Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and the most common cause of dementia in the elderly worldwide. The exact etiology of AD, particularly its genetic mechanisms, remains incompletely understood. Traditional genome-wide association studies (GWAS), which primarily focus on single-nucleotide polymorphisms (SNPs) with main effects, provide limited explanations for the “missing heritability” of AD, while there is growing evidence supporting the important role of epistasis. In this study, we performed a genome-wide SNP–SNP interaction detection using a linear regression model and employed multiple GPUs for parallel computing, significantly enhancing the speed of whole-genome analysis. The cerebrospinal fluid (CSF) phosphorylated tau (P-tau)/amyloid- (A) ratio was used as a quantitative trait (QT) to enhance statistical power. Age, gender, and clinical diagnosis were included as covariates to control for potential non-genetic factors influencing AD. We identified 961 pairs of statistically significant SNP–SNP interactions, explaining a high-level variance of P-tau/A level, all of which exhibited marginal main effects. Additionally, we replicated 432 previously reported AD-related genes and found 11 gene–gene interaction pairs overlapping with the protein-protein interaction (PPI) network. Our findings may contribute to partially explain the “missing heritability” of AD. The identified subnetwork may be associated with synaptic dysfunction, Wnt signaling pathway, oligodendrocytes, inflammation, hippocampus, and neuronal cells.
Subject terms: Epistasis, Quantitative trait, Alzheimer's disease, Alzheimer's disease
Subject terms: Epistasis, Quantitative trait, Alzheimer's disease, Alzheimer's disease
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by cognitive decline involving memory loss, judgment, reasoning, and impaired daily functioning. It is the most common cause of dementia among the elderly worldwide. The exact etiology of AD is not fully understood, especially the genetic mechanisms of AD, but it is believed to result from complex interactions between genetic and environmental factors1–5. For the last two decades, genome-wide association studies (GWAS) have been a successful approach of AD for exploring the correlation between different gene variations and different phenotypic changes. However, the traditional GWAS is mainly based on the association study of single-nucleotide polymorphisms (SNPs) with main effects in the whole genome, and these results only explained part of the “missing heritability” in AD6,7.
The “missing heritability” refers to the phenomenon where the heritability of a trait or disease cannot be entirely explained by the genetic variants identified through traditional GWAS. It suggests that there are additional genetic factors contributing to the trait or disease that have not been discovered yet8–10.
Epistatic interactions, also known as gene–gene interactions, refer to the phenomenon in genetics where the effect of one gene mask or modifies the effect of another gene. These interactions can influence the expression, phenotype, or inheritance patterns of traits8,11–13. Epistasis detection has attracted much attention in recent years, because mounting evidence has shown that epistatic interactions might partially explain the “missing heritability” of AD and will help us to better understand the genetic architecture that underlies complex variation of phenotypes for complex diseases such as AD14,15.
However, detecting epistatic interactions across the whole genome still remain twofold challenges of statistical and heavy computational burden. To combat these challenges, many machine learning methods and multifactor dimensionality reduction (MDR) methods were performed, but the results identified by which were challenged for “missing heritability”16,17. Furthermore, most of these methods are based on case-control designs, with limited research focusing on performing epistatic interactions detection for quantitative trait (QT)18,19.
Although the aforementioned methods have demonstrated their effectiveness on identifying epistasis from genome-wide data, the results identified by which were still challenged for “missing heritability”. One possible reason is that those methods typically used screening strategy, which were easy to missed some SNPs with marginal main effects. Another reason may be the advantages of QT over case–control. To our knowledge, QT analysis allows for the use of continuous measurements, providing more information and increasing statistical power compared to dichotomous outcomes used in case-control studies. In the capture of variation, QT capture the full spectrum of variation within a population, allowing for a more comprehensive understanding of genetic influences on complex traits. At the same time, QT analysis enables the examination of subtle differences in phenotype among individuals, facilitating the identification of marginal effect sizes and complex genetic interactions. Noteworthy, QT can represent intermediate phenotypes or endophenotypes, which may be closer to underlying biological mechanisms and provide insights into disease pathways8,20–22.
In recent years, the use of the cerebrospinal fluid (CSF) phosphorylated tau (P-tau)/amyloid- (A) ratio was an important intermediate endophenotype in AD research, which enabling the understanding of disease progression, diagnosis, and differentiation from other neurodegenerative disorders. It’s worth noting that there are many more studies available on this topic, and further research continues to explore the significance of these biomarkers in AD. The researchers found that this ratio was significantly higher in AD compared to frontotemporal dementia (FTD), suggesting its potential utility as a diagnostic biomarker23–26.
