Abstract
Background
Persistent Staphylococcus aureus bacteremia is associated with metastatic infection and adverse outcomes, whereas gram-negative bacteremia is normally transient and shorter course therapy is increasingly advocated for affected patients. Whether the prolonged detection of pathogen DNA in blood by culture-independent systems could have prognostic value and guide management decisions is unknown.
Methods
We performed a multicenter, prospective, observational study on 102 patients with bloodstream infection (BSI) to compare time to bloodstream clearance according to T2 magnetic resonance and blood cultures over a 4-day follow-up. We also explored the association between duration of detectable pathogens according to T2 magnetic resonance (magnetic resonance–DNAemia [MR-DNAemia]) and clinical outcomes.
Results
Time to bloodstream clearance according to T2 magnetic resonance was significantly longer than blood culture clearance (HR, .54; 95% CI, .39–.75) and did not differ according to the causative pathogen (P = .5). Each additional day of MR-DNAemia increased the odds of persistent infection (defined as metastatic infection or delayed source control) both in the overall population (OR, 1.98; 95% CI, 1.45–2.70) and in S. aureus (OR, 1.92; 95% CI, 1.12–3.29) and gram-negative bacteremia (OR, 2.21; 95% CI, 1.35–3.60). MR-DNAemia duration was also associated with no improvement in Sequential Organ Failure Assessment score at day 7 from infection onset (OR, 1.76; 95% CI, 1.21–2.56).
Conclusions
T2 magnetic resonance may help diagnose BSI in patients on antimicrobials with negative blood cultures as well as to identify patients with metastatic infection, source control failure, or adverse short-term outcome. Future studies may inform its usefulness within the setting of antimicrobial stewardship programs.
Keywords: bloodstream infection, rapid diagnostic tests, blood culture
Bloodstream infection (BSI) detection by T2 magnetic resonance (T2MR) persists longer than detection by blood culture. T2MR could help diagnose BSI in patients on antimicrobials. Prolonged pathogen detection by T2MR is associated with source control failure, metastatic infection, adverse short-term outcome.
The association between persistent bloodstream infection (BSI) and adverse clinical outcome is well known for some specific pathogens, mainly Staphylococcus aureus. Persistent bacteremia occurs in up to 39% of cases of S. aureus bacteremia [1–3] and has been associated with increased risk of complicated infection defined by the presence of metastatic spread, relapse, and attributable mortality [1, 2, 4]. Candida spp. can also cause persistent BSI with metastatic localizations [5]. Early differentiation between complicated and uncomplicated disease is a priority in the management of such infections to establish the most appropriate treatment duration and assess the need for source control. Follow-up blood cultures (BCs) represent a cornerstone of making therapeutic decisions in these cases [6, 7].
Less evidence exists relative to the clinical implications and prognostic value of the duration of gram-negative bacteremia. Increasingly, data from observational studies and randomized controlled trials (RCTs) support shorter course therapy (7 days) over traditional durations (14 days) for uncomplicated gram-negative BSI, without considering time to bloodstream clearance [8–10]. On the other hand, recent research has suggested improved clinical outcomes associated with the use of follow-up BCs also in gram-negative BSI [11, 12].
While current knowledge on time to bloodstream clearance is based on BC methods [1, 6, 7], several novel technologies are emerging for the rapid identification of pathogens from positive BC bottles or directly from whole blood, with the aim of shortening time to results and improving sensitivities of conventional methods [13, 14]. Among these, T2 magnetic resonance (T2MR) (T2 Biosystems, Lexington, MA, USA) is a technology able to detect pathogens from whole blood in only 3–8 hours. In T2MR, microbial DNA amplified by polymerase chain reaction (PCR) binds by hybridization to probes enriched with superparamagnetic nanoparticles, which allow the identification of amplicons in the magnetic field. Of note, this system enables the detection of intact cells rather than free-DNA [15]. A few studies have compared the performance of T2MR and conventional BCs, suggesting that T2MR has higher sensitivity [16, 17].
Despite showing good performance, data on the clinical impact of T2MR are scarce, and the best way to implement it in clinical practice is yet to be defined. Specifically, whether these novel technologies will have a role in the assessment of the duration of the BSI is largely unknown. Indeed, whether, in the setting of a BSI, the persistence in the bloodstream of microbial DNA, rather than of live culturable microorganisms, may have clinical and prognostic significance or play a role in determining the duration of antimicrobial therapy is yet to be established. In this study we assessed the performance of T2MR compared with BC for measuring the duration of the BSI and we investigated the association between the duration of BSI according to T2MR (magnetic resonance–DNAemia [MR-DNAemia]) and microbiological outcomes, including persistent infection, as well as short- and long-term clinical outcomes.
METHODS
Study Design
This was a multicenter, prospective, observational study conducted from January 2022 to March 2023 at 3 sites in Australia: The Royal Brisbane and Women's Hospital (RBWH), Redcliffe Hospital, and Caboolture Hospital.
Hospitalized adult patients with proven BSI due to any T2MR-on-panel pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, and Candida spp.) as detected by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) were deemed eligible for enrolment. Throughout the study period, BCs collected at the participating sites were screened daily by the investigators to ensure prompt patient enrollment after the release of pathogen speciation results. After obtaining consent, research samples were collected for 4 consecutive days, including daily BCs (1 set) and a daily whole-blood sample for T2MR assessment. Exclusion criteria included palliative care, inability to provide consent, and polymicrobial index BC. Patients were also excluded if discharge was expected within 24 hours, preventing blood collection to be performed for at least 2 days. According to the observational design, T2MR results were not used for clinical decision making.
Demographic and clinical data were collected from the medical charts and managed using REDCap electronic data-capture tools hosted at the University of Queensland [18, 19]. Long-term functional outcomes were collected either from the medical charts or by means of follow-up phone calls at 30, 90, and 180 days from the index BSI.
