Abstract
Live, attenuated bacteria are effective vectors for heterologous antigen delivery. However, loss of heterologous gene-bearing plasmids is problematic, and antibiotics and their resistance genes are not desirable for in vivo DNA vaccine delivery due to biosafety and regulatory concerns. To solve this problem, we engineered the first vaccine delivery strain that has no requirement for antibiotics or other selectable marker genes to maintain the recombinant plasmid. This model strain of Salmonella enterica serovar Typhimurium, SLDAPD, uses operator-repressor titration (ORT) technology, which requires only the short, nonexpressed lacO sequence for selection and maintenance. SLDAPD, recovered from the spleens and Peyer's patches of mice following oral inoculation, was shown to maintain a plasmid that, in contrast, was lost from parental strain SL3261. We also demonstrated successful application of this technology to vaccine development, since SLDAPD carrying a plasmid without an antibiotic resistance gene that expressed the Yersinia pestis F1 antigen was as efficacious in protecting vaccinated mice against plague as the parental SL3261 strain carrying an antibiotic-selected version of this plasmid. Protection of mice against plague by immunization with Salmonella expressing F1 has previously required two or more doses; here we demonstrated for the first time protective immunity after a single oral immunization. This technology can easily be used to convert any suitable attenuated strain to an antibiotic-free ORT strain for recombinant protein vaccine delivery in humans.
The use of live, attenuated bacteria to deliver recombinant protein vaccine antigens is a promising approach for overcoming the difficulties and cost of vaccine antigen purification, and it enhances presentation to the immune system. When this approach is used, genes encoding vaccine antigens are expressed from multicopy plasmids in the live attenuated bacterial vectors. An effective system depends on the transformation and stable maintenance in the bacterial cell of the plasmid carrying the antigen gene. Although some antigen-expressing plasmids are stable, segregational plasmid loss during cell division remains an important problem, and various balanced lethal approaches have been used to enhance plasmid stability (7, 8).
If a strain is auxotrophic for an essential metabolite due to a chromosomal gene mutation, the mutation can be complemented with a functional, plasmid-borne copy of the gene. This enables plasmid selection on agar plates if the metabolite is not present and maintenance in vivo if the metabolite cannot be scavenged from the recipient animal. The choice of the essential gene used for complementation of a host auxotrophy is critical. The thymidylate synthetase gene thyA has been used for plasmid maintenance in Salmonella (13), but growth in culture was restricted to minimal medium as thymidine is present in complex nutrient broth media (14). A similar approach was used for Vibrio cholerae with a plasmid complementing a mutation in the chromosomal glutamine synthetase gene glnA, although antibiotic resistance genes were present on the plasmid to enable selection in complex media (21). The strain was able to survive in vivo even without a plasmid by obtaining sufficient glutamine from the host. An alternative approach involves diaminopimelic acid (DAP), a bacterial amino acid that is not found in eukaryotes, so mutants with mutations in lysine-DAP biosynthetic pathway genes can be grown in complex media when they are complemented with a plasmid expressing the gene product. The asd (aspartate β-semialdehyde dehydrogenase) gene is a DAP biosynthetic gene that has been used to complement asd mutants of both Salmonella enterica serovar Typhimurium (5, 17) and S. enterica serovar Typhi (25).
When an auxotrophy is effectively complemented and the essential metabolite is not present in the culture medium or the recipient animal, plasmids can still be lost at a high rate, but cells die if they lose the plasmid. Nakayama et al. (17) and Galán et al. (5) cultured asd mutant strains for 50 generations and demonstrated that a significant proportion of the plasmid was lost with DAP present. However, the rate of plasmid loss would have been equivalent in the absence of DAP, but it could not be recorded since plasmid-free segregants lysed. Therefore, there is no increase in the total number of plasmid-containing cells when there is complementation of a host auxotrophy, but there is a transition from plasmid-free cells to dead cells. Gene dosage and metabolic burden are key factors in plasmid loss, since a single chromosomal gene is complemented on a multicopy plasmid. This is the most likely explanation for the instability of plasmids expressing asd (5, 17). As the cells synthesize excess DAP, coupled with the continual release of DAP into the growth medium from reprocessing of the cell wall, the selection pressure is effectively removed, and plasmid loss occurs. Another disadvantage of the approach of complementing auxotrophies is the limitation of the choice of the essential gene. The selective metabolite must be absent from the recipient animal cells and, ideally, from complex nutrient broth, but it must be possible to add it to the medium for growth prior to transformation. This rules out the majority of essential genes.
In an alternative approach that has been evaluated for plasmid stabilization the postsegregational killing mechanism is used. This approach relies on a stable toxin and labile antitoxin expressed from a plasmid; the toxin kills any cell that loses the plasmid after antitoxin degradation (2). The hok-soc postsegregational killing system has been tested in S. enterica serovar Typhi, but it proved to be ineffective for plasmid maintenance during prolonged culture, as plasmid-free segregants that escaped the lethal effects of the toxin rapidly proliferated in the culture (6). In comparison, integration of the vaccine antigen gene into the bacterial vector chromosome does address the genetic stability problem for recombinant vaccines (11). However, lower expression levels from a single gene could lead to a weak or ineffective immune response compared to that obtained when a multicopy plasmid is used. In addition, antibiotic gene-free integration is technically difficult compared to plasmid transformation.