Overall, this paper aims to detect the genome-wide SNP–SNP interaction by using P-tau/A ratio as QT, without disregarding the possibility of SNP-SNP interactions between the marginal main effects, which may be the crucial way for explaining the “missing heritability” of AD. In addition, a muti-GPU method was used for parallel computing to enhance the detection power of SNP–SNP interaction.
Results
Two-marker Interaction Study Results and Post Hoc Analysis
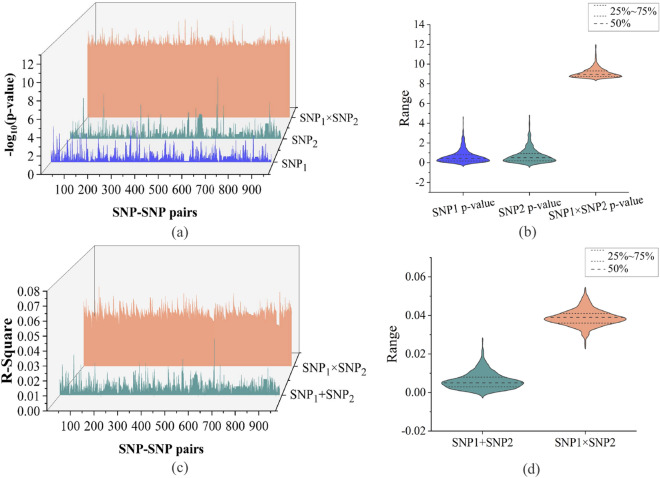
We used age, gender, and clinical diagnosis (Dx) as covariates in our two-marker interaction study. A total of 1469 SNPs (961 pairs of SNP-SNP interaction) passed the p-value threshold at P-tau/A ratio level. Linkage disequilibrium was assessed by querying the 961 pairs of SNP-SNP interaction using PLINK v1.90 and Haploview 4.1 tool with R2 threshold of 0.8. The results indicated that these SNPs are considered to be largely independent (see Supplementary Table S1 online). Considering the technical complexities of pairwise scan of SNP-SNP interaction across the whole genome, we used a muti-GPU tool named GEEpiQt to combat with the heavy computational burden27. In addition, the genotype data and participants information of the corresponding loci were extracted according to the SNP-SNP interaction combination information with significant interaction. The main effect of each SNP site was obtained by GWAS, which was used to compare with two-marker interaction results. In this paper, the main effect and interaction effect of 961 SNP-SNP interaction pairs at P-tau/A ratio level are statistically compared (Fig. 1a). The violin plot shows the numerical distribution and corresponding probability density of the main effect and interaction effect of SNP1 and SNP2 (Fig. 1b). As show in Fig. 1a, the blue and green areas respectively represent the main effects of SNP1 and SNP2 in the interaction combination, and the red part represents the interaction effect after the interaction of SNP1 and SNP2. Moreover, from the overall distribution, the average distribution of interaction effect was much higher than the distribution level of main effect. It suggests that the statistical significance of the interaction effect was much higher than the main effect. The results also showed that there was a significant interaction between SNPs with marginal main effect in terms of statistical implications.
Figure 1.
The main effect and interaction effect of 961 SNP-SNP interaction pairs at P-tau/A ratio level, and R square of the additive terms and the interaction terms. (a) The waterfall comparison plot of interaction effects and main effects of single SNP with −log10 adjusted p-value. The bule waterfall represents main effects of SNP1, the green waterfall represents main effects of SNP2, the orange waterfall represents the interaction effects of SNP1 SNP2. (b) The violin distribution of interaction effects (SNP1 SNP2) data and main effects (SNP1, SNP2) data. (c) The waterfall comparison plot of the R square of SNP1 SNP2 term (orange waterfall) and the R square of SNP1 + SNP2 term (green waterfall). (d) The violin distribution of R square: interaction (SNP1 SNP2) and additive terms (SNP1 + SNP2).
The top 10 R-squared values of SNP–SNP interaction pairs are shown in Table 1. The results of top 10 two-marker interaction were: rs2223610(MROH9)–rs12684467, rs11082417(SETBP1)–rs1966778(PITPNM3), rs17810546(IL12A-AS1)–rs17836920(LINC02074), rs9493790(TBPL1)–rs7955904(SSPN), rs17809756(IL12A-AS1)–rs17836920(LINC02074), rs9730519–rs12458881(NEDD4L), rs718222 (PRICKLE2)–rs437202(CSMD1), rs7617057(GXYLT2)–rs17051925(PARP4P2), rs3860070(KLB)–rs6912680(ANKRD6), rs1780555(RAP1A)–rs7074813(RAB18).