Definitions
“Metastatic infection” was defined according to Souli et al [3], based on the presence of any of the following: proven infective endocarditis, septic emboli, septic thrombophlebitis, vertebral osteomyelitis, septic arthritis, a metastatic abscess, or other deep tissue abscess. As also acknowledged by Souli et al [3], differentiating a metastatic localization from the source of BSI might be challenging and was out of the scope of this study. “Proven endocarditis” was defined according to Duke's criteria [20]. As this was an observational study, no additional investigations for metastatic infection were performed other than those ordered by treating clinicians. “Persistent clinically determined infection” was defined by the presence of either metastatic infection or delayed source control after the commencement of the research blood sample collection. “BSI relapse” was defined by a repeated positive BC for the index pathogen 2 weeks after the index culture. “Persistent MR-DNAemia” was defined by the presence of any T2MR samples positive for the index pathogen over the 4-day follow-up [4]. “Empirical treatment” was defined as effective if any antibiotic active in vitro against the final isolate was administered while pending speciation and sensitivity results on the index BC. A “successful short-term outcome” or “SOFA success” was defined as survival for the first 7 days from BSI onset with a stable or decreased Sequential Organ Failure Assessment (SOFA) score (for intensive care unit [ICU] patients) or modified SOFA score (mSOFA) (for non-ICU patients), defined as follows: if the baseline SOFA/mSOFA is greater than or equal to 3, a decrease of at least 30% in that score, if the baseline SOFA/mSOFA is less than 3, a stable or decreased SOFA/mSOFA score. Patients discharged before day 7 were assumed to have improved scores. The lack of “SOFA success” was defined “SOFA failure” [21, 22]. A “successful long-term outcome” was defined as survival for the first 6 months from the BSI onset and maintenance of the baseline functional performance status defined by the Functional Bloodstream Infection Score (FBIS) score 7 days prior to BSI onset [22].
Laboratory Methods
Blood culture bottles were incubated in the BacT/Alert Virtuo (bioMérieux). After flagging positive, a Gram stain was performed, and BC media was subcultured into agar plates. Microorganism identification was performed with MALDI-TOF MS (VITEK MS; bioMérieux). Whole-blood samples for T2MR assessment were collected into 4-mL EDTA tubes and were either run fresh or frozen upon collection (−65°C) and run retrospectively. Two milliliters of blood were pipetted into the T2MR cartridges.
Statistical Analysis
Categorical variables are presented as frequencies and proportion (%), and continuous variables as medians and interquartile range (IQR). The performance of T2MR versus BC was described in terms of agreement and McNemar's test. Chi-square and Fisher's exact test were used to compare categorical variables, Kruskal-Wallis test was used to compare non–normally distributed continuous variables, and analysis of variance (ANOVA) was used to compare normally distributed continuous variables. Kaplan-Meier curves were used to present time to bloodstream clearance according to T2MR and BC assuming that all patients had positive T2MR at infection onset. Hazard ratios (HRs) and P values were obtained from fitting a Cox proportional hazards model, with a shared frailty for a patient. Kaplan-Meier curves were also used to compare time to MR-DNAemia clearance according to the presence of persistent clinically determined infection. Logistic regression was performed to explore the association between persistent MR-DNAemia and MR-DNAemia duration with the outcomes of interest. When logistic regression was performed, patients who were censored before MR-DNAemia clearance were imputed as having +0.5 day of MR-DNAemia positivity. Statistical analysis was performed with Stata 17 [23].
Ethical Approval
Study approval was granted by the RBWH's Ethics Committee (approval number HREC/2021/QRBW/70126) and ratified by the University of Queensland (2021/HE000073).
RESULTS
T2 Performance
A total of 102 patients were included in the study. Of them, 47 of 102 (46%) had at least 1 T2MR follow-up sample positive for the index pathogen (persistent MR-DNAemia) versus 25 of 102 (25%) who had at least 1 positive follow-up BC (P < .001). Overall, 357 T2MR samples and 337 BCs were collected from the enrolled patients, including 329 paired collections of concurrent T2MR and BC samples, of which 102 of 329 (31.0%) were T2MR positive and 42 of 329 (12.8%) were BC positive (P < .001) (Table 1 and Supplementary Figure 1). In 6 of 329 cases (2%) T2MR detected a different pathogen from the index one, consistently with false-positive results (Supplementary Table 1).
Table 1.
Concordance of Paired T2MR and Blood Culture Samples Collected Over the 4-Day Follow-up in the 102 Enrolled Patients
| Follow-up BC | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| Follow-up T2MR | |||
| Positive (concordant with index BC) | 35 | 67a | 102 |
| Negative | 7 | 217 | 224 |
| Invalidb | 0 | 3 | 3 |
| Total | 42 | 286 | 329 |
Abbreviations: BC, blood culture; T2MR, T2 magnetic resonance.
aIncludes 6 samples where T2MR detected a different pathogen from the index one (false positives).
bInvalid = no results reported by the instrument due to failure of internal control.
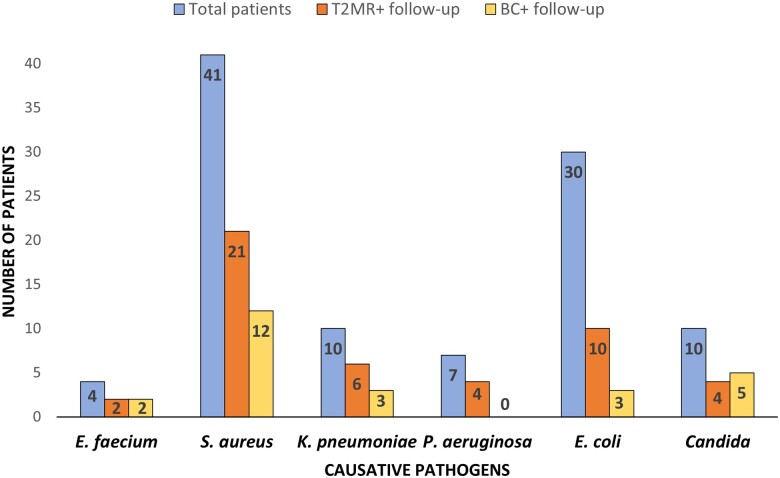
Interestingly, 12 of 47 (25%) patients had intermittent positivity of T2MR over the 4 days and 5 of 25 (20%) patients had intermittent BC positivity. Significantly more patients with gram-positive BSI (31%) or candidemia (50%) had positive follow-up BC compared with patients with gram-negative BSI (13%) (P = .029), while there was no difference in the likelihood of having persistent MR-DNAemia according to the BSI causative pathogen (P = .63) (Figure 1). All patients were receiving effective antimicrobials at the time of blood sample collection.
