Abstract
Background
Invasive fungal infections have been described throughout the COVID-19 pandemic. Cryptococcal disease after infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been reported in several isolated case reports and 1 larger case series. We sought to describe cryptococcal infections following SARS-CoV-2 through establishing a database to investigate underlying risk factors, disease manifestations, and outcomes.
Methods
We created a crowdsourced call for cases solicited through the Mycoses Study Group Education and Research Consortium, the Centers for Disease Control and Prevention Emerging Infectious Diseases Network, and infectious diseases Twitter groups. Data were collected in a web-based and secure REDCap survey without personal identifiers.
Results
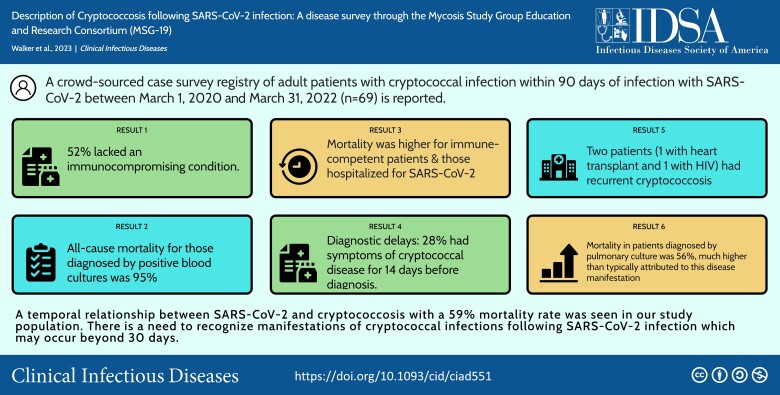
Sixty-nine cases were identified and submitted by 29 separate institutional sites. Cryptococcosis was diagnosed a median of 22 days (interquartile range, 9–42 days) after SARS-CoV-2 infection. Mortality among those with available follow-up was 72% (26/36) for the immunocompetent group and 48% (15/31) for the immunocompromised group (likelihood ratio, 4.01; P = .045). We observed a correlation between disease manifestation (central nervous system infection, proven/probable disseminated disease, and respiratory) and mortality (P = .002).
Conclusions
The mortality rate of 59% for patients with cryptococcosis following SARS-CoV-2 is higher than that of modern Cryptococcus cohorts. There was an association between immunocompromised status and cryptococcal disease manifestations as well as mortality. Moreover, our series emphasizes the need for clinical and laboratory assessment of opportunistic infections beyond 30 days when concerning symptoms develop.
Keywords: SARS-CoV-2, COVID-19, cryptococcal disease, cryptococcal meningitis, crowdsourced survey
We provide a description of cryptococcal infections following SARS-CoV-2 infection describing predisposing factors, symptoms, disease manifestations, and mortality. Findings were notable for high mortality, associated with immunocompetent status and hospitalization for viral illness.
Graphical Abstract
Graphical Abstract.
This graphical abstract is also available at Tidbit: https://tidbitapp.io/tidbits/description-of-cryptococcosis-following-sars-cov-2-infection-a-disease-survey-through-the-mycosis-studygroup-education-and-research-consortium-msg-19-1025e8ce-d229-408a-9712-b4946015aa6b
Cryptococcosis is an invasive yeast infection typically caused by Cryptococcus neoformans and Cryptococcus gattii. It is well described in all hosts but is most commonly seen in immunosuppressed patients. Current understanding of cryptococcosis is that the disease usually represents reactivation of quiescent infection from the lungs, which then disseminates into the bloodstream and other sites including the central nervous system (CNS) [1, 2].