Table 1.
Top 10 R square of SNP–SNP interaction pairs of the P-tau/A ratio.
| No | SNP–SNP | CHR | Gene | p-value | Explained variance (R square) | |||
|---|---|---|---|---|---|---|---|---|
| GWAS | Interaction | Age+gender+Dxa | SNP1+SNP2 b | SNP1 SNP2 c | ||||
| 1 |
rs2223610 rs12684467 |
1 9 |
MROH9 – |
0.496 0.281 |
1.20E−11 | 0.133 | 0.011 | 0.058 |
| 2 |
rs11082417 rs1966778 |
18 17 |
SETBP1 PITPNM3 |
0.025 0.061 |
3.08E−10 | 0.133 | 0.002 | 0.054 |
| 3 |
rs17810546 rs17836920 |
3 17 |
IL12A-AS1 LINC02074 |
0.087 0.169 |
1.25E−12 | 0.133 | 0.006 | 0.053 |
| 4 |
rs9493790 rs7955904 |
6 12 |
TBPL1 SSPN |
0.188 0.082 |
1.09E−9 | 0.133 | 0.003 | 0.053 |
| 5 |
rs17809756 rs17836920 |
3 17 |
IL12A-AS1 LINC02074 |
0.259 0.169 |
1.75E−12 | 0.133 | 0.006 | 0.052 |
| 6 |
rs9730519 rs12458881 |
1 18 |
– NEDD4L |
0.272 0.482 |
8.59E−11 | 0.133 | 0.009 | 0.051 |
| 7 |
rs718222 rs4372022 |
3 8 |
PRICKLE2 CSMD1 |
0.708 0.096 |
2.50E−09 | 0.133 | 0.006 | 0.051 |
| 8 |
rs7617057 rs17051925 |
3 13 |
GXYLT2 PARP4P2 |
0.117 0.004 |
2.12E−12 | 0.133 | 0.001 | 0.050 |
| 9 |
rs3860070 rs6912680 |
4 6 |
KLB ANKRD6 |
0.981 0.082 |
2.33E−11 | 0.133 | 0.018 | 0.050 |
| 10 |
rs1780555 rs7074813 |
1 10 |
RAP1A RAB18 |
0.072 0.378 |
3.57E−10 | 0.133 | 0.005 | 0.050 |
a Age+gender+Dx: the percent of variance in P-tau/A ratio level explained by age, gender, and Dx.
b SNP1+SNP2: in addition to accounting for three covariates of age, gender, and Dx, the percent of additional variance P-tau/A ratio level explained by the combined main effect of SNP1 and SNP2.
cSNP1 SNP2: In addition to accounting for age, gender, and Dx, SNP1 and SNP2, the percent of additional variance in P-tau/A ratio level explained by the interaction effect of SNP1 and SNP2.
Furthermore, Table 1 also shows the SNP-SNP interaction results of post hoc analysis on P-tau/A ratio level, and the additive term of SNP1+ SNP2, the interaction term of SNP1 SNP2 and the explained variance of covariates are listed in detail. In this paper, IBM SPSS 24.0 was used to explain variance of SNP-SNP pair with significant interaction on P-tau/A ratio. The R square of the additive terms (SNP1+ SNP2) and the interaction terms (SNP1 SNP2)are shown in Fig. 1c. The orange area and the green area represent the variance explained by interaction term and additive term at the P-tau/A ratio level respectively. In order to further obtain the overall distribution of R-square, the distribution state and corresponding probability density of R-square with additive term and interactive term are shown in Fig. 1d.
As shown in Table 1, all covariates (include age, gender, and Dx) accounted for 13.3% of variance on the P-tau/A level. The variance related to interaction term is determined by running a hierarchical linear regression model, that is, adding genetic main effect first and then adding genetic interaction term. Intriguingly, the variance proportion of the interaction term is much higher than the main effects for each identified SNP-SNP interaction pair as seen from the Table 1. As shown by the top three SNP pairs in the Table 1, the main effects of SNPs accounted for 1.1% of variance, and the interaction term accounted for 5.8% of variance, that is, the combination of the two accounts for 6.9% for the first-ranked rs2223610(MROH9)–rs12684467. And for the second-ranked rs11082417(SETBP1)–rs1966778(PITPNM3), the main effects accounted for 0.2% of variance, and the interaction term accounted for 5.4% of variance, that is, the combination of the two accounts for 5.6%. For the third-ranked rs17810546(IL12A-AS1)–rs17836920(LINC02074), the main effects accounted for 0.6% of variance, and the interaction term accounted for 5.3% of variance, that is, the combination of the two accounts for 5.9%. Our study revealed that the main effects of the identified interaction SNP pairs are all marginal, but they explained a relatively high-level of variance of P-tau/A ratio. The gene marked in bold indicates that it is directly related to AD.