Figure 1.
Overall number of patients (blue column), patients with positive T2MR samples at follow-up (persistent MR-DNAemia, orange column), and those with positive BCs (yellow column) according to the BSI causative pathogen. Only T2MR-concordant results for the index pathogen are included. Abbreviations: BC, blood culture; BSI, bloodstream infection; MR-DNAemia, magnetic resonance–DNAemia; T2MR, T2 magnetic resonance.
Time to Bloodstream Clearance
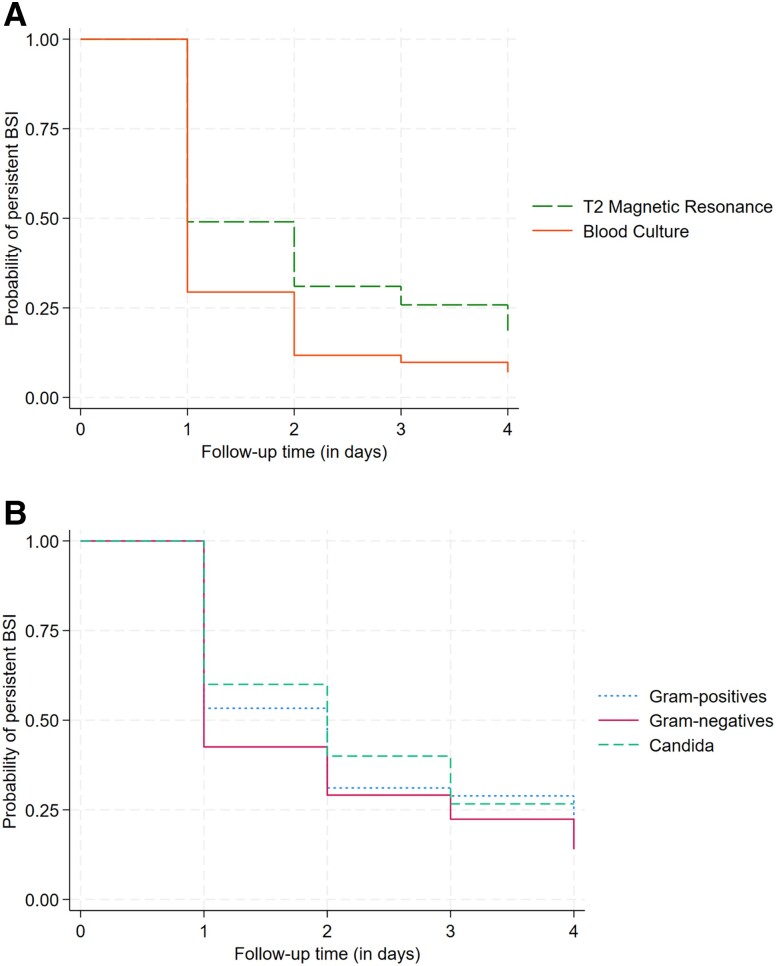
Bloodstream clearance over the 4-day follow-up was longer with T2MR than with BC (HR, .54; 95% confidence interval [CI], .39–.75; P < .001) (Figure 2A). No difference was observed in time to MR-DNAemia clearance according to the BSI causative pathogen (log-rank P = .5) (Figure 2B).
Figure 2.
A, Kaplan-Meier curves showing time to bloodstream clearance according to T2MR and BC (log-rank test, P = .002). B, Kaplan-Meier curves showing time to MR-DNAemia clearance according to the underlying BSI causative pathogen (log-rank test P = .5). Abbreviations: BC, blood culture; BSI, bloodstream infection; MR-DNAemia, magnetic resonance–DNAemia; T2MR, T2 magnetic resonance.
Duration of MR-DNAemia and Microbiological and Clinical Outcomes
Table 2 summarizes patient characteristics overall and according to persistent MR-DNAemia. Patients with and without persistent MR-DNAemia were similar in terms of number of samples collected during follow-up (median, 4; IQR, 3–4 in both groups; P = .57) and median times from index BC to research sample collection. Moreover, the groups did not differ in terms of BSI causative pathogen (P = .5), comorbidities (P = .2), or immunocompromised status (P = .1).
Table 2.
