Abstract
A lipoprotein diacylglyceryl transferase (lgt) deletion mutant of Staphylococcus aureus SA113 was constructed. The lipoprotein and prelipoprotein expression, the growth behavior, and the ability of the mutant to elicit an immune response in various host cells were studied. In the wild type, the majority of [14C]palmitate-labeled lipoproteins were located in the membrane fraction, although some lipoproteins were also present on the cell surface and in the culture supernatant. The lgt mutant completely lacked palmitate-labeled lipoproteins and released high amounts of some unmodified prelipoproteins, e.g., the oligopeptide-binding protein OppA, the peptidyl-prolyl cis-trans isomerase PrsA, and the staphylococcal iron transporter SitC, into the culture supernatant. The growth of the lgt mutant was hardly affected in rich medium but was retarded under nutrient limitation. The lgt mutant and its crude lysate induced much fewer proinflammatory cytokines and chemokines in human monocytic (MonoMac6), epithelial (pulmonary A549), and endothelial (human umbilical vein endothelial) cells than the wild type. However, in whole blood samples, the culture supernatant of the lgt mutant was equal or even superior to the wild-type supernatant in tumor necrosis factor alpha induction. Lipoprotein fractionation experiments provided evidence that a small proportion of the mature lipoproteins are released by the S. aureus wild type despite the lipid anchor and are trapped in part by the cell wall, thereby exposing the immune-activating lipid structure on the cell surface. Bacterial lipoproteins appear to be essential for a complete immune stimulation by gram-positive bacteria.
The N terminus of lipoproteins contains an N-acyl-S-diacylglyceryl cysteine moiety, resulting from the modification of the N-terminal cysteine residue by a diacylglyceryl group thioether linked to the side chain sulfhydryl group and by an additional acyl group amide linked to the amino group of this cysteine (22). Lipoproteins are synthesized as preproteins with a distinct signal sequence. The A consensus sequence, the lipobox (38), is found in the C-terminal portion of the signal sequence, -Leu−3-Ser/Ala−2-Ala/Gly−1-Cys+1 (16, 27). The first step in the biosynthesis of lipoproteins is the transfer of the diacylglyceryl group from phosphatidylglycerol to the sulfhydryl group of the invariant cysteine residue in the lipobox. This reaction is catalyzed by the phosphatidylglycerol-prolipoprotein diacylglyceryl transferase, encoded by the lgt gene (55). The lgt gene is highly conserved and appears to be ubiquitous in bacteria (48) and functionally exchangeable. When the lipoproteins are translocated through the cytoplasmic membrane, the specific signal peptidase II (Lsp) recognizes the diacylglyceryl modification and cleaves between the amino acid at position −1 and the lipid-modified cysteine residue (25). In several bacteria, but not all, the N terminus of the diacylglyceryl-modified cysteine residue is fatty acylated by an N-acyltransferase (Lnt) to form N-acyl diacylglyceryl cysteine (54). Lipoproteins are translocated across the cytoplasmic membrane through the Sec pathway (63). In Escherichia coli, the three enzymes involved in modification, processing, and N-acylation are encoded by the lgt, lsp, and lnt genes, respectively (55). The lipo-form is normally demonstrated by labeling with [3H]palmitate.
Microbial lipoproteins are a functionally diverse group of proteins that are active in the bacterial cell envelope. In gram-positive bacteria, many lipoproteins have substrate-binding activity, are a part of various ABC transport systems (67), are involved in sensing environmental signals (64), and play a role in protein secretion and folding of exoproteins (12, 29, 33), respiration (4), bacterial coaggregation (31), adherence to various surfaces (30, 32), attachment to and invasion of host cells (11, 51, 52), sporulation (44), antibiotic resistance (40), bacterial conjugation (68), and genetic competence (53).
The lipid modification of prelipoproteins appears to be essential in gram-negative bacteria, such as E. coli and Salmonella enterica serovar Typhimurium, since mutants defective in lgt, lsp, or lnt are temperature sensitive for growth, which suggests that one or more lipoproteins are required for normal growth, division, and viability of bacterial cells (16). The structural lipoproteins that bridge peptidoglycan with the outer membrane very likely play a crucial role. In gram-positive bacteria, such as Bacillus subtilis and Streptococcus pneumoniae, lipid modification of prelipoproteins appears not to be essential for cell growth in vitro (36, 47). However, lipoproteins play a role in virulence in S. pneumoniae and in protein secretion in B. subtilis (36). In gram-positive bacteria, the lipid-containing N terminus is presumed to anchor lipoproteins to the outer leaflet of the cell membrane, and lipoproteins can be compared topographically to the other major class of macroamphiphiles present in the gram-positive cell envelope, the lipoteichoic acids and lipoglycans (13).
In recent years, many studies have been aimed at elucidating the molecular basis of inflammatory responses elicited by gram-positive bacteria. Several components from different subcellular compartments of gram-positive bacteria have been reported to activate various host cells, such as components of the bacterial envelope (e.g., lipoproteins, peptidoglycan, teichoic acids, capsular polysaccharide), secreted components (e.g., enterotoxins or toxic shock syndrome toxin 1), and intracellular factors (e.g., unmethylated CpG DNA). Bacterial lipoproteins and synthetic lipopeptides bind to and activate toll-like receptor 2 (TLR2) (1, 6, 24), which leads to NF-κB activation (41, 50). TLR2 heterodimerizes with TLR1 or TLR6 depending on the source and type of the lipoproteins (9, 43, 65, 66).
Only limited information on the immune stimulatory activity of whole bacterial cells defective in the lipid modification of prelipoproteins is available. Most studies on lipoproteins have been carried out with isolated molecules or synthetic analogues. We therefore constructed a Staphylococcus aureus lgt::ermB mutant unable to carry out lipid modification of prelipoproteins and examined the contribution of staphylococcal lipoproteins to growth, adhesion to and invasion of host cells, cytotoxic effects, and host cell stimulation. As the crucial stimulatory lipid part of lipoproteins is normally embedded in the outer leaflet of the cytoplasmic membrane, we investigated the accessibility of these microbial components for recognition by receptors of the innate immune system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli was grown overnight at 37°C with constant shaking in Luria broth. S. aureus and Staphylococcus carnosus strains were grown at 37°C overnight with constant shaking in basic medium (BM) broth (Luria broth supplemented with 0.1% K2HPO4 and 0.1% glucose), diluted 1:100 into fresh medium, and cultivated for further investigations as indicated.
TABLE 1.
Strains and plasmids used in this work
| Strain | Plasmid | Description | Reference |
|---|---|---|---|
| S. aureus SA113 (ATCC 35556) | Wild type | 28 | |
| SA113 | PBT2 | Temperature-sensitive E. coli/Staphylococcus shuttle vector | 8 |
| SA113 | pRB474 | Derivative of pRB473 | 8 |
| SA113 lgt::ermB | lgt gene (SA0716) replaced with ermB gene | This study | |
| SA113 | pRBlgt | pRB474 carrying lgt | This study |
| SA113 | pTX16 | Control plasmid | 46 |
| SA113 | pTXsitC | pTX16 containing xylose-inducible sitC | This study |
| E. coli DH5-α | pUC18-lgt | Contains lgt | This study |
| E. coli DH5-α | pBT2-for | Contains lgt inactivation cassette with erm 5′→3′; Cm selection | This study |
| E. coli DH5-α | pBT2-rev | Contains lgt inactivation cassette with erm 3′→5′; Cm selection | This study |
For [14C]palmitate feeding, cells were cultivated to the mid-exponential phase in BM or MHB medium (Mueller-Hinton II cation-adjusted broth; Becton Dickinson) and fractionated as described below. For cytokine induction and the corresponding invasion assays, Dulbecco's modified Eagle medium (DMEM) supplemented with F-12 nutrient mixture and 2 mM l-glutamine (referred to as DMEM-F-12 medium; Gibco BRL) was used instead of complex medium to prevent attachment of potentially cell-activating agents to the bacterial surface. For cell activation studies with human umbilical vein endothelial cells (HUVEC), bacteria were grown in MHB, which gave results similar to those obtained when bacteria were cultivated in DMEM-F-12 medium. Bacteria were grown to the mid-exponential phase and washed three times in DMEM-F-12 medium. For infection assays, the cell numbers were adjusted using a Neubauer chamber to the same value in the medium used in the respective assays. When appropriate, the media were supplemented with 100 μg of ampicillin (Amp)/ml, 5 or 2.5 μg of erythromycin (Em)/ml, 10 μg of chloramphenicol (Cm)/ml, or 12.5 μg of tetracycline (Tc)/ml. Growth kinetics of S. aureus SA113 and SA113 lgt::ermB were compared in complex, BM, and MHB medium. The media were inoculated 1:100 with overnight cultures in the respective media. In order to assess growth under nutrient-poor conditions, either DMEM-F-12 or iron-depleted cell culture medium (RPMI-1640; Sigma R-5886) was used.
Disruption of the lgt gene by allelic replacement.