To solve the problems inherent in existing gene stabilization technologies, we adapted operator-repressor titration (ORT) for use in live vaccine vectors. ORT is an antibiotic-free plasmid maintenance system that was originally developed in Escherichia coli (3) and is currently used for the production of therapeutic recombinant proteins and plasmids (9). We engineered the attenuated S. enterica serovar Typhimurium strain SL3261 to generate the ORT strain SLDAPD. In SLDAPD, the essential gene dapD is controlled by the operator/promoter of the E. coli lactose operon. The LacI repressor binds to the chromosomal lac operator, and cell growth is prevented unless an inducer, such as isopropyl-β-d-thiogalactopyranoside (IPTG), is present. However, if a multicopy plasmid that also possesses the lac operator sequence is transformed into SLDAPD, it titrates the repressor away from lacO and enables gene expression and cell growth. Here we describe selection and stable in vivo maintenance of a high-copy-number plasmid by ORT in SLDAPD. Furthermore, in this study we demonstrated an application of the ORT technology by vaccination of mice with SLDAPD containing a plasmid expressing the F1 antigen of Yersinia pestis but no selectable marker gene in order to achieve protection against plague.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strain of Salmonella used as a control and modified to create an ORT version was S. enterica serovar Typhimurium strain SL3261 (10). This strain was kindly provided by B. A. D. Stocker, Stanford University School of Medicine, Palo Alto, Calif. Y. pestis GB is a virulent strain that was isolated from a fatal human case of plague (20). The E. coli strains used for plasmid construction were DH5α (Life Technologies), XL10-GoldKan (Stratagene, Amsterdam, The Netherlands), and DH1lacdapD (3). The pUC18I plasmid (4) is a modified version of pUC18 in which the alpha peptide has been deleted and replaced by an ideal palindromic lac operator (22, 23). pAH34L is a kanamycin-selected plasmid and expresses the caf operon from Y. pestis, comprising the F1 antigen (caf1) and the following three genes required for its assembly and export: caf1R (regulatory protein), caf1 M (molecular chaperone), and caf1A (outer membrane anchor) (26). All reagents and kits were obtained from commercial sources, and the manufacturers' instructions were followed. All bacterial cultures were grown in Luria-Bertani (LB) medium at 37°C, and the medium was supplemented when required with kanamycin (50 μg/ml), ampicillin (100 μg/ml for E. coli and 25 μg/ml for S. enterica serovar Typhimurium), chloramphenicol (20 μg/ml), DAP (80 μg/ml), IPTG (0.1 mM), and/or sucrose (0.5%, wt/vol).
Construction of ORT-VAC strain SLDAPD.
The following procedure was used to construct ORT-VAC strain SLDAPD (the term ORT-VAC is used to describe live vaccine delivery of the ORT technology). An expression cassette was constructed that contained lacIq and lacO/P-dapD replacing natural dapD in SL3261. pTrc99 (Pharmacia Biotech) (1) was cut with PflMI and blunt ended by using T4 DNA polymerase. The E. coli lacO/P-dapD expression cassette was removed from placPdapD6 (3) as an AflII-EarI fragment and blunt ended. These two fragments were ligated to create pTrc99dapD, in which dapD and lacIq were divergently oriented. The 5′ and 3′ flanks of the dapD chromosomal locus of SL3261 were amplified by PCR; the 5′ flank was amplified with primers 5STdapD (5′-CGAATGGCATGCGGCCGCAGCTATTTTTATCAGGATTTG-3′) and LST2dapD (5′-GCGAATGCATAGCGGGGGCGACAGCCCCCCTTTGC-3′), resulting in a 2,406-bp PCR product, and the 3′ flank was amplified with primers RSTdapD (5′-GCTTTTCACTCGATGCATCTACCCTTTATCGTTTGGATTGAGGGCC-3′) and 3STdapD (5′-TGTGGGGCATGCGGCCGCAGTGGTCGCACATCGTCGGAC-3′), resulting in a 1,361-bp PCR product. Primers 5STdapD and 3STdapD contained SphI and NotI sites, respectively (underlined); primers LST2dapD and RSTdapD contained NsiI sites (boldface type). Both flanks were cloned into pGEM-T Easy (Promega, Southampton, United Kingdom) to create pGEM-TeasydapDL(−) and pGEM-TeasydapDR(−). The right flank was cut out of pGEM-TeasydapDR(−) as an NsiI fragment, blunt ended, and ligated into Ecl136II-cut pTrc99dapD to create pDRdapD(+). Then the left flank was cut out of pGEM-TeasydapDL(−) as an NsiI fragment and ligated into NsiI-cut pDRdapD(+) to create pdapDB(+). The insert from pdapD(+) was cut out as a NotI fragment and ligated into NotI-cut pKO3 (12) to create the integration plasmid pKO3dapD. pKO3dapD was then used as described by Link et al. (12) to replace wild-type dapD with lacZ-dapD and lacIq in SL3261. The resulting strain was designated SLDAPD, and its genotype was hisG aroA554 Δ(dapD)::(lacIq Plac-dapD).
Construction of ORT-VAC plasmid pAHL.