Microbiological and Clinical Characteristics of Patients Included in the Study, Overall and According to Persistent MR-DNAemia
| Total (N = 102) | Persistent MR-DNAemia | P | ||
|---|---|---|---|---|
| No (n = 55) | Yes (n = 47) | |||
| Demographics | ||||
| Female at birth | 30 (29%) | 17 (31%) | 13 (28%) | .72 |
| Age, median (IQR), y | 66 (52–76) | 65 (53–77) | 67 (52–76) | .79 |
| Microbiological characteristics | ||||
| BSI causative pathogen | … | … | … | .49 |
| Enterococcus faecium | 4 (4%) | 2 (4%) | 2 (4%) | |
| Staphylococcus aureus | … | … | … | |
| PSSA | 12 (12%) | 6 (11%) | 6 (13%) | |
| MSSA | 20 (20%) | 10 (18%) | 10 (21%) | |
| MRSA | 9 (9%) | 4 (7%) | 5 (11%) | |
| Klebsiella pneumoniae | 10 (10%) | 4 (7%) | 6 (13%) | |
| Pseudomonas aeruginosa | 7 (7%) | 3 (5%) | 4 (4%) | |
| Escherichia coli | … | … | … | |
| ESBL− | 24 (24%) | 18 (33%) | 6 (13%) | |
| ESBL+ | 6 (6%) | 2 (4%) | 4 (9%) | |
| Candida spp. | 10 (10%) | 6 (11%) | 4 (9%) | |
| Index BC time to positivity, median (IQR), h | 12 (10–17) | 14 (10–19) | 11 (10–16) | .091 |
| Time from index BC to sample collection, median (IQR), h | ||||
| Day 1 | 61 (48–72) | 60 (50–72) | 63 (46–75) | .99 |
| Day 2 | 85 (70–97) | 84.2 (71–96) | 87 (68–100) | .67 |
| Day 3 | 110 (94–127) | 109 (99–122) | 112 (92–130) | .93 |
| Day 4 | 135 (122–154) | 136 (129–155) | 135 (117–152) | .75 |
| No. of follow-up samples collected, median (IQR) | 4 (3–4) | 4 (3–4) | 4 (3–4) | .57 |
| Comorbidities | ||||
| Charlson comorbidity score, median (IQR) | 4 (2–6) | 5 (3–7) | 4 (2–5) | .2 |
| Hemodialysis | 3 (3%) | 0 (0%) | 3 (6%) | .057 |
| Diabetes | 39 (38%) | 21 (38%) | 18 (38%) | .99 |
| Immunocompromised (any of the following) | 39 (38%) | 25 (46%) | 14 (30%) | .10 |
| Bone marrow transplant | 3 (3%) | 2 (4%) | 1 (2%) | |
| Solid-organ transplant | 2 (2%) | 2 (4%) | 0 (0%) | |
| Other immunosuppressiona | 8 (8%) | 5 (9%) | 3 (6%) | |
| Solid tumor (nonmetastatic) | 9 (9%) | 3 (6%) | 6 (13%) | |
| Metastatic solid tumor | 8 (8%) | 6 (11%) | 2 (4%) | |
| Leukemia | 8 (8%) | 6 (11%) | 2 (4%) | |
| Lymphoma/multiple myeloma | 5 (5%) | 4 (7%) | 1 (2%) | |
| Pacemaker/prosthetic heart valve | 5 (5%) | 1 (2%) | 4 (9%) | .12 |
| Central line | 17 (17%) | 8 (15%) | 9 (20%) | .50 |
| Vital signs and clinical severity at BSI onset | ||||
| Max temperature, median (IQR), °C | 38.5 (38.0–39.3) | 38.2 (37.7–39.0) | 38.9 (38.2–39.5) | .007* |
| Max HR, median (IQR), beats/min | 108 (95–116) | 101 (92–113) | 110 (100–122) | .030* |
| Max RR, median (IQR), breaths/min | 22 (18–27) | 21 (18–26) | 24 (19–27) | .31 |
| Afebrile 72 h from BSI onset | 84 (82%) | 50 (91%) | 34 (72%) | .014* |
| Normal HR 72 h from BSI onset | 89 (87%) | 51 (93%) | 38 (81%) | .073 |
| Normal RR 72 h from BSI onset | 73 (73%) | 41 (77%) | 32 (68%) | .30 |
| Inotropes requirement | 8 (8%) | 3 (6%) | 5 (11%) | .33 |
| mSOFA/SOFA, median (IQR) | 1 (0–3) | 1 (0–2) | 2 (1–4) | .022* |
| 0–2 | 72 (71%) | 43 (78%) | 29 (62%) | |
| 3–5 | 20 (20%) | 8 (15%) | 12 (26%) | |
| 6–8 | 6 (6%) | 2 (4%) | 4 (9%) | |
| 9–12 | 4 (4%) | 2 (4%) | 2 (4%) | |
| Length of stay post-index BSI, median (IQR), d | 10 (6–17) | 10 (6–16) | 10 (6–19) | .50 |
| Empirical treatment active | 73 (72%) | 39 (71%) | 34 (72%) | .87 |
| Source control | ||||
| Need of source control | 46 (45%) | 23 (42%) | 23 (49%) | .47 |
| Achievement of source control | ||||
| Achieved before or at day 1 follow-up | 25 (54%) | 15 (65%) | 10 (43%) | .15 |
| Achieved after day 1 follow-up | 15 (33%) | 7 (30%) | 8 (35%) | |
| Never achieved | 6 (13%) | 1 (4%) | 5 (22%) | |
| Type of source control | … | … | … | .22 |
| Line removal | 4 (10%) | 2 (9%) | 2 (11%) | |
| Open surgery | 13 (33%) | 10 (45%) | 3 (17%) | |
| Interventional/radiology procedure | 12 (30%) | 5 (23%) | 7 (39%) | |
| Otherb | 11 (28) | 5 (23%) | 6 (33%) | |
| Time from index BC to source control, median (IQR), d | 3 (1–5) | 3 (1–5) | 3 (1–5) | .70 |
| Microbiological outcomes | ||||
| Metastatic infection (any of the following) | 26 (26%) | 6 (11%) | 20 (43%) | <.001* |
| Vertebral osteomyelitis | 2 (2%) | 1 (2%) | 1 (2%) | .91 |
| Septic arthritis | 5 (5%) | 2 (4%) | 3 (6%) | .52 |
| Septic emboli | 6 (6%) | 1 (2%) | 5 (11%) | .059 |
| Septic thrombophlebitis | 0 (0%) | 0 (0%) | 0 (0%) | |
| Metastatic abscess | 1 (1%) | 0 (0%) | 1 (2%) | .28 |
| Any other deep tissue abscess | 8 (8%) | 1 (2%) | 7 (15%) | .014* |
| Endocarditis (proven) | 9 (9%) | 1 (2%) | 8 (17%) | .007* |
| Persistent infection (either metastatic infection or lack of source control after day 1 f.u.) | 39 (38%) | 12 (22%) | 27 (57%) | <.001* |
| BSI relapse during hospital admission | 2 (2%) | 0 (0%) | 2 (4%) | .13 |
| Clinical outcomes | ||||
| Short-term clinical outcome | … | … | … | .017* |
| SOFA success | 89 (87%) | 52 (94%) | 37 (79%) | |
| SOFA failure | 13 (13%) | 3 (6%) | 10 (21%) | |
| Long-term clinical outcome | … | … | … | .57 |
| Successful | 69 (76%) | 36 (73%) | 33 (79%) | |
| Unsuccessful | 22 (24%) | 13 (27%) | 9 (21%) | |
| In-hospital mortality | 5 (5%) | 2 (4%) | 3 (6%) | .52 |
| Day 30 mortality | 6 (6%) | 3 (6%) | 3 (6%) | .84 |
| Day 90 mortality | 8 (8%) | 3 (6%) | 5 (11%) | .33 |
| Day 180 mortality | 11 (11%) | 5 (9%) | 6 (13%) | .55 |
Data are presented as median (IQR) for continuous measures and n (%) for categorical measures.