For deletion of the lgt gene in the genome of S. aureus SA113, a 1-kb fragment including hprK was generated by PCR using Pwo polymerase (Hybaid). An EcoRI restriction site (underlined) was introduced into the forward primer (5′-TATTTTAGGAATTCAATGTAGATTGGTGTTATATTTTTG-3′), and the reverse primer (5′-AGTTAAATACAATACCCGGGCAACCTACTCCTCACTC-3′) contained a SmaI restriction site. The fragment was cut with EcoRI and SmaI and subcloned into pUC18 digested with the same restriction enzymes, yielding pUC18hprK. A 1.1-kb fragment including the entire yvoF gene and the 5′ end of the yvcD gene was amplified using 5′-GGCGCTTCCATGCCCGGGAAAAAAAGTGAAGTAGTGATAG-3′ as the forward primer and 5′-TATCTGTAGGATCCTCTTGTAATAGTGCTTCGTAC-3′ as the reverse primer, which contained a SmaI and a BamHI restriction site, respectively, and ligated into pUC18 hprK. The ermB gene from Tn551 was removed from pEC2 by 5′ cleavage with XbaI and 3′ cleavage with HindIII, the ends were filled in with Klenow enzyme, and ermB was blunt end ligated into the SmaI site between the hprK and yvoF genes in pUC18. The inserted fragments were sequenced and introduced into the temperature-sensitive shuttle vector pBT2 harboring the Cmr marker gene cat. The resulting inactivation plasmids were designated pBT-lgt for and pBT-lgt rev. After construction in E. coli, the plasmids were separately electroporated into S. aureus RN4220, purified after selection of transformants on Cm-containing BM agar plates, and electroporated into S. aureus SA113.
Insertional inactivation of lgt by homologous recombination was achieved as described previously (7). Single Em-resistant and Cm-sensitive colonies were presumed to have undergone double crossover and were obtained from cultures harboring the pBTlgt for plasmid. The lgt::ermB genotype of two colonies was confirmed by PCR analysis and sequencing. One clone, SA113 lgt::ermB, was used for further investigations and is referred to here as the lgt mutant.
Construction of expression plasmids.
For complementation of the lgt mutant, plasmid pRBlgt was constructed in E. coli by ligation of the lgt gene to the shuttle vector pRB474 (kindly provided by R. Brückner). lgt was amplified from S. aureus SA113 genomic DNA by PCR using the forward primer 5′-TGAAGAAAGGATCCAGAACAGTCATAAGAGTGAGGAGTAG-3′, which contained a BamHI site (underlined) and the original ribosomal binding site, and the reverse primer 5′-ACTCCCCCGGGGGATTGTATGATGATGTGTTTTTG-3′, which contained a SmaI site (underlined). Plasmid pRBlgt was identified by restriction analysis and was electroporated into S. aureus SA113lgt::ermB. S. aureus SA113lgt::ermB (pRBlgt), referred to as the complemented mutant, retained the wild-type phenotype.
A PCR product of sitC (encoding a 36-kDa lipoprotein) from genomic DNA of S. aureus SA113 carrying the ribosomal binding site was generated using the oligonucleotides 5′-TCACAAGATCTACGAATAGAAAGAAACGAGGAAG-3′ (forward primer, with the introduced BglII restriction site underlined) and 5′-TGTTGATGTGTGGCCTAAAATATTGGAGATACC-3′ (reverse primer, with the original HaeIII restriction site underlined) and ligated into the BamHI-SmaI-digested xylose-inducible expression vector pTX15. The plasmid was introduced into S. carnosus TM300 protoplasts by transformation as previously described (19), and Tc-resistant transformants were selected. pTXsitC was isolated from S. carnosus TM300 and transformed into S. aureus SA113 and the lgt mutant.
[14C]palmitic acid labeling of staphylococcal lipoproteins.
[U-14C]palmitic acid in toluene (Amersham Biosciences) was dried under a nitrogen stream and redissolved in 2% Tween 20. To MHB medium inoculated with an overnight culture of the same medium, 4 μCi of [14C]palmitic acid per ml was added. MHB was used since it is poor in competitive fatty acids. The bacteria were cultivated with shaking for 4.5 h at 37°C. The cells were harvested, and membrane proteins, surface-associated proteins, and supernatants were obtained as described below. Total protein (25 μg) was separated on a sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel and stained with Coomassie brilliant blue R-250. Gels were dried and autoradiographed for 4 to 5 days by using a phosphorimager for detection.
Preparation of staphylococcal crude extracts and isolation of membrane and supernatant proteins.
Bacteria were cultured in the appropriate medium and harvested in the desired growth phase as indicated in Results. For preparation of crude extracts and membrane proteins, cells were harvested by centrifugation for 15 min at 4°C and 5,000 × g and washed twice in ice-cold 20 mM Tris-HCl-100 mM NaCl, pH 8.0. After resuspension in 2.5 ml of buffer with 1 mM phenylmethylsulfonyl fluoride per gram of bacteria, glass beads (0.15 by 0.3 mm in diameter; Sigma) were added, and cells were disrupted mechanically by vortexing five times for 1 min at 4°C each. Contaminating lipopolysaccharide was inactivated by incubating glass beads at 200°C prior to their use in cell activation assays. Glass beads and nondisrupted bacteria were pelleted by centrifugation (3,500 × g for 15 min at 4°C). The crude extract was removed, and remaining cells were disrupted in the same volume of buffer. The pooled crude extract was used for stimulation of human cells or for isolation of membrane proteins. For membrane isolation, the crude extract was pelleted by ultracentrifugation (4°C for 1 h at 50,000 rpm in a Beckman Ti70 rotor) and washed in the same buffer. After a second ultracentrifugation step, membrane proteins were extracted with 2% Triton X-100 in the same buffer by rotation of the samples at 4°C for 1 h. The proteins were harvested by acetone precipitation and centrifugation for 30 min at 4°C and 13,000 × g and resuspended in 50 mM Tris-HCl-10 mM MgCl2-1 mM phenylmethylsulfonyl fluoride, pH 8.0.
Extraction of staphylococcal surface-associated proteins.
Noncovalent surface proteins were removed from staphylococci by LiCl treatment. Cells were grown in MHB for the indicated time period and washed twice with cold 20 mM Tris-HCl, pH 8.0, by vortexing gently and centrifugation for 10 min at 3,000 × g at 4°C. The pellet was resuspended in 1.5 M LiCl and shaken for 2 h at 37°C. Bacteria were pelleted by centrifugation for 15 min at 4,000 × g at 4°C, and the supernatants containing released surface proteins were dialyzed extensively against 10 mM Tris-HCl-20 mM NaCl-5 mM EDTA, pH 8.0, at 4°C. The extracts were stored at −80°C. Total protein (12 μg) was separated on an SDS-12.5% polyacrylamide gel according to Laemmli and stained with Coomassie brilliant blue R-250. Gels were dried and autoradiographed for 4 to 5 days by using a phosphorimager for detection.
Preparation of staphylococcal supernatants for cytokine induction studies.
DMEM-F-12 medium was inoculated with a small amount of bacterial cells grown on BM agar and incubated for 13 to 14 h. Bacteria were removed by centrifugation for 15 min at 5,000 × g at 4°C, and supernatants were filtered through pyrogen-free round filters (pore size, 0.2 μm; Schleicher and Schüll, Dassel, Germany). Freshly prepared supernatants were kept on ice until infection occurred and were adjusted to equal concentrations, corresponding to equal numbers of bacterial cells determined in a Neubauer chamber.
Protein identification by mass spectrometry.
Protein in gels was digested with trypsin as described previously (61) and modified as outlined below. Briefly, protein bands were excised from gels, fully destained, and digested for 3 h with porcine trypsin (sequencing grade, modified with 67 ng of 25 mM ammonium bicarbonate (pH 8.1)/μl; Promega, Madison, Wis.) at 37°C. Prior to peptide mass mapping and sequencing of tryptic fragments by tandem mass spectrometry, peptide mixtures were extracted from gels with 1% formic acid, followed by two changes of 50% acetonitrile. The combined extracts were vacuum dried until only 1 to 2 μl remained, and the peptides were purified by ZipTip according to the manufacturer's instructions (Millipore, Bedford, Mass.). Samples were analyzed by matrix-assisted laser desorption ionization-time of flight by using a Bruker Reflex III mass spectrometer (Bruker Daltonik, Bremen, Germany) equipped with an N2 337-nm laser and gridless pulsed-ion extraction from the matrix alpha-cyano-4-hydroxycinnamic acid-nitrocellulose prepared on the target by using the fast evaporation method (3).
The sequences of some fragments were verified by nanoelectrospray tandem mass spectrometry on a Q-Tof I mass spectrometer (Micromass, Manchester, England) equipped with a nanoflow electrospray ionization source. Gold-coated glass capillary nanoflow needles (Type Medium NanoES spray capillaries) were obtained from Proxeon (Odense, Denmark). Databases (National Center for Biotechnology Information nonredundant protein database [NCBInr]) were searched using MASCOT software from Matrix Science (45).
Cell culture.
MonoMac6 cells, a human monocytic cell line established from the peripheral blood of a patient with leukemia (71), were obtained from H. W. L. Ziegler-Heitbrock (University of Leicester, Leicester, United Kingdom) and cultured in RPMI-1640 (Sigma R-5886) supplemented with 2 mM l-glutamine, 1× minimal essential medium (MEM) nonessential amino acids (both Gibco BRL), oxaloacetate, pyruvate, and insulin (10 ml/liter, provided as OPI supplement mixture; Sigma), and 10% heat-inactivated fetal bovine serum (FBS, 0.25 of EU [Biochrom, Berlin, Germany]/ml). On the day of infection, 106 cells per 24-well microtiter plate were seeded in 750 μl of culture medium.
The human pulmonary epithelial cell line A549 (18) was maintained in DMEM-F-12 medium (Gibco-BRL) containing 2 mM l-glutamine and 10% heat-inactivated FBS (0.25 EU/ml). A549 cells were seeded in 24 wells 2 to 3 days prior to an experiment to reach confluency at the day of infection. All cells were cultured in a water-saturated atmosphere of 5% CO2 at 37°C without using any antibiotics.