Invasive fungal infections have been described throughout the coronavirus disease 2019 (COVID-19) pandemic. It is known that a hyperinflammatory state and cytokine release leads to a down-regulation in CD4 and CD8 cells, which is postulated to be the predisposing factor to concurrent fungal disease [3, 4]. Infections due to Mucorales, Aspergillus, and Candida species have been well described shortly following or concurrent with the viral infection [3–6]. Cryptococcus has been described in several isolated case reports following COVID-19 [7, 8] and 1 large case series based on electronic medical record (EMR) billing data [9], but generally timing has been within 2 weeks of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [9, 10]. However, cryptococcal disease in general has a slower incubation period than the other associated fungal infections, thus presenting a more unique challenge in establishing this temporal relationship [1, 2]. Moreover the mortality rates described in these series and case reviews were around 50% [9, 10], which is much higher than typically associated with cryptococcosis. Several patients with cryptococcal infection were observed from 1 of our institutions, most of whom were immunocompromised, but all of whom developed cryptococcosis within 90 days of proven SARS-CoV-2 infection. To gain a better understanding of this phenomenon, we surveyed several centers within the Mycoses Study Group Education and Research Consortium (MSGERC) network and beyond, using crowdsourcing outreach, to describe cryptococcal infection following SARS-CoV-2 infection.
METHODS
We designed a crowdsourced case survey registry of patients with cryptococcal infection within 90 days of infection with SARS-CoV-2. We accepted case patients who were diagnosed between 1 March 2020 and 31 March 2022.
Case Definitions
To be eligible for this survey, the cryptococcal infection must have occurred within 90 days of documented SARS-CoV-2 infection. Cryptococcal infection was defined as a positive serum and/or cerebrospinal fluid (CSF) cryptococcal antigen (CrAg) or a positive culture for Cryptococcus spp from any site. We limited our cases in this series to adults >18 years of age. We categorized cryptococcal infection as follows: CNS disease, disseminated cryptococcosis (proven/probable), and respiratory disease alone. The definition for CNS disease required either positive CSF culture or positive CSF CrAg. Disseminated cryptococcosis was defined based on serum and blood studies. Proven disseminated cryptococcosis required positive blood cultures for Cryptococcus spp, whereas probable disease was defined by positive serum CrAg with negative blood cultures. If a patient had CNS disease, they were classified as this regardless of other cultures. If patient had positive blood cultures or serum antigen without CNS involvement, then they were classified as having proven or probable disseminated disease. Respiratory cultures only were cases in whom diagnosis was made by positive respiratory cultures without any additional positive cultures or antigen results. Immunocompromised individuals were defined as those with human immunodeficiency virus (HIV), solid organ transplant (SOT), active malignancy on treatment, or chronic immunosuppressive medications.
Survey Development
The registry was developed to collect basic demographic information (age, sex, geographic region), known risk factors for cryptococcosis (immunocompromised states, diabetes, prior cryptococcal infection), description of SARS-CoV-2 infection (method of diagnosis, year of infection, hospitalization, therapies provided), and cryptococcosis disease (results of diagnostic tests, signs and symptoms of cryptococcosis, days between SARS-CoV-2 infection and cryptococcal infection, treatment, and 6-month outcomes). Contact details of site investigators were included to allow follow-up and clarification of all entries. All case data were to be collected through a secure REDCap database, and no identifiable protected health information was requested (Supplementary File 1). The University of Alabama at Birmingham institutional review board (IRB) determined that the registry was exempt from full review. Some participating sites were able to use our IRB exemption; others had policies requiring independent IRB submissions at the local sites before participating.
Survey Distribution
After responding to the call for cases meeting study-prespecified definitions, providers were emailed a web link to the study REDCap electronic data capture tools hosted at the University of Alabama at Birmingham [11, 12]. The survey instrument included contact information for the recording provider for the purpose of resolving missing or unclear data through direct email. Providers would complete a separate survey for each case they had identified. Our survey responses were collected from 11 February 2022 through May 31 2022.
We used several means to disseminate information about the availability of the survey. First, the MSGERC released a call for cases of cryptococcosis following SARS-CoV-2 infection through an independent listserv. We also disseminated the survey through the Centers for Disease Control and Prevention Emerging Infectious Diseases Network. Finally, we disseminated information about the survey through the Twitter infectious diseases community. Providers who were interested in participating were sent a direct email that included the REDCap survey web link. Four investigators (E. E. O., M. M., A. M. R., D. O.) identified cases from their institutions through locally approved searches of institutional EMR data to identify potential cases and then retrospectively reviewed the EMR to ensure they met inclusion criteria. This accounts for 26 of our total cases. Twelve additional cases were contributed by 2 centers (University of Alabama at Birmingham and Duke University) and reflect the collective of cases recognized by clinical faculty at those institutions. The remaining 31 cases were submitted by individual responses to the distributed survey.