Abbreviations: BC, blood culture; BSI, bloodstream infection; ESBL, extended-spectrum beta-lactamase; f.u., follow-up; HR, heart rate; IQR, interquartile range; Max, maximum; MR-DNAemia, magnetic resonance–DNAemia; MRSA, methicillin-resistant Staphylococcus aureus; mSOFA, modified Sequential Organ Failure Assessment; MSSA, methicillin-susceptible Staphylococcus aureus; PSSA, penicillin-susceptible Staphylococcus aureus; RR, respiratory rate; SOFA, Sequential Organ Failure Assessment.
aIncluding congenital and autoimmune diseases.
bIncluding laparoscopic or arthroscopic surgery, urinary catheter/ureteric stent/nephrostomy removal.
*Significant, P < .05.
Patients with persistent MR-DNAemia were significantly more likely to have metastatic infection (43% vs 11%, P < .001), especially endocarditis (17% vs 2%, P = .007) or deep tissue abscesses (15% vs 2%, P = .014) compared with patients without persistent MR-DNAemia. Out of 39 patients with persistent clinically determined infection, 27 of 39 (69%) had persistent MR-DNAemia versus 16 of 39 (41%) who had positive follow-up BC (P = .003), with 12 of 39 patients (31%) having persistent MR-DNAemia but no positive follow-up BCs (Supplementary Table 2). At infection onset, patients with persistent MR-DNAemia had significantly higher mSOFA/SOFA scores (P = .022) and more altered vital signs, including fever (P = .007) and heart rate (P = .030), compared with patients without persistent MR-DNAemia. They were also less likely to experience defervescence at 72 hours (P = .014) and SOFA success (79% vs 94%, P = .017).
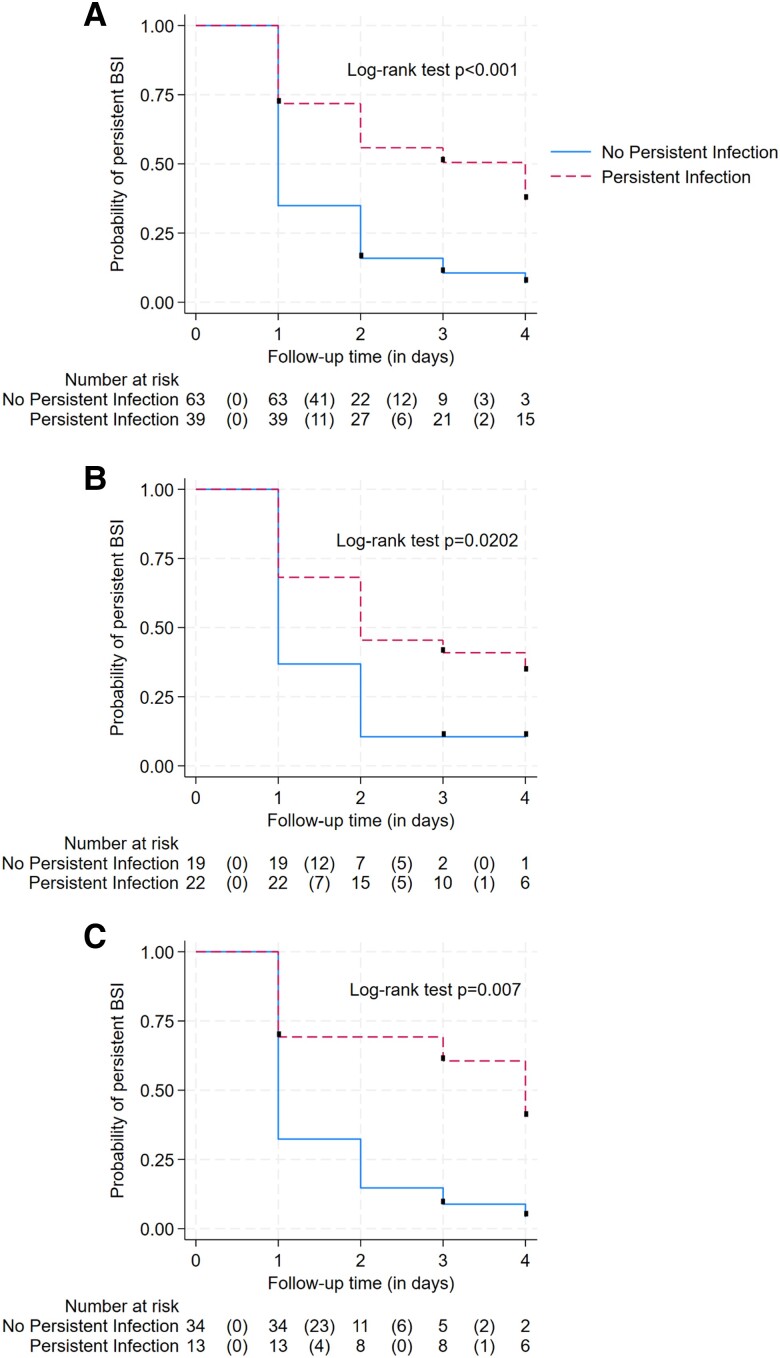
Figure 3 A shows time to MR-DNAemia clearance according to persistent clinically determined infection (P < .001). Five patients were censored while still having positive MR-DNAemia at day 3 and 3 patients at day 2, due to death/palliative care (n = 2), instrument failure (n = 2), missed collection (n = 3), and discharge (n = 1).