HUVEC and umbilical cords were a kind gift from K. E. Unertl (Department of Anaesthesiology and Critical Care Medicine, Tübingen University Hospital, Tübingen, Germany). Umbilical cords were stored no longer than 24 h at 4°C in endothelial growth medium (EGM) enriched with 0.4% endothelial cell growth supplement-heparin, 0.1 ng of epidermal growth factor/ml, 1 ng of basic fibroblast growth factor/ml, 1 μg of hydrocortisone/ml, and 2% heat-inactivated FBS (medium and supplements from Promo Cell, Heidelberg, Germany) containing penicillin, streptomycin, and amphotericin B (1:50 from antibiotic/antimycotic solution; Sigma A-5955). HUVEC were prepared from umbilical cords by limited collagenase perfusion. Briefly, cannulated interior sections from umbilical veins were rinsed with phosphate-buffered saline (PBS) containing antibiotic-antimycotic solution (1:100), filled with prewarmed (37°C) collagenase A solution (1 mg/ml; Roche, 103578), and incubated for 5 min at 37°C. HUVEC were collected by flushing the cells with medium 199 (Sigma M-4530) enriched with 10% heat-inactivated FBS and antibiotic-antimycotic solution (1:100). The cells were washed twice by centrifugation for 5 min at 250 × g in medium 199 containing 10% FBS. The primary cells were maintained in EGM in collagen-I-coated culture flasks (Becton Dickinson) overnight and washed with prewarmed PBS; the cultures were supplied with fresh EGM every 2 days. HUVEC were propagated by splitting after 3 to 4 days of incubation and were subsequently frozen at the second or third passage. After thawing, HUVEC were cultured in 2% gelatin-coated flasks (gelatin from bovine skin; Sigma G-1393) in EGM without antibiotics and antimycotics. For infection assays, the cells were seeded in gelatinized 24-well microtiter plates 2 to 3 days prior to an experiment to reach confluency at the day of infection. HUVEC were used for experiments up to the fourth passage.
Preparation of FITC-labeled bacteria.
Bacteria were grown in MHB to the mid-exponential phase, washed three times in PBS, and resuspended in the same volume of 0.1 M carbonate buffer (pH 9.0). Fluorescein isothiocyanate (FITC, 100 μg/ml; Sigma F-7250) was added, and staphylococci were labeled at 37°C in the dark with gentle shaking for 45 min. After four washes in DMEM-F-12 medium, the bacteria were resuspended in DMEM-F-12 medium and adjusted to appropriate titers in the desired medium.
Adherence assays.
Confluently grown A549 cells (approximately 7 × 105 cells/24 wells) were washed with prewarmed DMEM-F12 medium, and 250 μl of medium was added to each well. Cells were infected with 50 μl of FITC-labeled staphylococcal suspensions, resulting in a multiplicity of infection (MOI) of 5:1. After 1 h of incubation at 37°C under 5% CO2, monolayers were washed three times with DMEM-F-12 medium to remove nonadherent bacteria. Subsequently, cells were fixed with 3.5% paraformaldehyde in PBS, and fluorescent, adherent staphylococci were enumerated. Each experiment was carried out in triplicate, and fluorescent bacteria were counted in eight fields per well by using a Leica DM IRBE fluorescence microscope with a long-distance objective (PL Fluotar L63×/0.70 Corr LMC).
Invasion assays.
MonoMac6 cells were infected in a total volume of 750 μl of culture medium with 4 × 107 staphylococci, resulting in an MOI of 40:1, except as indicated in Results. After 1 h of incubation at 37°C, 250 μl of culture medium containing gentamicin (final concentration, 100 μg/ml) was added to kill extracellular bacteria, and cells were incubated for 1 h. Monocytes were then washed three times in RPMI-1640 supplemented with 2 mM l-glutamine, 1× MEM nonessential amino acids, and OPI mix by centrifugation for 8 min at 400 × g to remove gentamicin. After disruption of the cells in washing medium containing 0.25% Triton X-100, lysates were serially diluted and plated on MHB agar plates. Gentamicin-protected phagocytized bacteria were quantified by colony counting following overnight incubation.
A549 monolayers were washed with prewarmed DMEM-F-12 medium, and 250 μl of culture medium was added to each well. The cells were infected with 50 μl of staphylococci to yield a standard MOI of 40:1 in a total volume of 300 μl. After incubation for 1 h, gentamicin in 700 μl of culture medium was applied to kill extracellular bacteria, resulting in a final concentration of 100 μg/ml and a total volume of 1 ml. Monolayers were incubated for 1 h and washed three times with DMEM-F-12 medium, and cells were lysed with 0.25% Triton X-100 in 750 μl DMEM-F-12 medium. Lysates were plated for quantification of internalized bacteria.
Cell activation assays.
Epithelial monolayers were infected as described for the invasion assays. HUVEC monolayers were washed with prewarmed endothelial basal medium (Promo Cell) and infected with a standard MOI of 50:1 to 75:1 in 300 μl of endothelial cell growth medium. MonoMac 6 cells were infected in a total volume of 750 μl of culture medium with a standard MOI of 40:1. After 1 h of incubation at 37°C, 100 μg of gentamicin/ml was added to prevent bacterial overgrowth, resulting in a total volume of 1 ml, and cells were incubated for the indicated time periods. Gentamicin was applied to monocytes, epithelial cells, and endothelial cells throughout the entire incubation and did not alter cell morphology or viability, as determined microscopically or by using the trypan blue (PAA, Linz, Austria) exclusion method. Viability staining was carried out with MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]; MTT is cleaved by an enzyme in the respiratory chain in mitochondria if the cell is viable, thereby generating MTT formazan, a dark blue, highly visible product. Cells were observed with a light microscope.
Cell culture supernatants were collected by centrifugation for 8 min at 4°C and 600 × g (MonoMac 6) or at 1,000 × g (A549, HUVEC), diluted in 20 mM Tris-HCl-150 mM NaCl-0.1% bovine serum albumin fraction V-0.05% Tween 20, pH 7.4, and stored at −80°C until assayed for cytokine and chemokine concentrations.
Human whole blood infection with staphylococcal cells, staphylococcal culture supernatants, and synthetic lipopeptides.
Blood from healthy donors was collected by venipuncture into sterile pyrogen-free tubes. Anticoagulation was achieved with pyrogen-free heparin. Whole blood (250 μl) was incubated with 250 μl of staphylococcal cell suspensions containing 0.5 × 108 to 2 × 108 bacteria or staphylococcal supernatants at a final concentration of 12 to 50% (vol/vol) in 24-well microtiter plates at 37°C in a water-saturated 5% CO2 atmosphere.
For preparation of staphylococcal supernatants, DMEM-F-12 medium was inoculated with a small amount of bacteria grown on BM agar and incubated for 14 h. Bacteria were removed by centrifugation for 15 min at 5,000 × g and 4°C, and supernatants were filtered through pyrogen-free round filters with a pore size of 0.2 μm (Schleicher and Schüll). Freshly prepared supernatants were kept on ice until infection occurred and were adjusted to equal concentrations with DMEM-F-12 medium corresponding to the original bacterial titers by using a Neubauer chamber.
The staphylococcal culture supernatants, diluted 50% with medium, were supplemented with synthetic lipopeptides. The synthetic lipohexapeptides Pam2Cys-Ser-(Lys)4 {S-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-[R]cysteine; L2020}and Pam3Cys-Ser-(Lys)4 {N-palmitoyl-S-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-[R]-cysteine; L2000} were obtained from EMC Microcollections GmbH, Tübingen, Germany. The lipopeptides were diluted in pyrogen-free water, sonicated extensively, and added to the S. aureus culture supernatants prior to usage. DMEM-F-12 medium instead of staphylococcal suspensions or supernatants served as a negative control. Blood samples were removed for analysis after 5 h of infection, without addition of gentamicin, and centrifuged for 10 min at 4,000 × g at 4°C; plasma was stored at −80°C until used for enzyme-linked immunosorbent assay (ELISA). All experiments were performed in duplicate or triplicate as indicated.
Determination of cytokine and chemokine production by ELISA.
Extracellular release of cytokines and chemokines was determined by a sandwich ELISA technique using the Duo Set ELISA development systems (R&D Systems, Minneapolis, Minn.) according to the manufacturer's instructions. The ELISA detection limits were 15.6 pg/ml (tumor necrosis factor alpha [TNF-α]), 3.9 pg/ml (interleukin-1 [IL-1]), 4.7 pg/ml (IL-6), 31.2 pg/ml (IL-8/CXCL8), 62.5 pg/ml (IL-10), and 15.6 pg/ml (monocyte chemoattractant protein 1 [MCP-1]/CCL2). No corrections for background levels of cytokine and chemokine release were made. Enzyme immunoassay microtiter plates were purchased from Nunc (Roskilde, Denmark).
RESULTS
Screening of the S. aureus genome for genes encoding putative lipoproteins.
The genome of S. aureus N315 was screened for genes encoding putative lipoproteins by using the ParSeq program (58). The search criteria were as follows: methionine at position 1, followed by 2 to 9 amino acids with at least one positive charge, followed by a hydrophobic stretch of 7 to 36 amino acids, followed by the lipobox [LV][ASTVI][GAS][C]. More than 70 putative lipoproteins were identified, 55 of which contained a signal peptide of typical length. Most of these proteins were hypothetical, with unknown function. A number of the proteins, however, showed similarity to known proteins (Table 2), e.g., transporters (iron, zinc, amino acid, oligopeptide, glycine betaine, sugar, and teichoic acid), phage terminases, heme/copper-type cytochrome/quinol oxidase, protein-disulfide isomerase, peptidyl-prolyl cis-trans isomerase (PrsA), and pyruvate-formate-lyase-activating enzyme.
TABLE 2.