Data Analysis
Data from the registry REDCap database were analyzed using descriptive statistics. The χ2 test was used to compare groups, including host immune factors and mortality.
RESULTS
Sixty-nine cases were identified and submitted from 29 separate sites. Individual providers from 6 sites contributed a range of 4–9 cases each. Providers from the remaining sites contributed a range of 1–3 cases each. The majority of cases were reported from United States sites (Table 1). Median age of cases was 61 years and 68% were male; 36 (52%) cases had no significant immunocompromising condition. SARS-CoV-2 infection was documented across the time from pandemic onset in early 2020 through 2022. Fifty-six cases received some directed treatment for COVID-19 including 39 (57%) receiving antivirals, 15 (22%) antibody, and 51 (74%) additional immunosuppressants. The immunosuppressants used included glucocorticosteroids (n = 50), interleukin 6 inhibitor (IL-6; n = 7), and Janus kinase inhibitor (n = 5). Thirteen cases did not require hospitalization for SARS-CoV-2 infection and, of that group, only 2 received additional immunosuppressants as outpatient, both of which were glucocorticosteroids. Seven of the 13 cases not hospitalized for SARS-CoV-2 were immunocompromised due to active malignancy (n = 4), chronic immunosuppressants (n = 1), HIV (n = 1), and SOT (n = 1).
Table 1.
Patient Characteristics (N = 69)
| Characteristic | No. (%) |
|---|---|
| Age, y, median (IQR) | 61 (53–70) |
| Sex | |
| Male | 47 (68) |
| Female | 22 (32) |
| Country | |
| United States | 66 (96) |
| Southeast | 26 (38) |
| Midwest | 21 (30) |
| Southwest | 14 (20) |
| West | 4 (4) |
| Northeast | 1 (1) |
| International | 3 (4) |
| Immunocompromising condition | |
| None | 36 (52) |
| Solid organ transplant | 15 (22) |
| Malignancy on chemotherapy | 8 (12) |
| Other chronic immunosuppression | 7 (10) |
| HIV | 5 (7) |
| Other | 2 (3) |
| Comorbidity | |
| Diabetes | 28 (41) |
| Any chronic immunosuppressant | 27 (39) |
| Multiple agents | 16 (23) |
| Glucocorticoids | 17 (25) |
| Calcineurin inhibitors | 13 (19) |
| Antimetabolite | 11 (16) |
| T-cell co-stimulation blocker | 2 (3) |
| Alkylating agent | 2 (3) |
| Tyrosine kinase inhibitor | 2 (3) |
| Anti-CD20 | 1 (1) |
| mTOR inhibitor | 1 (1) |
| Year of COVID-19 diagnosis | |
| 2020 | 15 (22) |
| 2021 | 40 (58) |
| 2022 | 8 (12) |
| Unknown | 5 (7) |
| COVID-19 treatment | |
| Hospitalization for COVID-19 | 56 (81) |
| Supportive care only | 13 (19) |
| Any immunosuppressant | 51 (74) |
| Glucocorticosteroids | 50 (72) |
| IL-6 inhibitor | 7 (10) |
| JAK inhibitor | 5 (7) |
| Antiviral | 39 (57) |
| Remdesivir | 38 (55) |
| Paxlovid | 1 (1) |
| Antibody | 15 (22) |
| Convalescent plasma | 9 (13) |
| Monoclonal antibodies | 6 (9) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: COVID-19, coronavirus disease 2019; HIV, human immunodeficiency virus; IL-6, interleukin 6; IQR, interquartile range; JAK, Janus kinase; mTOR, mammalian target of rapamycin.