Figure 3.
Kaplan-Meier curves showing time to MR-DNAemia clearance according to persistent, clinically determined infection in the overall population (log-rank test, P < .001) (A) and in patients with Staphylococcus aureus (log-rank test, P = .02) (B) and gram-negative BSI (log-rank test, P = .007) (C). Abbreviations: BSI, bloodstream infection; MR-DNAemia, magnetic resonance–DNAemia
Persistent MR-DNAemia was associated with persistent, clinically determined infection (odds ratio [OR], 4.84; 95% CI, 2.04–11.5), with each additional day of MR-DNAemia doubling the odds of persistent infection (OR, 1.98; 95% CI, 1.45–2.70). Persistent MR-DNAemia was also associated with increased risk of SOFA failure (OR, 4.68; 95% CI, 1.21–18.2), with each additional day of MR-DNAemia increasing the odds of SOFA failure (OR, 1.76; 95% CI, 1.21–2.56). The association between MR-DNAemia duration with persistent infection and SOFA failure was confirmed also after adjusting for the duration of BC positivity (Supplementary Table 3). No association with long-term clinical outcomes was observed (OR, .99; 95% CI, .72–1.36).
Persistent MR-DNAemia in S. aureus and Gram-Negative Bloodstream Infection
Supplementary Table 4 shows patients' characteristics according to persistent MR-DNAemia stratified based on the 2 main groups of pathogens (S. aureus vs gram-negative BSI). Patients with persistent MR-DNAemia were more likely to have persistent, clinically determined infection in both groups. Times to MR-DNAemia clearance according to persistent, clinically determined infection in S. aureus (P = .02) and gram-negative BSI (P = .007) are reported in Figure 3B and 3C.
Logistic regression analysis confirmed the association between MR-DNAemia duration with persistent, clinically determined infection in both S. aureus (OR, 1.92; 95% CI, 1.12–3.29) and gram-negative BSI (2.21; 95% CI, 1.35–3.60), while the association with SOFA failure was significant only in S. aureus bacteremia (OR, 2.06; 95% CI, 1.18–3.61; gram-negative BSI: .96; 95% CI, .43–2.18).
Persistent MR-DNAemia in the Absence of Persistent Infection
Significantly, 6 of 21 (29%) patients with S. aureus BSI and 11 of 47 (23%) patients with gram-negative BSI had persistent MR-DNAemia despite not having persistent infection. These patients had shorter MR-DNAemia duration compared with patients with persistent infection, with a median of 1 single positive sample (IQR, 1–2) versus 3 positive samples (IQR, 2–4) (P = .009). To explore the significance of persistent MR-DNAemia in this setting, characteristics of patients without persistent infection were compared according to whether they had persistent MR-DNAemia (Supplementary Table 5). Interestingly, 50% (3/6) of patients with persistent S. aureus MR-DNAemia in the absence of persistent infection had an unknown source of BSI, while 82% (9/11) of patients with persistent gram-negative MR-DNAemia had a urinary source of infection. No differences in clinical outcomes were found according to persistent MR-DNAemia in patients without persistent infection.
DISCUSSION
Our study shows that the detection of BSI as measured by T2MR lasts longer compared with conventional BCs in patients on active treatment. These observations suggest the usefulness of T2MR in patients receiving antimicrobials, where conventional cultures may have limited usefulness. Nonetheless, some T2MR false-positive results were observed in our study, suggesting caution in using T2MR for early de-escalation of empiric therapy.
The second relevant finding of our study relates to the association between MR-DNAemia duration and persistent infection. Indeed, whether, in the setting of BSI, the persistence of microbial DNA in the bloodstream has clinical relevance is largely unknown. The T2MR claims to detect intact cells rather than free-DNA [15] and this might increase the likelihood of detecting microbial cells that are alive and still pathogenic. However, Eichenberger et al [24] recently demonstrated how, in patients with BSI, microbial cell-free DNA persists longer than conventional cultures and is associated with metastatic infection. These observations open interesting research questions about the potential prognostic role of culture independent technologies. Indeed, performing serial T2MR testing might be useful in patients with BSI with an unknown source, in particular those with prosthetic endovascular material who do not satisfy major echocardiographic criteria for infective endocarditis [20]. Our data suggest that the lack of MR-DNAemia clearance might be associated with a higher risk of metastatic infection in these cases. Interestingly, the association between persistent MR-DNAemia and persistent infection was independent of the causative pathogens, suggesting the prognostic role of T2MR also in gram-negative bacteremia, which is usually transient according to conventional systems [25]. MR-DNAemia duration was also shown to be associated with increased likelihood of SOFA failure, suggesting the usefulness of T2MR for patient risk stratification within the first week from the BSI onset.