Putative lipoproteins of S. aureus N315 with putative functions
| Similarity of proposed lipoproteins in S. aureus N315 | Primary locus namea |
|---|---|
| Metallo-β-lactamase/rhodanese-like domain protein/hydroxyacylglutathione hydrolase | SA0043 |
| Metallo-β-lactamase superfamily | SA0083 |
| ABC-type Fe3+-(hydroxamate, citrate, or hemin) transport system | SA0109 (sirC), SA0110 (sirB), SA0111 (sirA), SA0566, SA0691, SA2079, SA0891, SA0980, SA1979, SA2079 |
| ABC-type oligopeptide and amino acid transport system | SA0010, SA0201, SA0229, SA0849, SA0850, SA2255 |
| Maltose-binding periplasmic proteins/domains | SA0207 |
| ABC-type Fe3+ or thiamine transport system, or spermidine/putrescine-binding protein | SA0217 |
| ABC-type Zn2+ transport system, component/surface adhesin | SA2194, SA0587 |
| ModA, part of the ABC molybdate transporter | SA2074 |
| PBPb CD: diverse group of periplasmic transport receptors for lysine/arginine/ornithine (LAO), glutamine, histidine, sulfate, phosphate, molybdate, and methanol | SA2202 |
| ABC-type glycine betaine transport system | SA2235 |
| NLPA lipoprotein; family contains several antigens that might be involved in bacterial virulence; precise function unknown. | SA0422, SA0771 |
| Phage terminase, small subunit | SA1820, SA1820 |
| Heme/copper-type cytochrome/quinol oxidases, subunit 2. COX2 (cytochrome O ubiquinol oxidase 2) | SA0913 |
| Galactoside ABC transporter, permease protein | SA1361 |
| Teichoic acid transport protein TagG | SA1687 |
| Preprotein translocase subunit YidC: mediates membrane protein insertion in bacteria, essential for E. coli viability, homologues present in mitochondria and chloroplasts | SA1893 |
| Protein-disulfide isomerase | SA2197 |
| Peptidyl-prolyl cis-trans isomerase homolog (PrsA) | SA1659 |
| Radical SAM superfamily; pyruvate-formate lyase-activating enzyme | SA2409 |
The numbers refer to the annotated genome of S. aureus N315 (34).
The lgt operon region and construction of S. aureus SA113 lgt::ermB.
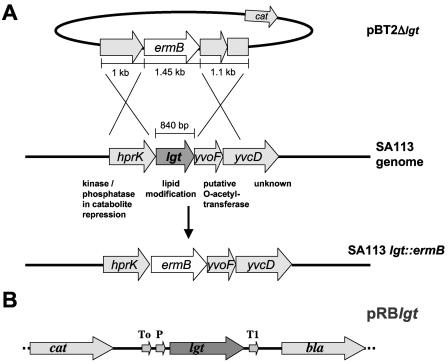
In the genomes of S. aureus strains NCTC8325 and N315, lgt (gene SA0716 in the N315 genome) is flanked upstream by hprK (HPr kinase/phosphatase) and downstream by a gene with similarity to O-acetyltransferase (Fig. 1A). The same gene organization has been found in Staphylococcus xylosus and Staphylococcus epidermidis (26) and is therefore likely conserved in staphylococci. The three genes are transcribed in the same direction and separated by only a few base pairs, which suggests that they are cotranscribed. Whether the two genes flanking lgt are involved in lipoprotein maturation is unknown. However, one can speculate that the O-acetyltransferase homolog could be involved in the acylation of phosphoglycerol to yield phosphatidylglycerol and that the HPr kinase (HPrK) could carry out the dephosphorylation of phosphatidylglycerol to yield the diacylglyceryl group that is transferred to the thiol group of the cysteine residue by Lgt. The lgt deletion mutant of strain SA113 was constructed by replacing the lgt gene with the erythromycin resistance cassette (ermB) without affecting the upstream and downstream genes, yielding SA113 lgt::ermB, referred to as the lgt mutant. This mutant could be complemented by plasmid pRBlgt, in which lgt transcription is driven by the constitutive vegII promoter from B. subtilis (Fig. 1B).
FIG. 1.
Allelic replacement of lgt in the genome of S. aureus SA113 (A) and map of the complementation plasmid pRBlgt (B). (A) A plasmid based on the shuttle vector pBT2 that allowed replacement of lgt by ermB through homologous recombination was constructed. The flanking regions of lgt, hprK upstream, and yvoF/yvcD′ downstream were amplified from the S. aureus SA113 genome. ermB contains its own promoter, which mediates moderate expression of downstream genes and does not contain a transcriptional termination signal. The ermB gene in the isogenic lgt::ermB mutant is in the same orientation as lgt in the wild type. The postulated functions of the proteins encoded by the four genes of the operon are indicated. (B) Only the relevant part of the complementation plasmid pRBlgt is shown. The lgt gene with its original initiation translational signal was cloned using the XbaI and SacI restriction sites of the multiple cloning site of the shuttle vector pRB474. Transcription of lgt is driven by the constitutive vegII promoter from B. subtilis. lgt, prolipoprotein diacylglyceryl transferase; P, vegII promoter; T, transcriptional terminators from phage λ (To) and from rrnB of E. coli (T1); cat, chloramphenicol acetyltransferase gene; bla, β-lactamase.
Lipoprotein monitoring and cellular distribution.
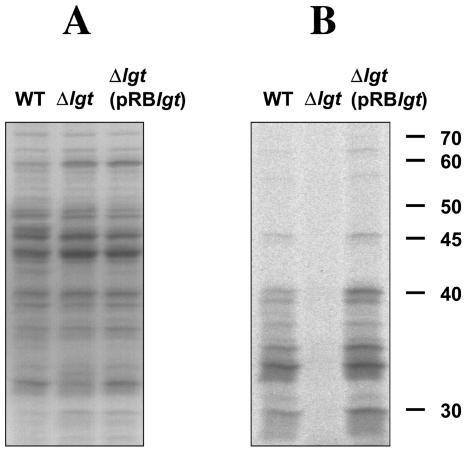
To determine whether lipoproteins were lipid modified in the lgt mutant, lipoproteins were labeled by cultivating cells in the presence of [14C]palmitic acid until the mid-exponential phase was reached. As shown in Fig. 2B, no labeled lipoproteins were observed in the membrane fraction of the mutant, whereas at least 20 labeled proteins (25 to 70 kDa) were observed in the membrane fractions of the wild type and the complemented mutant. In the Coomassie-blue-stained gel (Fig. 2A), no marked differences were observed. Interestingly, some lipoproteins were also present in the culture supernatant of the wild type and the complemented mutant cultivated for 4.5 h (not shown), with a pattern resembling that of the membrane fraction. This observation indicates that lipoproteins were released already in the mid-exponential phase.
FIG. 2.
Lack of lipid modification of lipoproteins in the lgt mutant. Coomassie blue staining of membrane proteins (A) and autoradiograph of membrane proteins in the same gel (B). Wild-type S. aureus, the lgt mutant, and the complemented mutant were cultivated in MHB medium with shaking for 4.5 h at 37°C. Proteins were labeled and isolated as described in Materials and Methods and separated by SDS-PAGE on 12.5% polyacrylamide gels. Right margin, size markers in kilodaltons.
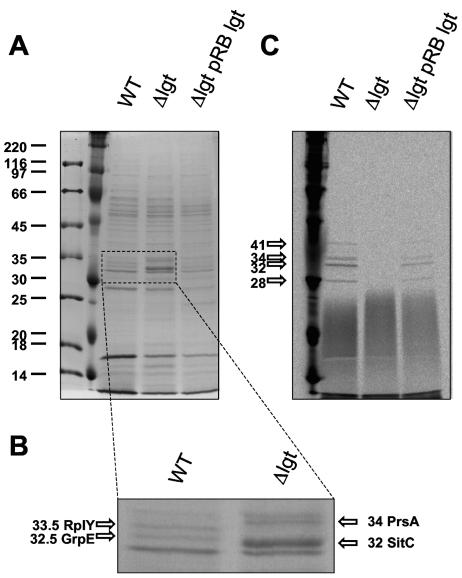
Cell-wall-associated proteins were isolated by treating the cells with 1.5 M LiCl. Separation of the proteins by SDS-polyacrylamide gel electrophoresis (PAGE) and subsequent matrix-assisted laser desorption ionization-time of flight analyses revealed the presence of GrpE (a cochaperon in the DnaK/DnaJ/GrpE system) and L25 (50S ribosomal protein L25) homologs in the wild type, but not in the lgt mutant. The prelipoprotein forms of PrsA and SitC were found only in the preparation of the cell-wall-associated proteins of the lgt mutant (Figs. 3A and B). Lipoproteins could be released from the cell wall, although in lesser amounts than from the membrane fraction (Fig. 3C). These results indicated that in S. aureus, the majority of the lipoproteins are in the membrane fraction and a small proportion are in the cell wall and the culture supernatant. The surface-bound and released staphylococcal lipoproteins could potentially interact with the host immune system.
FIG. 3.
Cell-wall-associated proteins. Cell-wall-associated lipoproteins were released from whole bacterial cells with 1.5 M LiCl. Bacteria were grown in MHB supplemented with [14C]palmitic acid. Total protein (12 μg) was separated on an SDS-12.5% polyacrylamide gel and stained with Coomassie brilliant blue (A and B). Gels were dried and autoradiographed for 4 or 5 days (C). WT, wild type.
The lgt mutant releases unmodified lipoproteins into the culture supernatant.