Cryptococcosis was diagnosed a median of 22 days (interquartile range, 9–42 days) after SARS-CoV-2 infection (Table 2). The symptoms were broad and overlapped with SARS-CoV-2 symptoms including cough (n = 30), fever (n = 26), confusion (n = 20), and lethargy (n = 18). Eleven cases of cryptococcal infection were diagnosed postmortem, primarily through blood or respiratory cultures. Evaluation for cryptococcal infection included blood cultures in 63 (91%), serum CrAg in 54 (78%), and CSF evaluation in 33 (47%). The site of infection was confirmed to be CNS cryptococcosis in 13 (19%), proven disseminated disease in 22 (32%), probable disseminated disease in 18 (26%), and respiratory disease only in 16 (23%) (Table 3). All of the 16 with respiratory disease had some evaluation for disseminated Cryptococcus, 11 by serum CrAg testing and the remaining 5 with blood cultures (Supplementary File 2 includes available testing information for all patients). Amphotericin B was used for induction therapy in 39 cases (57%). Of those 30 cases without amphotericin B induction, the reasons cited were diagnosis made postmortem (n = 11), disease severity did not warrant the use of this drug (n = 10), unknown reason (n = 4), drug not available (n = 3), and concerns for toxicity (n = 2).
Table 2.
Cryptococcosis Following Coronavirus Disease 2019
| Timing of Cryptococcal Infection Following COVID-19 | No. (%) |
|---|---|
| Days post-COVID-19 that Cryptococcus was diagnosed (n = 68), median (IQR) | 22 (9–42) |
| Symptoms of Cryptococcus | |
| Cough | 30 (43) |
| Fever | 26 (38) |
| Confusion | 20 (29) |
| Lethargy | 18 (26) |
| Headache | 17 (25) |
| Dyspnea | 17 (25) |
| Nausea | 6 (9) |
| Weakness | 5 (7) |
| Vomiting | 4 (6) |
| Vision changes | 3 (4) |
| None | 2 (3) |
| Unknown | 11 (16) |
| Confirmation of cryptococcal diseasea | |
| Serum CrAg performed | 54 (78) |
| Positive | 41 (59) |
| Blood cultures performed | 63 (91) |
| Positive | 30 (43) |
| CSF CrAg performed | 33 (47) |
| Positive | 13 (19) |
| CSF culture performed | 33 (47) |
| Positive | 9 (13) |
| Diagnosed by other culture | 16 (23) |
| BAL/tracheostomy aspirate | 10 (14) |
| Sputum | 5 (7) |
| Pleural effusion | 1 (1) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: BAL, bronchoalveolar lavage; COVID-19, coronavirus disease 2019; CrAg, cryptococcal antigen; CSF, cerebrospinal fluid; IQR, interquartile range.
aDiagnosis confirmed on the basis of data that were returned postmortem (n = 11).
Table 3.
Location of Cryptococcal Disease Diagnosis, Host Factors, and Outcomes
| Location of Cryptococcal Disease Diagnosis | Immunocompromised (n = 33) | Immunocompetent (n = 36) | 6-Month Outcome | ||
|---|---|---|---|---|---|
| Alive (n = 18) | Dead (n = 40) | Unknown | |||
| Respiratory cultures only | 3 | 13 | 7 | 9 | 0 |
| Probable disseminated cryptococcosis | 12 | 6 | 10 | 8 | 0 |
| Proven disseminated cryptococcosis | 9 | 13 | 1 | 19 | 2 |
| CNS involvement | 9 | 4 | 8 | 5 | 0 |
| χ2 | P = .0129 | P = .0020 | |||
Abbreviation: CNS, central nervous system.
Outcomes at 6 months were available in 67 of 69 patients (97%). Within that group, 41 (61%) patients had died within 6 months, representing 59% of our total population. Both patients for whom outcomes were unavailable were lost to follow-up. Mortality among those with available follow-up was 72% (26/36) for the immunocompetent group and 48% (15/31) for the immunocompromised group (likelihood ratio, 4; P = .046). Mortality outcomes were also higher for those hospitalized for SARS-CoV-2 (P < .001) and those with certain cryptococcal disease presentations (P = .002). Specifically, those with proven disseminated cryptococcosis and pulmonary cryptococcosis had the highest all-cause mortality at 95% (19/20) and 56% (9/16), respectively. By comparison, those with probable disseminated disease and CNS disease had all-cause mortality of 44% (8/18) and 38% (5/13), respectively. Disease category at time of diagnosis was also related to immunocompromised status, with immunocompromised patients having more frequent CNS disease (27% [9/33]) and probable disseminated disease (36% [12/33]), while immunocompetent patients were more likely to be diagnosed by positive blood culture (36% [13/36]) and respiratory culture (36% [13/36]). Among 13 patients with CNS disease, complications included cerebrovascular accident in 3, permanent shunting due to persistently elevated opening pressure in 2, and immune reconstitution–like syndrome in 2.