A third implication of our results might pertain to the possibility of using the time to MR-DNAemia clearance for optimizing antimicrobial treatment duration. Indeed, while the need of at least a 14-day treatment course for S. aureus bacteremia is well established, the optimal treatment duration for gram-negative bacteremia is more debated. Shorter courses have been advocated to reduce the risks associated with prolonged exposure to antimicrobials, including emergence of antimicrobial resistance. Recently, 2 RCTs have shown how a 7-day treatment course is noninferior to a 14-day course in patients with uncomplicated gram-negative BSI with respect to 30- and 90-day clinical outcomes [8, 9]. Further research has also suggested the use of biochemical tests such as C-reactive protein (CRP) to guide antibiotic discontinuation in this setting. Specifically, in a recent RCT involving 493 adults with gram-negative bacteremia, clinical failure for CRP-guided antibiotic treatment duration and fixed 7-day treatment was noninferior to 14-day treatment (2.4% vs 6.6% vs 5.5%, P < .001) [26]. Interestingly, in our study, up to 23% of patients with gram-negative BSI had positive T2MR samples at follow-up without having persistent infection. In these patients, MR-DNAemia duration was short and most had a urinary source. Overall, several factors may guide antimicrobial duration, including achievement of source control, metastatic complications, and timing of resolution of symptoms [27]. We speculate as to whether the persistence of microbial DNA in the bloodstream, as measured by T2MR, could also help establish the most appropriate timing for treatment discontinuation within the setting of antimicrobial stewardship programs, with patients having persistent MR-DNAemia being candidates for longer courses. An RCT on patients with uncomplicated gram-negative bacteremia comparing the effect on clinical outcomes of individualized T2MR-guided antibiotic duration compared with a 7-day duration may help answer this question. It also remains possible that patients with no MR-DNAemia after their initial positive result could be considered for “ultra-short” durations (<7 days). The health-economic implications of using T2MR for these purposes would need to be assessed in such evaluations.
Our study has several limitations. First, the sample size was small. Second, our study has some missing data due to early hospital discharge or missing blood collections, and this might have also influenced the significance of our results, although the median number of blood collections in patients with and without persistent MR-DNAemia was comparable. Third, we acknowledge that a follow-up longer than 4 days might have been more accurate, in particular considering that some patients had intermittent positivity of T2MR or BC. Fourth, the determination of metastatic/persistent infection was based on clinical judgement and no additional investigations were performed. We also acknowledge that we measured the duration of MR-DNAemia assuming that all patients had positive T2MR at infection onset; however, this might not have been the case. A last comment should be made about our definition of persistent MR-DNAemia as any positive T2MR sample over the 4-day follow-up rather than based on a specific cutoff duration. Nonetheless, recent evidence has suggested redefining the cutoff duration of persistent bacteremia to 2 days [4] and this is in line with our median time from index BC to first research sample collection (61 hours).
CONCLUSIONS
The increased sensitivity of T2MR compared with BC in detecting bloodstream pathogens during active treatment suggests the usefulness of T2MR in patients on antimicrobials, where the role of BC might be limited. Moreover, the association between the persistence of detectable pathogens by T2MR with persistent infection and SOFA failure suggests the usefulness of T2MR for identifying patients at risk of source control failure, metastatic infection, or adverse short-term clinical outcome. Well-designed studies are needed to define the best role of T2MR in clinical practice.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Anna Maria Peri, UQ Centre for Clinical Research, The University of Queensland, Brisbane, Queensland, Australia.
Kevin O’Callaghan, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Nastaran Rafiei, Infectious Diseases Unit, Caboolture Hospital, Caboolture, Queensland, Australia.
Bianca Graves, Herston Infectious Diseases Institute, Herston, Brisbane, Queensland, Australia.
Holly Sinclair, Infectious Diseases Unit, Royal Brisbane and Women’s Hospital, Brisbane, Queensland, Australia.
Anna Brischetto, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Karen Lim, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Jill Parkes-Smith, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Matthew Eustace, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Natalie Davidson, Infectious Diseases Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Alexis Tabah, Intensive Care Unit, Redcliffe Hospital, Redcliffe, Queensland, Australia.
Adam Stewart, Central Microbiology, Pathology Queensland, Royal Brisbane and Women's Hospital, Brisbane, Queensland, Australia.
Mark D Chatfield, UQ Centre for Clinical Research, The University of Queensland, Brisbane, Queensland, Australia.
Patrick N A Harris, UQ Centre for Clinical Research, The University of Queensland, Brisbane, Queensland, Australia; Herston Infectious Diseases Institute, Herston, Brisbane, Queensland, Australia; Central Microbiology, Pathology Queensland, Royal Brisbane and Women's Hospital, Brisbane, Queensland, Australia.
David L Paterson, UQ Centre for Clinical Research, The University of Queensland, Brisbane, Queensland, Australia; Infectious Diseases Unit, Royal Brisbane and Women’s Hospital, Brisbane, Queensland, Australia; ADVANCE-ID, Saw Swee Hock School of Public Health, National University of Singapore, Singapore; Infectious Diseases Translational Research Program, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Notes
Author contributions. D. L. P. and A. M. P. designed the study. A. M. P., K. O., N. R., B. G., H. S., A. B., K. L., J. P.-S., M. E., N. D., A. T., A. S. and P. N. A. H. contributed to screening and enrolling eligible patients. A. M. P. performed data analysis and wrote the main version of the manuscript. M. D. C. supervised statistical analysis. D. L. P. and P. N. A. H. supervised data interpretation and the manuscript drafting. All authors substantially contributed to revising the manuscript and approved its final version.
Acknowledgments. The authors acknowledge Ms. Tiffany Au for assistance with the study submission to the Human Research Ethics Committee and with obtaining site-specific approval; Ms. Megan Ratcliffe, Ms. Maree Duroux, Ms. Samantha Shone, Ms. Kylie Jacobs, and Ms. Julia Affleck for assistance with the study coordination as well as data collection at Redcliffe and Caboolture Hospitals; Ms. Michelle Bauer for laboratory support and assistance with the maintenance of the T2 instrument; and Mr. Haakon Bergh for the support with samples' handling at the Pathology Queensland Central Laboratory.
Disclaimer. T2 Biosystems had no role in the study design, samples' testing, data analysis, results interpretation, nor manuscript drafting.