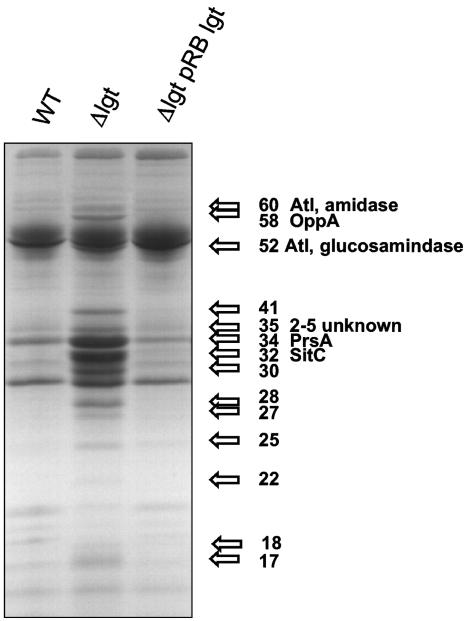
The protein pattern of the culture supernatant of the lgt mutant, cultivated aerobically for 14 h, resembled that of the wild-type membrane fraction (Fig. 4). However, a number of protein bands were also observed in the supernatant of the lgt mutant that were not found in the wild type or complemented mutant. To identify some of the most predominant proteins in the lgt supernatant, the proteins were excised from Coomassie-blue-stained SDS-polyacrylamide gels and identified by mass spectrometry. The location of the encoding genes in the genome of S. aureus 8325, the parental strain of SA113, was determined and compared with that of S. aureus N315, whose genome is annotated (34), by using the BLAST microbial genome database.
FIG. 4.
Release of proteins into the culture supernatant. Proteins from culture supernatants of stationary phase cultures (grown in BM for 14 h with shaking) of S. aureus wild type, the lgt mutant, and the complemented mutant were separated by SDS-PAGE and stained with Coomassie brilliant blue. At least 14 additional or more highly expressed protein bands (arrows) were visually detected in the supernatant of the lgt mutant. Some of the proteins were cut out and partially sequenced. Autolysin (Atl), oligopeptide-binding protein (OppA) and autolysin homolog protein comigrate; an unidentified protein and 1 to 4 homologous proteins comigrate at the 35-kDa position. PrsA, peptidyl-prolyl cis-trans isomerase; SitC, binding protein of staphylococcal iron transporter.
Most of the examined proteins were derived from lipoproteins; two were not lipoproteins. The following proteins were identified. (i) SitC (encoded by SA0587 of strain N315), a 32-kDa (309-amino acid [aa]) binding protein of the staphylococcal iron transporter SitABC, which exhibited the most predominant protein band. This lipoprotein contains an ATP/GTP-binding motif A (P-loop) and shows sequence similarity to the streptococcal adhesin PsaA. (ii) The 34-kDa peptidyl-prolyl cis-trans isomerase PrsA (encoded by SA1659 of strain N315). The 320-aa lipoprotein also appeared as a predominant band and is involved in protein folding. (iii) The 58-kDa oligopeptide-binding protein OppA (encoded by SA0894 of strain N315). The 551-aa lipoprotein is part of an oligopeptide ABC transporter system. (iv) A 35-kDa (317 aa) lipoprotein of unknown function (encoded by SA1056 of strain N315). (v) The 35-kDa protein band, which also contained another protein that corresponds to four homologous lipoproteins of unknown function: encoded by SA1317 (305 aa), SA1318 (316 aa), SA1319 (305 aa), and SA1321 (309 aa) of strain N315 (the latter is a lipoprotein in SA113, but lacks the leader sequence in N315). Therefore, we do not know which of these homologs are expressed. (vi) Surprisingly, also two nonlipoprotein enzymes derived from the AtlE major autolysin (encoded by SA0905 of strain N315), the 60-kDa amidase, and the 52-kDa N-acetylglucosaminidase (23), which were present in higher amounts in the lgt mutant. Both enzyme domains of AtlE are usually cell wall bound and processed by proteolytic cleavage. An explanation for the greater release by the mutant could be that the cell wall integrity in the lgt mutant is affected and that, therefore, also cell-wall-associated nonlipoproteins are more readily released.
The four lipoprotein genes sitC, prsA, SA1056, and oppA are conserved in the other staphylococcal genomes determined. We assume that many of the lipoproteins of the lgt mutant are released into the culture supernatant.
Transient anchoring of lipoproteins by unprocessed signal sequences to the mutant membrane.
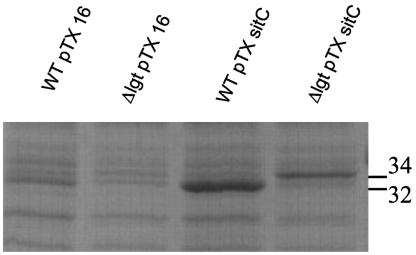
The sitC gene was cloned in the xylose-inducible vector pTX15, yielding pTXsitC, which was introduced into the wild type and lgt mutant by transformation. Bacteria were grown overnight, and proteins from the membrane fraction were separated by SDS-PAGE and stained (Fig. 5). The apparent molecular mass of SitC in the membrane fraction of the wild type was 32 kDa, whereas it was 34 kDa in the lgt mutant. This difference in size can be explained by the unprocessed leader peptide in the lgt mutant protein; the leader peptide anchors the protein, at least transiently, to the cytoplasmic membrane. On the other hand, the secreted form of SitC in the lgt mutant had a size of 32 kDa. We assume that the majority of the SitC protein lacking lipid is released from the membrane by proteolytic processing, even though a typical Ala-X-Ala motif is lacking.
FIG. 5.
Localization of SitC to the membranes of S. aureus SA113 (pTXsitC) and S. aureus SA113 lgt::ermP (pTXsitC). Expression of sitC was induced by the addition of xylose after 1 h of growth. Bacteria were grown in BM for 13 h, and proteins from the membrane fraction were separated by SDS-PAGE and stained with Coomassie brilliant blue. pTX16 is the control plasmid. In the wild-type (WT) strain carrying pTXsitC, SitC migrates at the 32-kDa position, whereas in the lgt mutant carrying pTXsitC, SitC migrates at the 34-kDa position, very likely because it contains a leader sequence.
Lipoproteins lacking lipids become phenotypically apparent only under substrate limitation or stress situations.
The lgt mutant had a pleiotropic phenotype. More than 50 lipoproteins lacked lipids, and many were released into the culture supernatant. One would expect that this alteration would be reflected in a general retardation in growth.
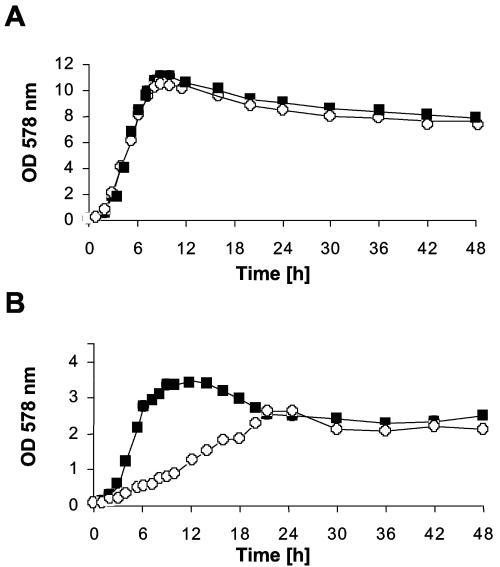
However, in complex media, such as BM, there was hardly any difference between the wild-type and the lgt mutant with respect to growth rate, maximal growth level, and CFU (Fig. 6A). The growth of the lgt mutant was impaired only in the culture medium RPMI-1640, which is nutrient poor (Fig. 6B). Anaerobic growth of the lgt mutant in the absence of nitrate was reduced (results not shown). Again, the results obtained indicated that the lack of lipid modification of lipoproteins becomes phenotypically apparent only under substrate limitation or stress situations.
FIG. 6.
Growth of S. aureus wild-type and lgt mutant in different growth media. Fresh medium was inoculated to an optical density (OD) at 578 nm of 0.1 with staphylococcal overnight cultures that had been grown for 18 h in the respective medium. (A) BM; (B) RPMI-AV1. Filled squares, wild type; open circles, lgt mutant.
The lgt mutant is severely affected in activation of inflammatory immune responses.
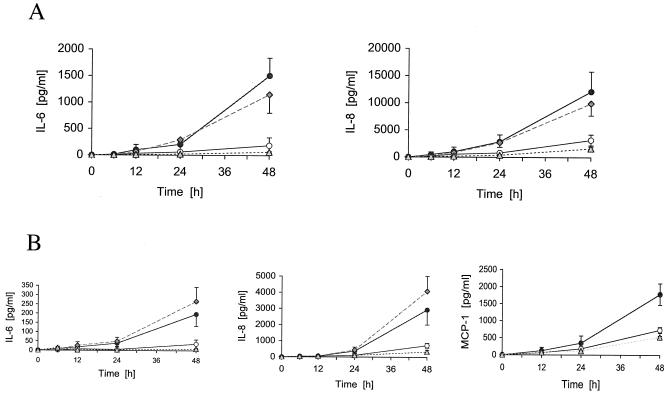
Activation of A549 pulmonary epithelial and endothelial cells. Epithelial cells, as an important component of mucosal defenses, provide both barrier and signaling functions to protect against bacterial pathogens. They release cytokines and chemokines in order to activate phagocytic cells. We used A549 pulmonary type II epithelial cells in our assays since S. aureus is an important respiratory pathogen of immunocompromised and hospitalized people and cystic fibrosis patients. Epithelial monolayers were infected with the S. aureus wild type, the lgt mutant, or the complemented mutant, and the induction of IL-6, an important mediator of the acute-phase response and activator of systemic inflammation, and IL-8, a chemokine and activator of neutrophils, was assessed. Over time, the wild type and the complemented lgt mutant induced many more cytokines than the lgt mutant (Fig. 7A). The optimal dose response (MOI) was 25 to 50 bacteria per cell. At a higher MOI, the monolayer was disrupted owing to cytotoxic effects, as observed by microscopy and quantified by staining A549 cells with MTT (data not shown). Therefore, we assume that the decreased cell activation by the lgt mutant is not due to an elevated cytotoxic effect.