We observed a few unusual presentations of Cryptococcus among these subjects that should be highlighted. Two patients had recurrent cryptococcal infection. One patient had a history of heart transplant and CNS cryptococcosis 9 years previously and had completed therapy 7 years prior to diagnosis of COVID-19. The patient had minimally symptomatic SARS-CoV-2 infection managed with supportive care only, and subsequently presented with recurrent CNS cryptococcal disease including positive blood and CSF cultures. This patient responded to reinduction antifungal therapy with amphotericin B and flucytosine, then transitioned to lifelong suppressive fluconazole. Another patient had uncontrolled HIV and CNS cryptococcal infection 2 years prior to COVID-19 diagnosis and was no longer receiving antiretroviral or antifungal therapy when they presented with symptoms of cryptococcal CNS infection and consistent CrAg results. Finally, a patient with rheumatoid arthritis on chronic immunosuppressive medications had positive blood cultures for C. neoformans but simultaneously had negative serum CrAg, suggesting a prozone phenomenon or less likely an acapsular strain.
DISCUSSION
Invasive mycoses associated with severe SARS-CoV-2 infections are being increasingly recognized, especially pulmonary aspergillosis and invasive candidiasis. Herein we report the characteristics of subjects who developed CNS, disseminated, or respiratory cryptococcosis shortly following SARS-CoV-2 infection. There have been numerous case reports and small reviews describing cryptococcal infection following COVID-19 [8, 10, 13–15]. In this series of 69 cases from 29 centers, we attempt to better describe the characteristics of this population including disease manifestations, underlying comorbidities, treatment, and outcome. The largest series to date described 65 patients who were identified via International Classification of Diseases, Tenth Revision (ICD-10) codes from anonymous data captured through a federated research network [9]. This series was notable for high rates of HIV (32%), organ transplant (28%), and immunodeficiency (47%) which, alongside other autoimmune disease, diabetes, and end-organ failure, were all found to be risk factors for cryptococcosis when compared to other patients identified in their system with COVID-19 [9]. Our disease survey adds significant detail to this body of literature as each case was confirmed by both standardized testing and individualized provider review. Additionally, our cohort includes more patients traditionally considered immunocompetent and demonstrates an overall high mortality.
In immunocompromised states such as SOT, cryptococcosis is generally seen as a reactivation of quiescent disease residing in lungs, which may disseminate into bloodstream and the CNS. This fact and the low baseline incidence of infection make it challenging to establish a causal association between SARS-CoV-2 and cryptococcal diagnosis. Our series identified a few groups of individuals that provide important clues to this disease phenomenon.
We identified 11 cases postmortem through blood (n = 7) and respiratory cultures (n = 4). The median time between diagnosis of SARS-CoV-2 infection and cryptococcosis was 22 days (range, 5–42 days). Nine of these patients were immunocompetent preceding SARS-CoV-2 infection, all were hospitalized for COVID-19 treatment, and most received immunosuppressive medications for COVID-19 (6 received glucocorticoids and 1 received both glucocorticoids and an IL-6 inhibitor). Overall, there were 36 immunocompetent individuals who most commonly manifested disease through positive cultures either from respiratory system or blood. This pattern fits with our known epidemiology of cryptococcosis, and the rapid blood dissemination suggests a profoundly immunosuppressed state as has been described in critical illness with COVID-19 [3]. The immunocompetent group had a very high mortality at 72%, significantly higher than the 48% mortality observed in immunocompromised patients, with likelihood ratio of 4. A trend toward higher mortality in immunocompetent individuals was also suggested in a prior study [16]. Overall mortality among patients with respiratory disease alone was >56%, significantly higher than reported in any modern case series, where pulmonary cryptococcosis is consistently associated with lower mortality risks, usually <10% [16]. Further exploration is required to describe this phenomenon, but we suspect the profound lung injury and immunodeficiency seen with severe SARS-CoV-2 infection could predispose to rapid quiescent reactivation with organism burden sufficient for culture-proven disease. Although the retrospective nature of this study limited testing, 69% defined with respiratory disease did have negative serum CrAg testing. We can only speculate about the impact of pulmonary cryptococcal infection on overall mortality in our series, but at a minimum it appears to be a prognostic marker in hospitalized patients with SARS-CoV-2.