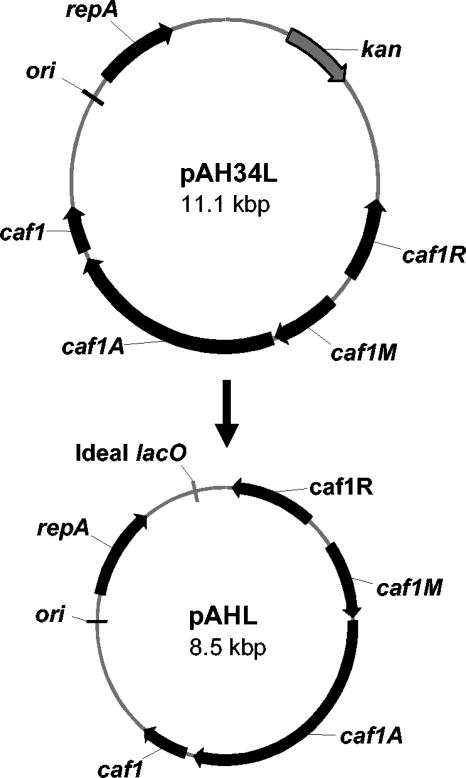
Plasmid pAH34L was digested with NcoI and BssHII to remove the kan gene. The fragment was then ligated to a synthetic oligonucleotide encoding an ideal palindromic lac operator. This plasmid, designated pAH34LORT, was selected in DH1lacdapD, but it was not possible to transform it into SLDAPD due to the methylation barrier between E. coli and S. enterica serovar Typhimurium. To overcome this problem, a kan gene was cloned back into a unique SalI site of pAH34LORT, generating plasmid pAHLK, which was transformed into SLDAPD and selected on LB agar containing kanamycin and IPTG. DNA with a Salmonella methylation pattern was then prepared from SLDAPD/pAHLK and digested with SalI to remove kan and create pAHL. This DNA was transformed into SLDAPD and selected by ORT on LB agar with no additives. The sequence of pAHL is identical to that of pAH34LORT, but these plasmids differ in genealogy and methylation.
Growth analysis in culture.
Single colonies were inoculated into 5 ml of LB broth and grown overnight at 37°C in an orbital shaker at 200 rpm. These precultures were used to inoculate 50 ml of LB broth in 250-ml baffled Erlenmeyer flasks to a starting optical density at 600 nm of 0.01, and readings were taken hourly. All experiments were performed in triplicate.
Detection and localization of F1 antigen expression.
F1 antigen expression was detected by Western blotting. Cells were resuspended in Tris-HCl (50 mM, pH 8) containing RNase A (100 μg/ml) and benzonase (1 U/μl) and then lysed with alkaline sodium dodecyl sulfate. The cell lysates were incubated at 37°C for 20 min to obtain sufficient degradation of the nucleic acids. Samples of the cell lysates were then mixed with a reducing agent and sample loading buffer, heated for 5 min at 95°C, loaded on a sodium dodecyl sulfate-polyacrylamide gel, and transferred to a nitrocellulose membrane. The membrane was then treated as described by Oyston et al. (18) by using mouse monoclonal anti-F1 antigen and alkaline phosphatase-labeled polyclonal rabbit anti-mouse antibody (Abcam, Cambridge, United Kingdom). Bound antibodies were detected by using nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indoyl phosphate (toluidine salt). To demonstrate surface expression of the F1 antigen on SLDAPD/pAHL, 1 × 108 CFU of bacteria was placed into a glass-bottom WillCo-dish (Intracel, Hertfordshire, United Kingdom) and allowed to dry. The bacteria were fixed in acetone for 2 min or left unfixed and then probed with rabbit polyclonal anti-F1 antigen serum at 37°C for 1 h, and binding was detected by incubation with fluorescein isothiocyanate-labeled goat anti-rabbit antibody at 37°C for 1 h. The bacteria were washed with phosphate-buffered saline (PBS) between steps. Fluorescence was visualized with an Olympus IX70 confocal laser scanning microscope, and images were captured and processed by using the Fluoview image software (Olympus, London, United Kingdom). Similar results were obtained for fixed and unfixed cells, although unfixed cells were localized in clumps around the edges of the coverslip as a result of the washing procedure.
In vitro plasmid maintenance.
Single colonies were inoculated into 50 ml of LB broth (containing the appropriate antibiotic) and incubated overnight at 37°C in an orbital shaker at 200 rpm. The optical density was determined on day zero, and then 50 ml of fresh LB broth was inoculated to an A600 of 0.001 and incubated as described above to obtain day 1 values. This process was repeated until day 5. The cells grew approximately 12 generations per day for 5 days, so there were 60 generations during the experiment. A number of cells that gave an A600 (i.e., optical density at 600 nm) in 1.0 ml of culture of 2.0 was extracted daily, and plasmids were extracted by the mini-prep procedure (QIAGEN, West Sussex, United Kingdom) and examined by agarose gel electrophoresis. To determine plasmid maintenance, volumes equivalent to an A600 value of 10−7 were plated onto permissive and nonpermissive media, and the colonies were counted.
Inoculations and in vivo plasmid maintenance.
Strains were cultured statically overnight at 37°C and then centrifuged (6,000 × g, 20 min, 4°C), washed once in PBS, recentrifuged, and resuspended in PBS to a final density of approximately 1 × 1010 to 5 × 1010 CFU/ml (for colonization studies) or 1 × 1011 to 5 × 1011 CFU/ml (for protection studies). Viable bacteria were enumerated on LB agar plates containing IPTG, ampicillin, and kanamycin when appropriate. Subsequently, groups of 30 female BALB/c mice (Charles-River Laboratories, Kent, United Kingdom) were orally inoculated with approximately 1 × 109 to 5 × 109 CFU of Salmonella/100-μl dose by using an intragastric gavage needle. On days 8, 12, and 16 postinoculation, 10 mice were culled by cervical dislocation, and the spleen and six Peyer's patches of each mouse were removed. The spleen or the six Peyer's patches were homogenized in 1 ml of sterile PBS by using 50-μm-pore-size cell strainers (Becton Dickinson Labware, Franklin Lakes, N.J.), and suitable dilutions were plated onto suitable agar for enumeration. Specifically, SL3261 was plated onto LB agar only, SL3261/pUC18I was plated onto LB agar and LB agar containing ampicillin, SLDAPD/pUC18I was plated onto LB agar containing IPTG or ampicillin, SL3261/pAH34L was plated onto LB agar and LB agar containing kanamycin, and SLDAPD/pAHL was plated onto LB agar and LB agar containing IPTG.