FIG. 7.
Production of IL-6, IL-8, or MCP-1 by A549 (A) and HUVEC (B) upon stimulation with S. aureus wild-type and lgt mutant cells. For time kinetic studies, A549 cells were infected with a ratio of 40:1; HUVEC were infected with a ratio of 50:1 to 75:1. Dose-dependent stimulation was determined after 36 h (results not shown). In some experiments, a basal IL-6 or IL-8 secretion was observed. Data represent means ± standard deviations of three different experiments, each carried out in duplicate. Filled circles, wild type; open circles, lgt mutant; gray diamonds, complemented lgt mutant; gray triangles, medium (control).
When local infections cannot be controlled, dissemination of S. aureus into the bloodstream can occur, which enables the colonization of other tissues. Adhesion to and transcytosis of intact and damaged endothelia play a crucial role for metastatic infections. HUVEC were infected with the S. aureus wild type, the complemented lgt mutant, or the lgt mutant, and the induction of IL-6, IL-8, and MCP-1 was assessed (Fig. 7B). As with the epithelial cells, only the wild type and complemented lgt mutant induced IL-6, IL-8, and MCP-1 significantly. The MOI was 50 to 75 bacteria per cell. The onset of activation started after 24 h of incubation.
While there was a clear difference in the induction of cytokines and chemokines, there was no difference between the wild type and the lgt mutant in attachment to or invasion of epithelial and endothelial cells (data not shown). We assume that the lipoproteins present in the wild-type cells are most likely responsible for cytokine and chemokine induction.
Activation of monocytes.
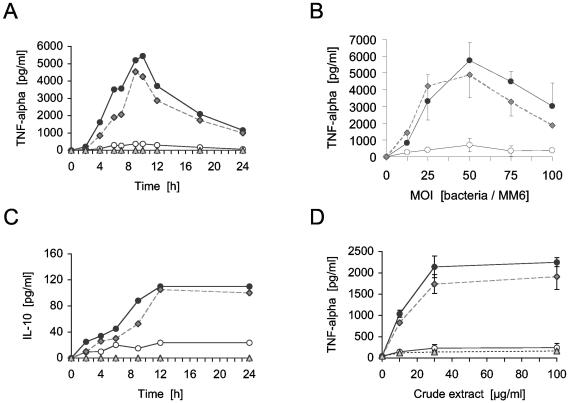
Induction of the proinflammatory cytokines TNF-α and IL-1β in MonoMac6 cells was investigated at an MOI of 40:1. Only the wild type and the complemented lgt mutant triggered a significant TNF-α response (Fig. 8A and B) and an IL-1β response (results not shown). Crude extracts (containing intracellular, membrane, and cell wall components) of the wild type and the complemented mutant provoked a concentration-dependent TNF-α production (Fig. 8D). In contrast, cells and extracts of the lgt mutant failed to induce any TNF-α response (Fig. 8A, B, and D), whereas a moderate activation of the anti-inflammatory cytokine IL-10 was observed (Fig. 8C). Culture supernatants of overnight cultures of all strains did not induce cytokine responses in the assays (results not shown). These results suggest that the MonoMac6 activating factor in the wild type is membrane or cell wall associated, which is supportive of the involvement of bacteria-associated lipoproteins.
FIG. 8.
Induction of TNF-α or IL-10 in MonoMac 6 cells by S. aureus wild type and the lgt mutant (A through C) or crude extract (D). For time-dependent studies with bacteria, MonoMac 6 cells were infected with a ratio of 40:1. Dose-dependent stimulation was determined after 8 h. Panels A and C show one representative experiment of three, each carried out in duplicate. In panel B, data of three different experiments carried out in duplicate are shown. Panel D illustrates one representative experiment carried out with three separate crude extract preparations. Crude extract was derived from overnight cultures in DMEM-F-12, and the indicated amounts were added to 106 monocytes; 10 μg of the crude extract (wet weight)/ml corresponds to 2.6 × 106 bacteria. Error bars indicate standard deviation. Filled circles, S. aureus SA113 wild type; open circles, the isogenic lgt::erm mutant; gray diamonds, complemented lgt mutant; gray triangles, medium.
Activation of whole blood.
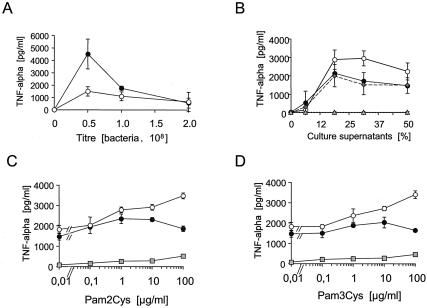
Whole blood was used instead of blood monocytes or peripheral blood mononuclear cell preparations to minimize preactivation of immune cells and to mimic the in vivo situation as closely as possible. When whole blood was used for stimulation with S. aureus cells and culture supernatants, S. aureus wild type induced higher TNF-α concentrations than the lgt mutant, but in contrast to single cell types, a significant cytokine induction by the lgt mutant could be detected (Fig. 9A). Also, in this assay system, the culture supernatants were able to induce TNF-α production, and surprisingly, the supernatant of the lgt mutant was superior to that of the wild type in a distinct concentration range (Fig. 9B).
FIG. 9.
Cytokine induction in human whole blood by S. aureus cells, supernatants, and synthetic lipopeptides. Fresh blood samples were treated for 5 h with either bacteria (A) or bacterial culture supernatants (B through D). (C and D) Synthetic lipopeptides were added to the bacterial supernatants (diluted 50% with medium) at the indicated concentrations prior to stimulation of the blood samples. Values are means of three (A) or four (B) different experiments, each carried out in duplicate. Panels C and D show one representative experiment of three, carried out in triplicate. Error bars indicate standard deviations. Filled circles, S. aureus SA113 wild type; open circles, the isogenic lgt::ermP mutant; gray diamonds, complemented lgt mutant; gray triangles, medium (control).
Synthetic lipopeptides alone were only poor inducers of cytokines even at high concentrations (100 μg/ml) (Fig. 9C and D). However, different effects were observed when lipopeptides were added to wild-type or mutant supernatants. The addition of Pam2Cys or Pam3Cys to wild-type culture supernatant diluted 50% with medium had little effect. At higher concentrations (>10 μg/ml), stimulation was even repressed. Apparently, the wild-type culture supernatant was saturated with mature lipoproteins. With the culture supernatant of the lgt mutant, a dose-dependent increase in stimulation was observed when either Pam2Cys or Pam3Cys was added, which indicated that the culture supernatant was not saturated with lipoproteins (Fig. 9C and D). Therefore, addition of synthetic lipopeptides led to a further increase in TNF-α stimulation. Our results also indicate that human blood cells respond equally well to diacylated or triacylated lipoproteins, even in combination with other soluble inducing factors.
DISCUSSION
Bacterial lipoproteins from different species have recently been shown to play a role in virulence and to be potent inducers of cytokines (14, 70). Most of these studies, however, have been carried out using isolated molecules or synthetic lipopeptides. Results of these investigations might provide only a rough insight into the significance of bacterial lipoproteins since the physiological backgrounds of other stimuli, such as peptidoglycan, teichoic acids, CpG DNA, or toxins, have not been considered. Furthermore, any isolated components could include trace contaminants that could mask or affect the function of the isolated component to an unknown magnitude and quality. We therefore compared S. aureus wild type with its lgt mutant, in which the lipidation of all prelipoproteins is affected, with respect to activation of inflammatory host responses.
More than 300 distinct bacterial lipoproteins are listed in the DOLOP database of bacterial lipoproteins (38; http://www.mrc-lmb.cam.ac.uk/genomes/dolop/). By screening the S. aureus genome, we identified more than 50 genes displaying a typical type II signal sequence, 35 of which can be associated with a known or predicted function (Table 2). At least 14 protein bands derived from the supernatant of an lgt mutant overnight culture were visualized in a Coomassie-blue-stained SDS-polyacrylamide gel, representing released proteins not present in the wild-type supernatant (Fig. 4). This protein pattern resembled that of the radiolabeled lipoprotein bands derived from the membrane fraction of S. aureus wild type in the autoradiographs (Fig. 3B). Therefore, the additional proteins released by the lgt mutant can be assumed to be unlipidated prelipoproteins which could not be retained in the bacterial membrane. Indeed, when SitC was overexpressed, the majority of this protein was not retained in the membrane of the lgt mutant (Fig. 5). We estimate approximately 20 to 25% of the unlipidated SitC molecules to be retained in the membrane of the lgt mutant. We assume that generally unmodified and unprocessed hydrophobic leader sequences dock the corresponding prelipoprotein at least transiently to the cytoplasmic membrane. This explains the ability of the lgt mutant to acquire nutrients and to behave like the wild type, provided that a substantial substrate concentration is available. Therefore, a possible defect in the lipoprotein maturation pathway can be tolerated by S. aureus under optimal growth conditions.
Indeed, despite the large number of lipoproteins in S. aureus, our results show that the lack of lipid modification had no apparent effect on the growth of S. aureus in rich medium, a finding that is consistent with results from other authors obtained for B. subtilis (36) and S. pneumoniae (47). However, by using different media and by nutrient supplementation, we demonstrated that growth of the mutant was much more strongly affected in nutrient-poor medium than that of the wild type, which indicated an impaired ability of the mutant to acquire nutrients.
In both the LiCl extract and the culture supernatant of the lgt mutant, SitC and PrsA were found in higher concentrations than in the wild type and the complemented mutant (Fig. 4). These proteins probably interact with cell wall components, as has been shown for PsaA, a streptococcal SitC homolog, which is retained in the cell wall of Lactobacillus casei when heterologously expressed. In contrast, PspA, another streptococcal protein, is released into the medium (42).