We identified 13 cases with CNS cryptococcosis, which is typically considered the latest stage and the most severe disease manifestation. This was seen more commonly in patients with immunosuppressed conditions (69%) but not necessarily severe COVID-19. Indeed, 31% did not require hospitalization for COVID-19 and 24% were managed with supportive care only. The development of this severe manifestation in absence of critical illness (defined as hospitalization) and without additional immunosuppressants suggests some direct impact from viral illness on the immune system. Although this group of patients with CNS disease had lower mortality in our series than other disease sites, it remains high compared to literature with mortality rate at 38%. The overlap of symptoms for COVID-19 and Cryptococcus are difficult to distinguish and a few cases described a diagnostic delay due to symptoms such as headache, nausea, and fatigue being attributed to “long COVID.” Thus it is important to consider cryptococcal disease including meningitis in the 3- to 6-week period following SARS-CoV-2 infection, particularly in those with additional risk factors such as immunocompromised states.
The most compelling case example to suggest a causative relationship between viral infection and subsequent cryptococcal disease was from a heart transplant recipient with recurrent cryptococcal disease. This patient had completed cryptococcal treatment >7 years prior without recurrence. The patient's SARS-CoV-2 infection was mild, and the patient was managed as an outpatient without any change in baseline immunosuppressants (sirolimus and tacrolimus) or additional therapy. At time of cryptococcal diagnosis, both CNS and blood cultures were positive. This case provides support toward an additional risk to infection following viral infection, although further study is required to confirm and define the level of risk and understand how it varies based on presentation of viral infection from SARS-CoV-2. For example, the predisposition for pulmonary mold infections is well described in influenza [3, 17], but Cryptococcus has been described only in a few reports [7, 8, 18].
Many studies looking at concurrent infections with SARS-CoV-2, particularly in immunocompromised persons such as SOT recipients, have focused on nosocomial infections and short-term outcomes with average follow-up <1 month [5, 6]. The prior series utilizing ICD-10 codes found that most cases occurred within 10 days, but we found a longer delay with median of 22 days between diagnosis, and 25 cases (36%) had diagnosis confirmed after 30 days. This difference could be related to our methodology, as physician case surveys would be more likely to link outpatient SARS-CoV-2 infections to inpatient cryptococcal infections. In addition to later presentations, we also observed diagnostic delays with 19 (28%) of our cases having symptoms of cryptococcal disease for ≥14 days prior to diagnosis of fungal infection. This delay has been described frequently in cryptococcal disease along with more severe disease manifestations and complications [2, 16]. Thus, our data suggest a need for assessment of opportunistic infections beyond 30 days for patients with SARS-CoV-2 infection, especially in subsequent studies.
This report is limited to retrospective data collection entered as a survey by medical providers and thus some data points are incomplete due to these tests not being completed or unavailable. We did require documentation of laboratory confirmation for both diagnoses to ensure case confirmation. Although this approach prevents us from being able to review other EMR data not included in our survey (Supplementary File 1), it did allow us to expand reach to 29 centers. Although we utilized listservs with global contacts for our “call for cases,” we only had 3 cases submitted from outside the United States. Given that Cryptococcus is a cosmopolitan organism, it would be expected that this limitation speaks to the reach of the professional networks utilized rather than a geographic predisposition, although this cannot be stated with certainty. As our case series is heterogenous including many types of immunosuppression, as well as patients not traditionally considered immunosuppressed, we are unable to provide a sufficient comparator group and thus we are limited to case description rather than identifying causation. Finally, the retrospective design of the study may promote recruitment of memorable cases, which could contribute to the higher mortality observed. However, more than half of our cases represented a broader institutional experience rather than individual anecdote and higher mortality has been demonstrated in other descriptions of invasive fungal infections following severe respiratory illness including SARS-CoV-2 [9, 18, 19]. The strengths of our study include the number of cases and complexity of information regarding risk factors, disease manifestations, and outcomes.