Protection against Y. pestis and immune response analysis.
Groups of four to six female BALB/c mice were orally inoculated with approximately 1 × 1010 to 5 × 1010 CFU of the relevant Salmonella strain/100 μl, either once or twice (days 0 and 14). Mice were individually labeled with microchips with unique reference numbers so that their immune responses could be monitored throughout the experiment. Prior to challenge with Y. pestis, tail vein serum samples were taken from the mice and used in an enzyme-linked immunosorbent assay (ELISA) to detect the presence of immunoglobulin G (IgG) against F1 antigen, as described previously (15). All samples were assayed in duplicate. The end point antibody titer was expressed as the maximum dilution of sample that gave an absorbance at 414 nm of more than 0.1 U after subtraction of the absorbance due to nonspecific binding detected in control sera from mice immunized with the control strain SLDAPD or SL3261, as appropriate. The ELISA results were expressed as log10 of the reciprocal of the dilution and the mean of duplicate values for each sample. Fifty days postimmunization, mice were challenged subcutaneously with approximately 102 (185) CFU or 105 (1.85 × 105 or 8.8 × 104) CFU of Y. pestis GB. Y. pestis GB has a median lethal dose (MLD) of <1 CFU in BALB/c mice when the subcutaneous route is used (20). The mice were observed for 20 days with strict observance of humane end points, and the time to death was recorded. At the end of this period two surviving mice from each group were dissected, and blood, spleen, lung, liver, and inguinal and mediastinal lymph node samples were homogenized and plated onto Congo red agar to detect any remaining Y. pestis. The remaining animals were humanely culled.
Statistical analyses.
The mean and standard error of the mean were calculated. Student's t tests were used to determine the significance of differences between means for groups, and probability values of ≤0.05 were considered significant.
RESULTS
Construction of the ORT-VAC strain.
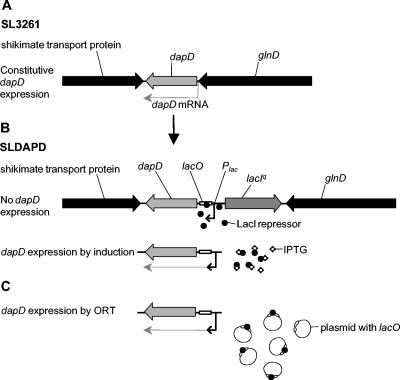
The ORT antibiotic-free plasmid maintenance system has been adapted for use in live vaccine vectors. In this study, the ORT-VAC strain SLDAPD was constructed from S. enterica serovar Typhimurium SL3261 by replacing the essential chromosomal gene dapD with a cassette containing dapD under the control of the lac operator-promoter and lacIq, which overexpressed the lac repressor LacI. It was necessary to include lacIq to avoid constitutive expression from the lac operator-promoter, as S. enterica serovar Typhimurium does not possess a natural lac operon. dapD expresses tetrahydrodipicolinate N-succinyltransferase, which catalyzes a step in the lysine/diaminopimelate pathway (19). DAP is a component of the cell wall, and cell lysis occurs in its absence. DAP is not found in the rich nutrient media used to grow bacteria, so an inducer, such as IPTG, must be added to enable growth on agar plates. However, when SLDAPD was transformed with a plasmid that contained a lac operator sequence (lacO), the lac repressor protein was titrated from lacO regulating dapD, thus increasing dapD expression sufficiently to allow cell growth and providing a mechanism for plasmid selection and maintenance (Fig. 1).
FIG. 1.
Conversion of SL3261 to SLDAPD and plasmid selection by ORT. (A) Wild-type dapD locus in SL3261. (B) Replacement of dapD with the lacO/P-dapD-lacIq cassette to create SLDAPD. Induction with IPTG is required for dapD expression in the absence of a plasmid. (C) When the organism is transformed with a plasmid containing a lacO sequence, the repressor is titrated and dapD is expressed.
Growth analyses showed that SLDAPD was not affected by the chromosomal modifications, since the growth of SLDAPD in vitro closely matched that of parental strain SL3261 (data not shown). The basal, leaky expression of dapD proved to be sufficient to allow cell growth in liquid medium in the absence of IPTG, but uninduced dapD expression was not sufficient to allow the growth of single colonies on solid agar plates, thus enabling selection of transformants by ORT.
Plasmid maintenance with ORT.
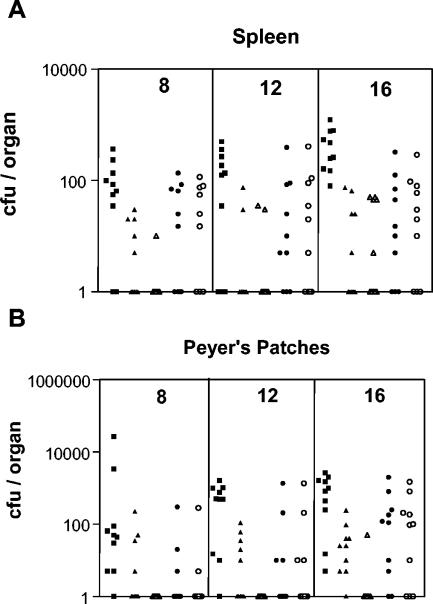
To demonstrate the ORT technology for live vaccine vectors, we investigated plasmid maintenance in SLDAPD containing pUC18I, a derivative of pUC18 with the lacO1 and lacO3 operators replaced with the ideal lacO (4). Serial subculture in LB broth in the absence of antibiotics showed that equivalent colony numbers were obtained on LB agar plates for total viable cells (with IPTG) and plasmid-containing cells, indicating that there had been no plasmid loss over the 5 days of culture. Subsequently, maintenance of plasmid pUC18I in SLDAPD was evaluated in vivo. Following intragastric inoculation of BALB/c mice with 1 × 109 to 5 × 109 CFU of SLDAPD/pUC18I or SL3261/pUC18I, recombinant bacteria could be detected in the spleens and Peyer's patches of all groups of mice (individual samples are shown in Fig. 2; mean results are shown in Table 1). However, significantly lower numbers of SL3261/pUC18I cells were detected on LB agar plates containing ampicillin than on LB agar plates, indicating that plasmid pUC18I was lost from the bacteria. In comparison, similar numbers of SLDAPD/pUC18I cells were detected on LB agar plates containing IPTG and LB agar plates containing ampicillin, showing that the plasmid was stable. In some cases, SL3261/pUC18I and SLDAPD/pUC18I were detected at significantly lower levels than SL3261 in both spleens and Peyer's patches. This may have reflected the burden on Salmonella of harboring the high-copy-number plasmid pUC18I.