Interestingly, LiCl treatment also released GrpE and L25 (RplY) from the cell surface of wild-type S. aureus; these proteins were thought to be located in the cytoplasm (Fig. 3A and B). GrpE is a component of the DnaK (Hsp70)/DnaJ (Hsp40)/GrpE (Hsp20) chaperone system, which has a broad array of biological functions, including protein folding and disaggregation, mediating the accessibility to proteolysis, and heat shock regulation (reviewed in reference 17). GrpE (Hsp20) acts as a nucleotide exchange factor that accelerates the rate of ADP dissociation from high-affinity ADP-DnaK, thus enabling ATP binding and the transition of DnaK to the low-affinity state (39) and promoting the efficient release of the substrate (5). GrpE also functions as a thermosensor that initiates the heat shock response (20). In principle, GrpE is a typical cytoplasmic protein; however, in some bacterial species, it is located at the surface along with DnaK (Hsp70) (49, 57). The large ribosomal protein L25 (RplY) binds specifically to loop E of the 5S rRNA (37) and shows strong sequence similarity to the general stress protein Ctc of B. subtilis, which also specifically binds 5S rRNA (56).
We have no clear explanation for the mechanism by which GrpE (Hsp20) and L25 are brought to the S. aureus cell surface. Whether GrpE and L25 become surface localized by autolysis or by a specific secretion mechanism is unknown. It is, however, remarkable that GrpE and L25 are found only on the surfaces of wild-type S. aureus cells and not on those of the lgt mutant (Fig. 3A and B). They are possibly associated with the lipid moiety of the lipoproteins and in this way become exposed to the outer surface, since many prokaryotic and eukaryotic heat shock proteins have been shown to interact noncovalently with lipids and fatty acids (2, 10, 21).
The infection assays with several cell types revealed that only the S. aureus wild type induces a strong inflammatory response with time, whereas the lgt mutant fails to provoke a considerable host cell activation. Only a moderate decrease in the production of anti-inflammatory IL-10 was observed. Based on these results, the lack of lipid modification has consequences for an efficient immune response to S. aureus since a whole set of proinflammatory cytokines and chemokines can be induced only when lipidation of lipoproteins occurs. We assume for two reasons that the decreased cell activation by the lgt mutant is not due to metabolic side effects such as a decreased growth rate but rather to specific molecular interactions between host cell receptors and the lipoproteins. First, crude extracts derived from the wild type, but not from the lgt mutant, were able to induce TNF-α in MonoMac6 cells (Fig. 8D) and IL-8 in A549 cells (data not shown) when used in equal amounts. Second, similar results were obtained from infection experiments when MonoMac6 cells were treated with bacteria that had been killed either by heat, gentamicin, or lysostaphin prior to infection (results not shown).
S. aureus is taken up essentially by phagocytes via Fc-γ receptors or complement receptors. The pathogen also invades epithelial, endothelial, and fibroblast cells, where fibronectin-binding proteins and the host fibronectin receptor integrin α5β1 play a crucial role (62). Some phagocytic receptors themselves trigger cytokine production, and phagocytic and inflammatory receptors often functionally cooperate, albeit these processes appear not to be necessarily coupled (69). The ability of the lgt mutant to adhere to or to invade various types of host cells was not affected (results not shown). Therefore, the lgt deletion does not affect staphylococcal surface structures or the engagement of host receptors that play a crucial role in the uptake processes. We assume that the decreased stimulating activity of the lgt mutant is caused by its inability to produce lipoproteins and that the cell-activating properties of lipoproteins might be due to the specific activation of inflammatory pathways in different host cells, mediated by inflammatory host cell receptors at the cell surface or in endosomal compartments.
The result that culture supernatants of the lgt mutant were equal or even superior to wild-type culture supernatants in the activation of whole blood samples (Fig. 9B) was somewhat surprising since some lipoproteins in the culture supernatant of the wild type and complemented lgt mutant grown for 4.5 h were detected (results not shown). It remains to be elucidated whether the lack of lipidated proteins in the culture supernatant of the lgt mutant is overcompensated by the released unlipidated prelipoproteins, or whether the mutant releases slightly higher concentrations of other pathogen-associated molecular patterns, such as peptidoglycan, lipoteichoic acid, unmethylated cytosine-guanine dinucleotide sequences, N-formyl-methionine peptides, and/or phospholipids; however, an increased autolysis rate of the mutant was not observed. Perhaps there is some disordering of the peptidoglycan or the teichoic acids, possibly caused by an affected teichoic acid transport protein, TagG, which is a lipoprotein. It has been shown that the tagGH operon of B. subtilis encodes a two-component ABC transporter involved in the metabolism of wall teichoic acids (35).
We think that the free mature lipoproteins in the S. aureus wild-type supernatant play only a minor role in immune activation or rather modulate the responses triggered by other soluble factors in a concentration-dependent manner. This assumption is based on our observation that synthetic lipopeptides alone have only an inadequate stimulating potential and that the low TNF-α-inducing activity of the supernatant of the lgt mutant can to some extent be increased by increasing concentrations of synthetic lipopeptides, whereas wild-type supernatants appear to be saturated with soluble lipoproteins (Fig. 9C and D). Pam2Cys and Pam3Cys acted in a similar manner, which indicated that an additional aminoacylation of the lipidated cysteine residue of the lipoproteins is not crucial for the stimulatory potency of S. aureus supernatants. We did not determine the structure of the lipoproteins, but we screened the published genome sequences of strains of S. aureus for a gene encoding a lipoprotein N-acyl transferase (Lnt) homolog and found no such protein, which suggests that lipoproteins in S. aureus are only diacylated. We therefore expect an involvement of TLR2 and/or TLR6 heterodimers in such a receptor complex, which is necessary for the recognition of diacylated lipoproteins (65).
The majority of the released wild-type lipoproteins might be organized in aggregates, and therefore, the concentration of freely accessible lipid structures might be too low to induce a strong activation in the tested cells. The mechanisms of human cells in mediating proinflammatory responses depend on the type and aggregation state of bacterial inducers in homotypic or heterotypic complexes and also depend on whether these factors are soluble or particle bound (15). Generally, a synergistic action of different microbial components, including lipoproteins, seems to be necessary to enable a full inflammatory outcome (59, 60).
In conclusion, our findings contribute to an emerging picture of molecular mechanisms at the host/bacterial interface and broaden the understanding of the physiological significance of bacterial lipoproteins in innate immune activation. We showed that the lack of lipid modification of lipoproteins in S. aureus has a clear effect in the immune response to S. aureus but has only a subtle effect on cell growth and is not vital in gram-positive bacteria. Bacterial-induced inflammation is a two-edged sword and can lead either to clearance of the pathogens or to severe systemic inflammation. The lgt mutant can be regarded as a suitable tool for in vivo investigations of the role of microbial lipoproteins on gram-positive infections. Whether the S. aureus lgt mutant displays attenuated virulence in vivo remains to be investigated.
Acknowledgments
We thank Andreas Peschel and Regine Landmann for stimulating discussions and Alfred Nordheim (Interfakultäres Institut für Zellbiologie) for use of the MALDI equipment; Klaus Unertl, Torsten Schröder, and Alice Mager (Universitätsklinik für Anaesthesiologie und Transfusionsmedizin) for helping in the isolation of HUVEC from umbilical cords; H. W. L. Ziegler-Heitbrock (University of Leicester) for providing us with MonoMac6 cells; and Karen A. Brune for editing the manuscript.
This work was supported by the BMBF (01GG9804/9) and the DFG, Graduate College “infection biology” 685.