Financial support. A. M. P. received a scholarship from the University of Queensland in support of her PhD candidature. P. N. A. H. was supported by an Early Career Fellowship from the National Health and Medical Research Council (GNT1157530). Cartridges for T2MR testing were supplied by T2 Biosystems.
References
- 1. Fowler VG Jr, Olsen MK, Corey GR, et al. Clinical identifiers of complicated Staphylococcus aureus bacteremia. Arch Intern Med 2003; 163:2066–72. [DOI] [PubMed] [Google Scholar]
- 2. Khatib R, Johnson LB, Fakih MG, et al. Persistence in Staphylococcus aureus bacteremia: incidence, characteristics of patients and outcome. Scand J Infect Dis 2006; 38:7–14. [DOI] [PubMed] [Google Scholar]
- 3. Souli M, Ruffin F, Choi SH, et al. Changing characteristics of Staphylococcus aureus bacteremia: results from a 21-year, prospective, longitudinal study. Clin Infect Dis 2019; 69:1868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuehl R, Morata L, Boeing C, et al. Defining persistent Staphylococcus aureus bacteraemia: secondary analysis of a prospective cohort study. Lancet Infect Dis 2020; 20:1409–17. [DOI] [PubMed] [Google Scholar]
- 5. Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med 2015; 373:1445–56. [DOI] [PubMed] [Google Scholar]
- 6. Kimmig A, Hagel S, Weis S, Bahrs C, Loffler B, Pletz MW. Management of Staphylococcus aureus bloodstream infections. Front Med (Lausanne) 2020; 7:616524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pappas PG, Kauffman CA, Andes DR, et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 2016; 62:e1–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yahav D, Franceschini E, Koppel F, et al. Seven versus 14 days of antibiotic therapy for uncomplicated gram-negative bacteremia: a noninferiority randomized controlled trial. Clin Infect Dis 2019; 69:1091–8. [DOI] [PubMed] [Google Scholar]
- 9. Molina J, Montero-Mateos E, Praena-Segovia J, et al. Seven-versus 14-day course of antibiotics for the treatment of bloodstream infections by Enterobacterales: a randomized, controlled trial. Clin Microbiol Infect 2022; 28:550–7. [DOI] [PubMed] [Google Scholar]
- 10. Nelson AN, Justo JA, Bookstaver PB, Kohn J, Albrecht H, Al-Hasan MN. Optimal duration of antimicrobial therapy for uncomplicated gram-negative bloodstream infections. Infection 2017; 45:613–20. [DOI] [PubMed] [Google Scholar]
- 11. Thaden JT, Cantrell S, Dagher M, et al. Association of follow-up blood cultures with mortality in patients with gram-negative bloodstream infections: a systematic review and meta-analysis. JAMA Netw Open 2022; 5:e2232576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gatti M, Bonazzetti C, Tazza B, et al. Impact on clinical outcome of follow-up blood cultures and risk factors for persistent bacteraemia in patients with gram-negative bloodstream infections: a systematic review with meta-analysis. Clin Microbiol Infect 2023: 29:1150–8. [DOI] [PubMed] [Google Scholar]
- 13. Peker N, Couto N, Sinha B, Rossen JW. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect 2018; 24:944–55. [DOI] [PubMed] [Google Scholar]
- 14. Peri AM, Harris PNA, Paterson DL. Culture-independent detection systems for bloodstream infection. Clin Microbiol Infect 2022; 28:195–201. [DOI] [PubMed] [Google Scholar]
- 15. Neely LA, Audeh M, Phung NA, et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med 2013; 5:182ra54. [DOI] [PubMed] [Google Scholar]
- 16. Clancy CJ, Pappas PG, Vazquez J, et al. Detecting Infections Rapidly and Easily for Candidemia Trial, part 2 (DIRECT2): a prospective, multicenter study of the T2Candida panel. Clin Infect Dis 2018; 66:1678–86. [DOI] [PubMed] [Google Scholar]
- 17. Nguyen MH, Clancy CJ, Pasculle AW, et al. Performance of the T2Bacteria panel for diagnosing bloodstream infections: a diagnostic accuracy study. Ann Intern Med 2019; 170:845–52. [DOI] [PubMed] [Google Scholar]
- 18. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li JS, Sexton DJ, Mick N, et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis 2000; 30:633–8. [DOI] [PubMed] [Google Scholar]
- 21. Grissom CK, Brown SM, Kuttler KG, et al. A modified sequential organ failure assessment score for critical care triage. Disaster Med Public Health Prep 2010; 4:277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McNamara JF, Harris PNA, Chatfield MD, Lorenc P, Paterson DL. Measuring patient-centered long-term outcome following a bloodstream infection: a pilot study. Clin Microbiol Infect 2020; 26:257.e1–.e4. [DOI] [PubMed] [Google Scholar]
- 23. StataCorp . Stata statistical software: release 17. College Station, TX: StataCorp LLC, 2021. [Google Scholar]
- 24. Eichenberger EM, de Vries CR, Ruffin F, et al. Microbial cell-free DNA identifies etiology of bloodstream infections, persists longer than conventional blood cultures, and its duration of detection is associated with metastatic infection in patients with Staphylococcus aureus and gram-negative bacteremia. Clin Infect Dis 2022; 74:2020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiggers JB, Xiong W, Daneman N. Sending repeat cultures: is there a role in the management of bacteremic episodes? (SCRIBE study). BMC Infect Dis 2016; 16:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Dach E, Albrich WC, Brunel AS, et al. Effect of C-reactive protein-guided antibiotic treatment duration, 7-day treatment, or 14-day treatment on 30-day clinical failure rate in patients with uncomplicated gram-negative bacteremia: a randomized clinical trial. JAMA 2020; 323:2160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heil EL, Bork JT, Abbo LM, et al. Optimizing the management of uncomplicated gram-negative bloodstream infections: consensus guidance using a modified Delphi process. Open Forum Infect Dis 2021; 8:ofab434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.