Gene set enrichment analysis results
For illustrating the correlation between the SNP-SNP interaction pairs and AD from different angles and using different databases, a total of 961 SNP-SNP interaction pairs were mapped onto 286 gene–gene pairs (include 505 different genes) by Homo sapiens genome assembly GRCh37. First, by searching in two gene set libraries of Enrichr: PhenGenI Association 2021 and HDSigDB Human 2021, we found that 432 genes out of 505 genes were included in the items related to AD, and 85 genes were not included in the items related to AD, which indicated that 432 genes were reported as AD-related genes, and 85 genes were not yet reported as AD-related genes.
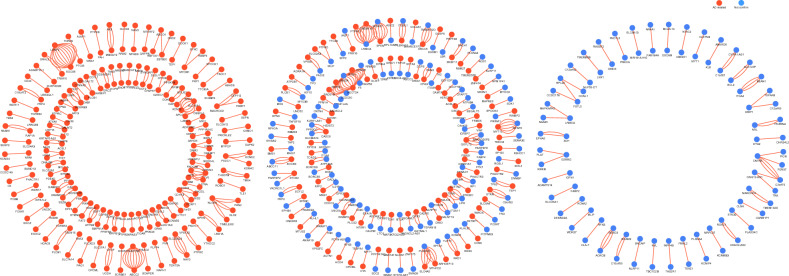
Given that two genes in a gene–gene interaction pair have three relationships with AD: both genes are related to AD, only one gene is related to AD, and neither gene of the two in a gene–gene pair is related to AD, so we divide these interaction pairs into three groups (G1, G2, and G3), as shown in Fig. 2. Specifically, G1 represents all gene pairs in this group that have entries containing the keyword “AD”. We consider these gene pairs to be reported as associated with AD. G2 represents gene pairs in this group where only one gene has an entry containing the keyword “AD”. G3 represents gene pairs in this group where neither gene has an entry containing the keyword “AD”, indicating that these genes have not been reported as associated with AD.
Figure 2.
The circle diagram of gene–gene interaction pairs based on the Enrichr database: shows three relationships between genes and AD. All genes related to AD are shown in red dots. The genes that not yet been reported as AD-related are shown in bule dots. G1 include 122 pairs of gene–gene interaction (221 different genes); G2 include 117 pairs of gene–gene interaction (225 different genes); G3 include 47 pairs of gene–gene interaction (85 different genes).
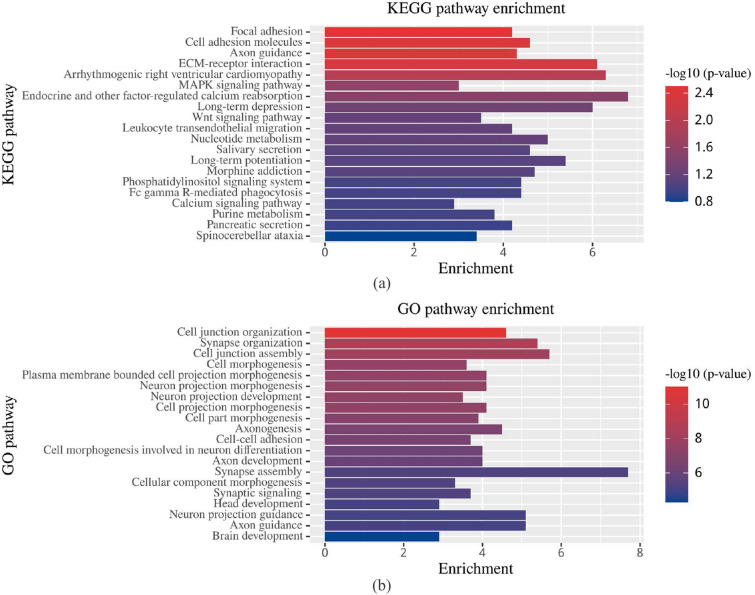
In addition, the biological significance of our results was analyzed further based on two well-known biological pathway databases including KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology)28–30. The top 20 biological processes based on the corresponding p-value of KEGG/GO are shown in Fig. 3.
Figure 3.
Results of KEGG pathway enrichment analysis and GO functional enrichment analysis. (a) Enriched KEGG terms ranked top 20 based on −log10 adjusted p-values. (b) Enriched GO terms ranked top 20 based on −log10 adjusted p-values. Numbers of genes contributing to each term is displayed as bars.