Editor: J. B. Bliska
REFERENCES
- 1.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 2.Arispe, N., M. Doh, and A. De Maio. 2002. Lipid interaction differentiates the constitutive and stress-induced heat shock proteins Hsc70 and Hsp70. Cell Stress Chaperones 7:330-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnott, D., K. L. O'Connell, K. L. King, and J. T. Stults. 1998. An integrated approach to proteome analysis: identification of proteins associated with cardiac hypertrophy. Anal. Biochem. 258:1-18. [DOI] [PubMed] [Google Scholar]
- 4.Bengtsson, J., H. Tjalsma, C. Rivolta, and L. Hederstedt. 1999. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J. Bacteriol. 181:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehmer, D., C. Gassler, W. Rist, M. P. Mayer, and B. Bukau. 2004. Influence of GrpE on DnaK-substrate interactions. J. Biol. Chem. 279:27957-27964. [DOI] [PubMed] [Google Scholar]
- 6.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 7.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Brückner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 9.Bulut, Y., E. Faure, L. Thomas, O. Equils, and M. Arditi. 2001. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167:987-994. [DOI] [PubMed] [Google Scholar]
- 10.De Bruyn, J., K. Soetaert, P. Buyssens, I. Calonne, J. L. De Coene, X. Gallet, R. Brasseur, R. Wattiez, P. Falmagne, H. Montrozier, M. A. Laneelle, and M. Daffe. 2000. Evidence for specific and non-covalent binding of lipids to natural and recombinant Mycobacterium bovis BCG Hsp60 proteins, and to the Escherichia coli homologue GroEL. Microbiology 146:1513-1524. [DOI] [PubMed] [Google Scholar]
- 11.Elsner, A., B. Kreikemeyer, A. Braun-Kiewnick, B. Spellerberg, B. A. Buttaro, and A. Podbielski. 2002. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect. Immun. 70:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, W. 1994. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med. Microbiol. Immunol. (Berlin) 183:61-76. [DOI] [PubMed] [Google Scholar]
- 14.Fisette, P. L., S. Ram, J. M. Andersen, W. Guo, and R. R. Ingalls. 2003. The Lip lipoprotein from Neisseria gonorrhoeae stimulates cytokine release and NF-kappaB activation in epithelial cells in a Toll-like receptor 2-dependent manner. J. Biol. Chem. 278:46252-46260. [DOI] [PubMed] [Google Scholar]
- 15.Flo, T. H., L. Ryan, L. Kilaas, G. Skjak-Braek, R. R. Ingalls, A. Sundan, D. T. Golenbock, and T. Espevik. 2000. Involvement of CD14 and beta2-integrins in activating cells with soluble and particulate lipopolysaccharides and mannuronic acid polymers. Infect. Immun. 68:6770-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan, K., S. D. Gupta, K. Sankaran, M. B. Schmid, and H. C. Wu. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in prolipoprotein modification. J. Biol. Chem. 268:16544-16550. [PubMed] [Google Scholar]
- 17.Georgopoulos, C., K. Liberek, M. Zylicz, and D. Ang. 1994. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Giard, D. J., S. A. Aaronson, G. J. Todaro, P. Arnstein, J. H. Kersey, H. Dosik, and W. P. Parks. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51:1417-1423. [DOI] [PubMed] [Google Scholar]
- 19.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 20.Groemping, Y., and J. Reinstein. 2001. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J. Mol. Biol. 314:167-178. [DOI] [PubMed] [Google Scholar]
- 21.Guidon, P. T., Jr., and L. E. Hightower. 1986. The 73 kilodalton heat shock cognate protein purified from rat brain contains nonesterified palmitic and stearic acids. J. Cell. Physiol. 128:239-245. [DOI] [PubMed] [Google Scholar]
- 22.Hantke, K., and V. Braun. 1973. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur. J. Biochem. 34:284-296. [DOI] [PubMed] [Google Scholar]
- 23.Heilmann, C., M. Hussain, G. Peters, and F. Götz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 24.Hirschfeld, M., C. J. Kirschning, R. Schwandner, H. Wesche, J. H. Weis, R. M. Wooten, and J. J. Weis. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 163:2382-2386. [PubMed] [Google Scholar]
- 25.Hussain, M., S. Ichihara, and S. Mizushima. 1982. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257:5177-5182. [PubMed] [Google Scholar]
- 26.Huynh, P. L., I. Jankovic, N. F. Schnell, and R. Brückner. 2000. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol. 182:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inouye, S., S. Wang, J. Sekizawa, S. Halegoua, and M. Inouye. 1977. Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc. Natl. Acad. Sci. USA 74:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs, M., J. B. Andersen, V. Kontinen, and M. Sarvas. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8:957-966. [DOI] [PubMed] [Google Scholar]
- 30.Jenkinson, H. F., R. McNab, D. M. Loach, and G. W. Tannock. 1995. Lipoprotein receptors in oral streptococci. Dev. Biol. Stand. 85:333-341. [PubMed] [Google Scholar]
- 31.Kolenbrander, P. E. 1993. Coaggregation of human oral bacteria: potential role in the accretion of dental plaque. J. Appl. Bacteriol. 74(Suppl.):79S-86S. [DOI] [PubMed] [Google Scholar]
- 32.Kolenbrander, P. E., R. N. Andersen, R. A. Baker, and H. F. Jenkinson. 1998. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J. Bacteriol. 180:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 35.Lazarevic, V., and D. Karamata. 1995. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol. Microbiol. 16:345-355. [DOI] [PubMed] [Google Scholar]
- 36.Leskela, S., E. Wahlstrom, V. P. Kontinen, and M. Sarvas. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol. Microbiol. 31:1075-1085. [DOI] [PubMed] [Google Scholar]
- 37.Lu, M., and T. A. Steitz. 2000. Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8-A resolution. Proc. Natl. Acad. Sci. USA 97:2023-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madan Babu, M., and K. Sankaran. 2002. DOLOP-database of bacterial lipoproteins. Bioinformatics 18:641-643. [DOI] [PubMed] [Google Scholar]
- 39.Mally, A., and S. N. Witt. 2001. GrpE accelerates peptide binding and release from the high affinity state of DnaK. Nat. Struct. Biol. 8:254-257. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, J. B., and J. O. Lampen. 1982. Membrane-bound penicillinases in Gram-positive bacteria. J. Biol. Chem. 257:4490-4495. [PubMed] [Google Scholar]
- 41.Norgard, M. V., L. L. Arndt, D. R. Akins, L. L. Curetty, D. A. Harrich, and J. D. Radolf. 1996. Activation of human monocytic cells by Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides proceeds via a pathway distinct from that of lipopolysaccharide but involves the transcriptional activator NF-κB. Infect. Immun. 64:3845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira, M. L., V. Monedero, E. N. Miyaji, L. C. Leite, P. Lee Ho, and G. Perez-Martinez. 2003. Expression of Streptococcus pneumoniae antigens, PsaA (pneumococcal surface antigen A) and PspA (pneumococcal surface protein A) by Lactobacillus casei. FEMS Microbiol. Lett. 227:25-31. [DOI] [PubMed] [Google Scholar]
- 43.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 45.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 46.Peschel, A., B. Ottenwälder, and F. Götz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 47.Petit, C. M., J. R. Brown, K. Ingraham, A. P. Bryant, and D. J. Holmes. 2001. Lipid modification of prelipoproteins is dispensable for growth in vitro but essential for virulence in Streptococcus pneumoniae. FEMS Microbiol. Lett. 200:229-233. [DOI] [PubMed] [Google Scholar]
- 48.Qi, H. Y., K. Sankaran, K. Gan, and H. C. Wu. 1995. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. J. Bacteriol. 177:6820-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raulston, J. E., C. H. Davis, D. H. Schmiel, M. W. Morgan, and P. B. Wyrick. 1993. Molecular characterization and outer membrane association of a Chlamydia trachomatis protein related to the hsp70 family of proteins. J. Biol. Chem. 268:23139-23147. [PubMed] [Google Scholar]
- 50.Rawadi, G., J. Garcia, B. Lemercier, and S. Roman-Roman. 1999. Signal transduction pathways involved in the activation of NF-kappa B, AP-1, and c-fos by Mycoplasma fermentans membrane lipoproteins in macrophages. J. Immunol. 162:2193-2203. [PubMed] [Google Scholar]
- 51.Reglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 278:49469-49477. [DOI] [PubMed] [Google Scholar]
- 52.Romero-Steiner, S., T. Pilishvili, J. S. Sampson, S. E. Johnson, A. Stinson, G. M. Carlone, and E. W. Ades. 2003. Inhibition of pneumococcal adherence to human nasopharyngeal epithelial cells by anti-PsaA antibodies. Clin. Diagn. Lab. Immunol. 10:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudner, D. Z., J. R. LeDeaux, K. Ireton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sankaran, K., S. D. Gupta, and H. C. Wu. 1995. Modification of bacterial lipoproteins. Methods Enzymol. 250:683-697. [DOI] [PubMed] [Google Scholar]
- 55.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269:19701-19706. [PubMed] [Google Scholar]
- 56.Schmalisch, M., I. Langbein, and J. Stulke. 2002. The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J. Mol. Microbiol. Biotechnol. 4:495-501. [PubMed] [Google Scholar]
- 57.Schmiel, D. H., J. E. Raulston, E. Fox, and P. B. Wyrick. 1995. Characterization, expression and envelope association of a Chlamydia trachomatis 28 kDa protein. Microb. Pathog. 19:227-236. [DOI] [PubMed] [Google Scholar]
- 58.Schmollinger, M., I. Fischer, C. Nerz, S. Pinkenburg, F. Götz, M. Kaufmann, K. J. Lange, R. Reuter, W. Rosenstiel, and A. Zell. 2004. ParSeq: searching motifs with structural and biochemical properties. Bioinformatics 20:1459-1461. [DOI] [PubMed] [Google Scholar]
- 59.Seifert, R., G. Schultz, M. Richter-Freund, J. Metzger, K. H. Wiesmüller, G. Jung, W. G. Bessler, and S. Hauschildt. 1990. Activation of superoxide formation and lysozyme release in human neutrophils by the synthetic lipopeptide Pam3Cys-Ser-(Lys)4. Involvement of guanine-nucleotide-binding proteins and synergism with chemotactic peptides. Biochem. J. 267:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sela, M. N., A. Bolotin, R. Naor, A. Weinberg, and G. Rosen. 1997. Lipoproteins of Treponema denticola: their effect on human polymorphonuclear neutrophils. J. Periodontal Res. 32:455-466. [DOI] [PubMed] [Google Scholar]
- 61.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 62.Sinha, B., P. Francois, Y.-A. Que, M. Hussain, C. Heilmann, P. Moreillon, D. Lew, K.-H. Krause, G. Peters, and M. Herrmann. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68:6871-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugai, M., and H. C. Wu. 1992. Export of the outer membrane lipoprotein is defective in secD, secE, and secF mutants of Escherichia coli. J. Bacteriol. 174:2511-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutcliffe, I. C., and R. R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeuchi, O., T. Kawai, P. F. Muhlradt, M. Morr, J. D. Radolf, A. Zychlinsky, K. Takeda, and S. Akira. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13:933-940. [DOI] [PubMed] [Google Scholar]
- 66.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R. L. Modlin, and S. Akira. 2002. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10-14. [DOI] [PubMed] [Google Scholar]
- 67.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanimoto, K., and D. B. Clewell. 1993. Regulation of the pAD1-encoded sex pheromone response in Enterococcus faecalis: expression of the positive regulator TraE1. J. Bacteriol. 175:1008-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825-852. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H., D. W. Niesel, J. W. Peterson, and G. R. Klimpel. 1998. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziegler-Heitbrock, H. W., E. Thiel, A. Futterer, V. Herzog, A. Wirtz, and G. Riethmüller. 1988. Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int. J. Cancer 41:456-461. [DOI] [PubMed] [Google Scholar]