FIG. 2.
Colonization of spleens and Peyer's patches following oral inoculation of mice (n = 10) with SLDAPD or SL3261 carrying plasmid pUC18I. Mice were inoculated with 5 × 109 CFU of SLDAPD/pUC18I (circles), SL3261/pUC18I (triangles), or control strain SL3261 (squares) by using a gavage needle. Spleens (A) and Peyer's patches (B) were removed 8, 12, or 16 days after inoculation and homogenized in PBS before preparations were plated onto LB agar (solid symbols) or LB agar containing ampicillin (open symbols) for enumeration of bacteria.
TABLE 1.
Stability of plasmids following intragastric inoculation of micea
| Group | % of bacteria retaining plasmid
|
|||||
|---|---|---|---|---|---|---|
| Day 8
|
Day 12
|
Day 16
|
||||
| Spleen | Peyer's patches | Spleen | Peyer's patches | Spleen | Peyer's patches | |
| SLDAPD/pUC18I | 92 | 88 | 100 | 100 | 100 | 83 |
| SL3261/pUC18I | 12 | 0 | 62 | 0 | 100 | 10 |
| SLDAPD/pAHL | 84 | 100 | 99 | 100 | 88 | 96 |
| SL3261/pAH34L | 100 | 95 | 97 | 83 | 94 | 100 |
Groups of BALB/c mice (n = 10) were orally inoculated with SLDAPD/pUC18I, SL3261/pUC18I, SLDAPD/pAHL, SLDAPD, SL3261/pAH34L, or SL3261. Spleens and six Peyer's patches per mouse were removed 8, 12, and 16 days after inoculation and homogenized in PBS before preparations were plated onto selective or nonselective agar for enumeration of the bacteria retaining plasmids. The values are the percentages of organisms that retained plasmids to the nearest 1%.
Construction of a Salmonella-based plague vaccine by using ORT.
We previously showed that SL3261/pAH34L, which surface expresses the Y. pestis F1 antigen, provides protection against plague in mice (26). The pAH34L plasmid contains the thermoregulated caf operon that includes the gene encoding F1 antigen and other genes involved in the export and assembly of F1 antigen on the bacterial cell surface in Y. pestis. To adapt pAH34L for use in SLDAPD, the kanamycin resistance gene was replaced with the ideal lac operator to create the ORT-VAC plasmid pAHL, but no modifications of the caf operon or pSC101 origin of replication were made (Fig. 3). pAHL was subsequently transformed into SLDAPD for evaluation of plasmid maintenance.
FIG. 3.
Conversion of the kanamycin-resistant plasmid pAH34L to the ORT plasmid pAHL by replacement of kan with ideal lacO.
Similar to the results obtained with pUC18I, serial subculture in LB broth in the absence of antibiotics showed that pAHL was not lost from SLDAPD over 5 days. In vivo, recombinant bacteria could also be detected in the spleens and Peyer's patches of BALB/c mice orally inoculated with 1 × 109 to 5 × 109 CFU of SLDAPD/pAHL or SL3261/pAHL. However, unlike pUC18I, pAH34L was relatively stable (84 to 100%) in SL3261 (Table 1). The ORT-VAC plasmid pAHL was also stable (83 to 100%) in SLDAPD (Table 1). There was no significant difference between the numbers of SL3261 and SLDAPD cells colonizing tissues (data not shown), suggesting that SLDAPD was able to grow well in vivo. There was also no significant difference between the numbers of SLDAPD and SLDAPD/pAHL cells or between the numbers of SL3261 and SL3261/pAH34L cells (data not shown), indicating that plasmids pAHL and pAH34L had no deleterious effects on the colonization abilities of their Salmonella host strains.
ORT Salmonella strain expressing F1 antigen provides protection against plague.
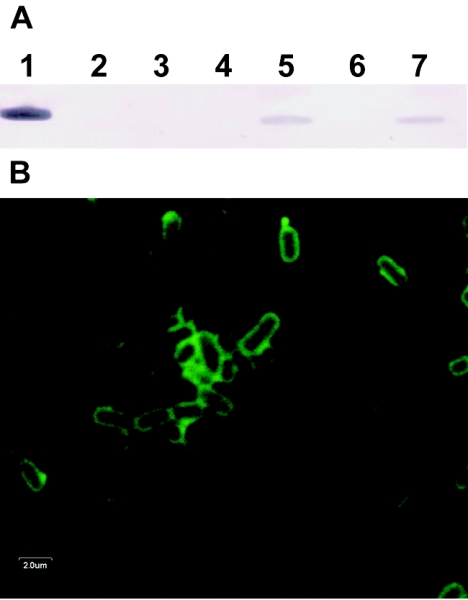
In Y. pestis, F1 antigen is expressed at 37°C but not at 28°C. Here, Western blotting was used to detect expression of F1 antigen in SL3261/pAH34L and SLDAPD/pAHL after induction at 37°C, and immunofluorescence confocal microscopy was used to demonstrate the surface location of expressed F1 antigen on SLDAPD/pAHL (Fig. 4).