This case series highlights a temporal relationship between SARS-CoV-2 and cryptococcosis and reveals a concerning mortality rate of 59% in our overall population that is significantly higher than other cryptococcal infection case series in the modern era. Although a theoretical framework exists to explain this association, it has not been previously described in this amount of detail. Notably, several of our cases were delayed in diagnosis, in part due to attributions of symptoms to long-term effects of COVID-19. This demonstrates the need for careful consideration for clinicians when caring for this population.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Jeremey Walker, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
Todd McCarty, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
Gerald McGwin, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
Eloy E Ordaya, Mayo Clinic, Infectious Diseases, Rochester, Minnesota, USA.
Paschalis Vergidis, Mayo Clinic, Infectious Diseases, Rochester, Minnesota, USA.
Luis Ostrosky-Zeichner, University of Texas Health Sciences Center, Infectious Disease, Houston, Texas, USA.
Mehriban Mammadova, University of Texas Health Sciences Center, Infectious Disease, Houston, Texas, USA.
Andrej Spec, Washington University, Division of Infectious Diseases, St Louis, Missouri, USA.
Adriana M Rauseo, Washington University, Division of Infectious Diseases, St Louis, Missouri, USA.
John Perfect, Duke University, Infectious Diseases, Durham, North Carolina, USA.
Julia Messina, Duke University, Infectious Diseases, Durham, North Carolina, USA.
Gabriel Vilchez, University of Kentucky, College of Medicine, Lexington, Kentucky, USA.
Rachel McMullen, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
Carolynn T Jones, The Ohio State University, College of Nursing, Columbus, Ohio, USA.
Peter G Pappas, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
for the Mycoses Study Group Education and Research Consortium (MSGERC) Cryptococcal Registry Cohort:
Zachary Yetmar, Nigo Masayuki, Julie Steinbrink, Lizbeth Cahuayme-Zuniga, Shobha Vootukuri, Chizaram Onyeaghala, Tuan V Ta, Pratibha Kale, Alexander Franklin, Ravi Gandhi, Darin Ostrander, Marisa Miceli, Nathaniel Warner, Lora Thomas, Karam Obeid, Ricardo M La Hoz, Ada Sochanska, Benjamin Klausing, Rima El-Herte, Amir Tirmizi, Edward C Traver, George R Thompson, Chelsea Gorsline, Geetha Sivasubramanian, Rebecca Osborn, and Mark Mounajjed
Notes
Author Contributions. J. W., P. G. P., T. M., R. M., C. T. J., and J. M. contributed to the design of the study, case report form and database development, data analyses, and initial manuscript outline. The remaining authors contributed to manuscript development and editing, in addition to contributing cases.
Acknowledgments. Multiple additional physicians and clinicians contributed cases to this registry and constitute the remaining members of the MSGERC Cryptococcal Registry Cohort: Zachary Yetmar, Mayo Clinic, Rochester, Minnesota; Nigo Masayuki, University of Texas Health Sciences Center, Houston; Julie Steinbrink, Duke University, Durham, North Carolina; Lizbeth Cahuayme-Zuniga, Baylor Scott & White Medical Center, Temple, Texas; Shobha Vootukuri, Our Lady of the Lake Physicians Group, Baton Rouge, Louisiana; Chizaram Onyeaghala, University of Port Harcourt Teaching Hospital, Port Harcourt, Nigeria; Tuan V. Ta, Orange County Global Medical Center, Santa Ana, California; Pratibha Kale, Institute of Liver and Biliary Sciences, New Delhi, India; Alexander Franklin, Baylor College of Medicine, Houston, Texas; Ravi Gandhi, Jeannina Smith, and Lucas Schultz, University of Wisconsin, Madison; Darin Ostrander, Johns Hopkins University, Baltimore, Maryland; Marisa Miceli, University of Michigan, Ann Arbor; Nathaniel Warner and Lora Thomas, Virginia Commonwealth University, Richmond; Karam Obeid, University of Minnesota, Minneapolis; Ricardo M. La Hoz, University of Texas Southwestern Medical Center, Dallas; Ada Sochanska, University of Arkansas, Little Rock; Benjamin Klausing, Baptist Health Medical Group, Louisville, Kentucky; Rima El-Herte, Creighton University, Omaha, Nebraska; Amir Tirmizi, Indiana University, Indianapolis; Edward C. Traver, University of Maryland, Baltimore; George R. Thompson, University of California, Davis; Chelsea Gorsline, University of Kansas Medical Center, Kansas City; Geetha Sivasubramanian, University of California, San Francisco at Fresno; Rebecca Osborn; Northwestern University, Chicago, Illinois; Mark Mounajjed, SSM Health, Madison, Wisconsin.
