabdomyosarcoma with epithelioid features and NSD3::FOXO1 fusion: evidence for reconsideration of previously reported FOXO1::FGFR1 fusion. Int J Surg Pathol. 2023;31:213-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-pat-10.1177_2632010X241230262 for RNASeq Analysis for Accurate Identification of Fusion Partners in Tumor Specific Translocations Detected by Standard FISH Probes in Hematologic Malignancies by Prasad Koduru, Weina Chen, Franklin Fuda, Gurbakhash Kaur, Farrukh Awan, Samuel John, Rolando Garcia and Jeffrey Gagan in Clinical Pathology