FIG. 4.
Analysis of Y. pestis F1 antigen expression. (A) Western blot probed with an anti-F1 antigen mouse monoclonal antibody. Lane 1, recombinant F1 antigen reference; lane 2, SL3261 grown at 37°C; lane 3, SLDAPD grown at 37°C; lane 4, SL3261/pAH34L grown at 28°C; lane 5, SL3261/pAH34L grown at 37°C; lane 6, SLDAPD/pAHL grown at 28°C; lane 7, SLDAPD/pAHL grown at 37°C. (B) Expression of F1 antigen on the surface of SLDAPD/pAHL grown at 37°C. Immunofluorescently labeled SLDAPD/pAHL fixed cells were detected by confocal microscopy.
To evaluate the protection provided by the ORT Salmonella vaccine strain that surface expressed F1 antigen, BALB/c mice were orally immunized with one or two doses consisting of 1 × 1010 to 5 × 1010 CFU of SLDAPD/pAHL or the control Salmonella strain SLDAPD. For comparison, other groups of mice were not immunized or were immunized with the original SL3261/pAH34L vaccine strain or control strain SL3261. Following challenge with 185 MLD of Y. pestis, five of six mice vaccinated with a single dose of SLDAPD/pAHL or SL3261/pAH34L were protected against plague (Table 2). These mice had high serum IgG responses to F1 antigen, whereas mice that did not survive the challenge had low or undetectable IgG responses to F1 antigen (Fig. 5). In comparison, 14 of 15 control mice did not survive. The surviving control mouse had been inoculated with SLDAPD alone, did not show detectable levels of F1 antigen-specific IgG, and showed symptoms of Y. pestis infection following the challenge. Furthermore, Y. pestis was detected in the spleen of the surviving control mouse but not in tissue samples from other survivors. This is the first example of complete protection against plague provided by a single dose of a Salmonella-based vaccine.
TABLE 2.
Survival of immunized mice after Y. pestis challengea
| Group | No. of survivors/total no. with the following no. of vaccine doses (challenge inoculum):
|
||
|---|---|---|---|
| One (185 MLD) | One (1.9 × 105 MLD) | Two (8.8 × 104 MLD) | |
| SLDAPD/pAHL | 5/6 | 3/6 | 5/5 |
| SLDAPD | 1/6b | 0/6 | 0/4 |
| SL3261/pAH34L | 5/6 | 4/6 | 4/4 |
| SL3261 | 0/6 | 0/6 | 0/4 |
| Naïve | 0/6 | 0/6 | |
Groups of BALB/c mice were orally inoculated with one or two doses of SLDAPD/pAHL, SLDAPD, SL3261/pAH34L, or SL3261. Fifty days after the last dose, immunized mice and naïve control mice were challenged subcutaneously with approximately 2 × 102 or 105 MLD of Y. pestis strain GB and observed for 20 days.
The surviving control mouse showed signs of Y. pestis infection.
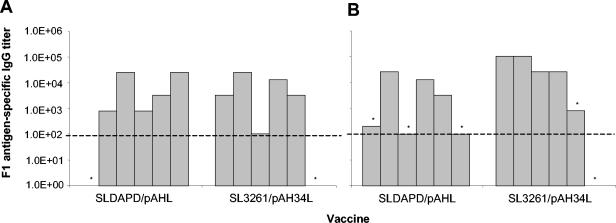
FIG. 5.
F1 antigen-specific IgG responses induced in individual mice following immunization with Salmonella expressing F1 antigen. Groups of BALB/c mice (n = 6) were orally immunized with a single dose of 1 × 1010 to 5 × 1010 CFU of ORT strain SLDAPD/pAHL or original vaccine strain SL3261/pAH34L. Serum samples were removed 41 days after inoculation, and IgG responses were measured by ELISA. The results are expressed as the reciprocal of the end point dilution for each animal group. The limit of detection is indicated by the horizontal dashed line. Mice were subsequently challenged with 185 MLD (A) or 1.85 × 105 MLD (B) of Y. pestis GB. The asterisks indicate mice that did not survive the Y. pestis infection.
When mice were given a high challenge dose of Y. pestis (1.85 × 105 MLD), a single dose of SLDAPD/pAHL or SL3261/pAH34L was not sufficient to provide complete protection (Table 2). However, mice that did not survive the challenge had low or undetectable IgG responses to F1 antigen (Fig. 5). In comparison, equal or greater IgG responses to F1 antigen were detected in all mice that survived the Y. pestis challenge. In a second challenge experiment, mice orally vaccinated with two doses of either of the F1 antigen-expressing Salmonella strains were completely protected against a high challenge dose of Y. pestis (8.8 × 104 MLD). The finding that SLDAPD/pAHL and SL3261/pAH34L provided similar levels of protection against plague in mice demonstrates that application of the ORT technology to the Salmonella-based plague vaccine does not have any deleterious effects on its efficacy. Moreover, utilization of the ORT technology enabled development of an effective antibiotic-free Salmonella-based vaccine against plague.