Disclaimer. The Centers for Disease Control and Prevention (CDC) is an agency within the Department of Health and Human Services (HHS). The contents of this article do not necessarily represent the policy of CDC or HHS and should not be considered an endorsement by the federal government.
Financial support. This work was supported in part by a cooperative agreement with the Centers for Disease Control and Prevention (CFD-RFA-CK20-2003) to the University of Alabama at Birmingham. The University of Alabama at Birmingham is collaborating with the Mycoses Study Group Education and Research Consortium on this initiative.
References
- 1. Baddley JW, Forrest GN. Cryptococcosis in solid organ transplantation—guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 2019; 33:e13543. [DOI] [PubMed] [Google Scholar]
- 2. Maziarz EK, Perfect JR. Cryptococcosis. Infect Dis Clin North Am 2016; 30:179–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhatt K, Agolli A, Patel MH, et al. High mortality co-infections of COVID-19 patients: mucormycosis and other fungal infections. Discoveries (Craiova) 2021; 9:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 2020; 81:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azzi Y, Bartash R, Scalea J, Loarte-Campos P, Akalin E. COVID-19 and solid organ transplantation: a review article. Transplantation 2021; 105:37–55. [DOI] [PubMed] [Google Scholar]
- 6. Kates OS, Haydel BM, Florman SS, et al. COVID-19 in solid organ transplant: a multi-center cohort study. Clin Infect Dis 2020; 73:e4090–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghanem H, Sivasubramanian G. Cryptococcus neoformans meningoencephalitis in an immunocompetent patient after COVID-19 infection. Case Rep Infect Dis 2021; 2021:5597473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khatib MY, Ahmed AA, Shaat Said B, Mohamed AS, Nashwan AJ. Cryptococcemia in a patient with COVID-19: a case report. Clin Case Rep 2021; 9:853–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chastain DB, Kung VM, Golpayegany S, et al. Cryptococcosis among hospitalised patients with COVID-19: a multicentre research network study. Mycoses 2022; 65:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Regalla D, VanNatta M, Alam M, Malek AE. COVID-19-associated Cryptococcus infection (CACI): a review of literature and clinical pearls. Infection 2022; 50:1007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap): a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thota DR, Ray B, Hasan M, Sharma K. Cryptococcal meningoencephalitis during convalescence from severe COVID-19 pneumonia. Neurohospitalist 2021; 12:96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Passerini M, Terzi R, Piscaglia M, Passerini S, Piconi S. Disseminated cryptococcosis in a patient with metastatic prostate cancer who died in the coronavirus disease 2019 (COVID-19) outbreak. Cureus 2020; 12:e8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Traver EC, Malavé Sánchez M. Pulmonary aspergillosis and cryptococcosis as a complication of COVID-19. Med Mycol Case Rep 2022; 35:22–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brizendine KD, Baddley JW, Pappas PG. Predictors of mortality and differences in clinical features among patients with cryptococcosis according to immune status. PLoS One 2013; 8:e60431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alshabani K, Haq A, Miyakawa R, Palla M, Soubani AO. Invasive pulmonary aspergillosis in patients with influenza infection: report of two cases and systematic review of the literature. Expert Rev Respir Med 2015; 9:89–96. [DOI] [PubMed] [Google Scholar]
- 18. Huang J, Li H, Lan C, et al. Concomitant severe influenza and cryptococcal infections: a case report and literature review. Medicine 2019; 98:e15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shishido AA, Mathew M, Baddley JW. Overview of COVID-19-associated invasive fungal infection. Curr Fungal Infect Rep 2022; 16:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.