DISCUSSION
A frequent problem associated with the use of live, attenuated bacteria to deliver heterologous antigens is the instability of the genes encoding the antigens. In this study, we investigated plasmid maintenance by ORT using plasmids with two different types of origin of replication.
It is known that plasmids with the high-copy-number pMB1 origin of replication, such as pUC18I, are unstable in Salmonella (24), so we first wanted to compare stability using this plasmid in a conventional Salmonella vaccine strain and an ORT-VAC strain. We based our ORT-VAC strain (SLDAPD) on S. enterica serovar Typhimurium SL3261, an auxotrophic, avirulent, live vaccine model strain that has previously been used to immunize mice against virulent strains of Salmonella (10), as well as for vaccine delivery against a range of other infections, including influenza A virus (27) and Y. pestis (26).
When pUC18I was transformed into SLDAPD, the organism was maintained in liquid culture with no indication of plasmid loss. With alternative plasmid stabilization systems, plasmid loss is often masked by the death of cells that lose the plasmid. Untransformed SLDAPD does grow well in liquid culture in the absence of DAP or IPTG, but it is unable to grow in the same medium on agar plates. Thus, plasmid loss in culture should be detectable by plating onto LB agar containing IPTG, but such plasmid loss was not seen in this study. In vivo, pUC18I was unstable in SL3261 but was successfully stabilized in SLDAPD. The degree of plasmid loss in SL3261/pUC18I was apparent when we compared the number of total viable cell counts with the number of plasmid-containing cell counts in organs removed from orally inoculated mice; the latter number was generally lower, indicating that the plasmid was indeed being lost but the plasmid-free segregants were surviving. This plasmid loss was more acute in the spleen than in the Peyer's patches. This may reflect the greater metabolic burden on bacteria that reach the spleen than on bacteria that do not penetrate further than the Peyer's patches. The untransformed SL3261 cells were present in tissues in larger numbers than the equivalent plasmid-containing cells of both strains, indicating that there was a metabolic burden on replicating pUC18I even though the only expressed gene was bla (encoding β-lactamase for ampicillin resistance). This illustrates that for recombinant protein delivery, an antibiotic or other expressed gene is an undesirable element that may reduce the fitness of the live vaccine vector strain. A further advantage of the ORT-VAC technique over plasmid maintenance systems that rely upon complementation of a host auxotrophy is that the selective gene is present as a single, chromosomal copy, thus preventing overexpression of the complementary gene product and loss of selection pressure, which are inherent in the use of plasmid-borne selective genes.
We also investigated whether a plasmid that has been shown to give a protective immune response, pAH34L, could be successfully adapted to create an antibiotic gene-free vaccine delivery system. A Salmonella vaccine strain containing this plasmid, SL3261/pAH34L, was previously shown to protect against a high challenge dose of Y. pestis, the causative agent of plague, following two oral immunizations. Thus, we transformed the adapted plasmid, pAHL, into SLDAPD for further evaluation. In vitro experiments showed that the plasmid was maintained in liquid culture with no indication of any plasmid loss. pAHL was also stable in SLDAPD in vivo, like pAH34L in SL3261.
The pSC101 origin of replication in pAHL gives a copy number of approximately six per chromosome (24), yet our results show that this is sufficient to cause titration of the lac repressor in both ORT strain DH1lacdapD (see Material and Methods) and ORT-VAC strain SLDAPD. It has previously been shown that medium-copy-number plasmids based on pBR322 (39 to 55 copies per cell) can be selected by ORT in DH1lacdapD with LacI at 20 copies per cell (4). However, pAHL is the lowest-copy-number plasmid that has been selected by ORT so far, and it is the first plasmid selected with a copy number lower than that of the repressor protein. This is more remarkable in SLDAPD, in which LacI expressed from lacIq provides approximately 200 copies per cell in E. coli (16), although the value may be lower in S. enterica serovar Typhimurium.
We used the BALB/c mouse model of Y. pestis infection to evaluate whether the antibiotic gene-free ORT-VAC strain SLDAPD/pAHL provided a level of protection against plague similar to that conferred by SL3261/pAH34L. Following single or double oral vaccination with either SLDAPD/pAHL or SL3261/pAH34L, high titers of serum F1 antigen-specific IgG were detected. A single oral inoculation of either vaccine protected five of six mice against 185 MLD of virulent Y. pestis. This is the first demonstration that a single oral inoculation of Salmonella expressing F1 antigen can protect against plague. Furthermore, partial protection against a high-level challenge (approximately 105 MLD of Y. pestis) was observed in mice given a single oral inoculation of SLDAPD/pAHL (three of six mice) or SL3261/pAH34L (four of six mice). All survivors of Y. pestis infection showed F1 antigen-specific IgG responses equivalent to or greater than those in nonsurvivors, suggesting that antibody to F1 antigen provides an indicator of protection, as described previously (15). Administering two doses of either vaccine could produce complete protection against this high-level challenge. These results show promise for future development of a highly protective, orally delivered, antibiotic-free vaccine against plague.
In conclusion, we constructed S. enterica serovar Typhimurium strain SLDAPD, which allows antibiotic-free plasmid selection and maintenance in a complex medium of high- and very-low-copy-number plasmids by ORT. This strain allows stable in vivo maintenance of pUC18I in mice, whereas this plasmid was lost from parental strain SL3261. In addition, we applied ORT-VAC technology to a Salmonella-based vaccine against plague. The ORT-VAC technology should therefore permit construction of versatile strains of recombinant bacteria that can be used as live vaccine vectors. These strains will not need antibiotic resistance or any other genes for stable plasmid maintenance and will have the advantage of potentially higher levels of antigen expression than would be possible by single chromosomal integration of an antigen gene.
Acknowledgments
We gratefully acknowledge technical assistance provided by Debbie Rogers and Dave Rawkins.
Editor: J. D. Clements
REFERENCES
- 1.Amann, E., B. Ochs, and K.-J. Abel. 1988. Tightly regulated tac promoter vectors for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cranenburgh, R. M., J. A. J. Hanak, S. G. Williams, and D. J. Sherratt. 2001. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 29:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cranenburgh, R. M., K. S. Lewis, and J. A. J. Hanak. 2004. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 7:197-203. [DOI] [PubMed] [Google Scholar]
- 5.Galán, J. E., K. Nakayama, and R. Curtiss III. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94:29-35. [DOI] [PubMed] [Google Scholar]
- 6.Galen, J. E., J. Nair, J. Yuang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garmory, H. S., K. A. Brown, and R. W. Titball. 2002. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol. Rev. 26:339-353. [DOI] [PubMed] [Google Scholar]
- 8.Hanak, J. A. J., and R. M. Cranenburgh. 2001. Antibiotic free plasmid selection and maintenance in bacteria, p. 111-124. In Recombinant protein production with prokaryotic and eukaryotic cells. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 9.Hanke, T., and A. J. McMichael. 2000. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat. Med. 6:951-955. [DOI] [PubMed] [Google Scholar]
- 10.Hoiseth, S. K., and B. A. D. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238. [DOI] [PubMed] [Google Scholar]
- 11.Hone, D., S. Attridge, L. van den Bosch, and J. Hackett. 1988. A chromosomal integration system for stabilization of heterologous genes in Salmonella based vaccine strains. Microb. Pathog. 5:407-418. [DOI] [PubMed] [Google Scholar]
- 12.Link, A. J., D. R. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNeill, H. V., K. A. Sinha, C. E. Hormaeche, J. J. Lee, and C. M. A. Khan. 2000. Development of a nonantibiotic dominant marker for positively selecting expression plasmids in multivalent Salmonella vaccines. Appl. Environ. Microbiol. 66:1216-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morona, R., J. Yeadon, A. Considine, J. K. Morona, and P. A. Manning. 1991. Construction of plasmid vectors with a non-antibiotic selection system based on the Escherichia coli thyA+ gene: application to cholera vaccine development. Gene 107:139-144. [DOI] [PubMed] [Google Scholar]
- 15.Morton, M., H. S. Garmory, S. D. Perkins, A. M. O'Dowd, K. F. Griffin, A. K. Turner, A. M. Bennett, and R. W. Titball. 2004. A Salmonella enterica serovar Typhi vaccine expressing Yersinia pestis F1 antigen on its surface provides protection against plague in mice. Vaccine 22:2524-2532. [DOI] [PubMed] [Google Scholar]
- 16.Müller-Hill, B., L. Crapo, and W. Gilbert. 1968. Mutants that make more lac repressor. Proc. Natl. Acad. Sci. USA 59:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama, K., S. M. Kelly, and Roy Curtiss III. 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Bio/Technology 6:693-697. [Google Scholar]
- 18.Oyston, P. C. F., E. D. Williamson, S. E. Leary, S. M. Eley, K. F. Griffin, and R. W. Titball. 1995. Immunization with live recombinant Salmonella typhimurium aroA producing F1 antigen protects against plague. Infect. Immun. 63:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richaud, C., F. Richaud, C. Martin, C. Haziza, and J. C. Patte. 1984. Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J. Biol. Chem. 259:14824-14828. [PubMed] [Google Scholar]
- 20.Russell, P., S. M. Eley, S. E. Hibbs, R. J. Manchee, A. J. Stagg, and R. W. Titball. 1995. A comparison of plague vaccine USP and EV76 vaccine induced protection against Yersinia pestis in a murine model. Vaccine 13:1551-1556. [DOI] [PubMed] [Google Scholar]
- 21.Ryan, E. T., T. I. Crean, S. K. Kochi, M. John, A. A. Luciano, K. P. Killeen, K. E. Klose, and S. B. Calderwood. 2000. Development of a ΔglnA balanced lethal plasmid system for expression of heterologous antigens by attenuated vaccine vector strains of Vibrio cholerae. Infect. Immun. 68:221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadler, J. R., H. Sasmor, and J. L. Betz. 1983. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl. Acad. Sci. USA 80:6785-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons, A., D. Tils, B. von Wilcken-Bergmann, and B. Müller-Hill. 1984. Possible ideal lac operator: Escherichia coli lac operator like sequences from eukaryotic genomes lack the central G · C pair. Proc. Natl. Acad. Sci. USA 81:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoker, N. G., N. F. Fairweather, and B. G. Spratt. 1982. Versatile low copy-number plasmid vectors for cloning in Escherichia coli. Gene 18:335-341. [DOI] [PubMed] [Google Scholar]
- 25.Tacket, C. O., S. M. Kelly, F. Schödel, G. Losonsky, J. P. Nataro, R. Edelman, M. M. Levine, and R. Curtiss III. 1997. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the Asd-balanced lethal vector system. Infect. Immun. 65:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Titball, R. W., A. M. Howells, P. C. F. Oyston, and E. D. Williamson. 1997. Expression of the Yersinia pestis capsular antigen (F1 antigen) on the surface of an aroA mutant of Salmonella typhimurium induces high levels of protection against plague. Infect. Immun. 65:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tite, J. P., X.-M. Gao, C. M. Hughes-Jenkins, M. Lipscombe, D. O'Callaghan, G. Dougan, and F. Y. Liew. 1990. Anti-viral immunity induced by recombinant nucleoprotein of influenza A virus. Immunity 70:540-546. [PMC free article] [PubMed] [Google Scholar]