The bar plot of KEGG pathway enrichment analysis showed that the significant biological processes include focal adhesion (FA), cell adhesion molecules, axon guidance, Wnt signaling pathway, and calcium signaling pathway, etc31–34. Foremost among these, As shown in Fig. 3a the most significant biological process was FA, which may be an important participator in the AD process. Moreover, FA signaling might regulate neuronal viability and synaptic loss during AD35,36.
Using GO functional enrichment analysis, we found that the significant pathway enrichments were related to AD, such as cell junction organization, synapse organization, cell junction assembly, neuron projection morphogenesis, neuron projectionc development, and cell–cell adhesion37–41. As aforementioned, the analysis of function and pathway enrichments improve the biological interpretability of our results.
Network analysis results
It is noticeable that protein–protein interaction (PPI) networks provide valuable information for predicting genotype-phenotype associations. They aid in understanding the relationship between genetic variations and phenotypic traits. Therefore, we used 286 pairs of gene–gene interaction from G1, G2 and G3 data sets for PPI network analysis to further interpret biological significance of our results. As shown in Fig. 4, 11 pairs of gene–gene interaction were identified to be overlapping with the PPI network by using the STRING tool (https://string-db.org/, Version 12, accessed on 31 July 2023). Nodes and edges in the PPI network represent the genes and interactions between two genes, respectively. The 22 nodes respectively represent 22 genes, which are: CSNK1A1, PTK7, HNRNPU, NEDD4, CD80, PLCG2, IGFBP7, VWF, ERBB4, ETV6, DNM3, USP25 and MYT1L, NYAP2, TESPA1, OPCML, SCD5, SORBS1, LBR, PRPF4B, PKP1, and SPOCK1. The interactions between them warrant further discussion in terms of biological implications.
Figure 4.
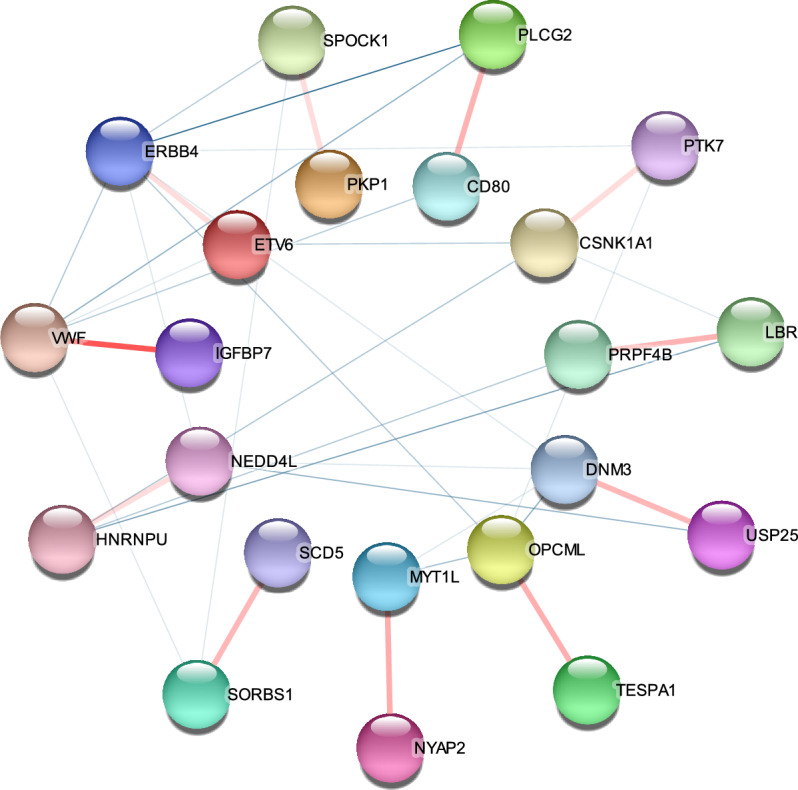
Plot of PPI network with 22 nodes and 11 edges construct by Cytoscape software. The nodes represent the identified genes (protein), and the pink edges represent the overlapped gene–gene interactions.
Discussion
This is a first genome-wide SNP-SNP interaction study using P-tau/A ratio as an AD imaging QT based on linear regression framework. Our study found 961 SNP–SNP interaction pairs to be statistically significant. Additionally, all these interaction pairs had marginal main effects and explained a relatively high-level variance at the P-tau/A ratio level. Two gene set libraries of Enrichr: HDSigDB Human 2021 and PhenGenI Association 2021 showed that 320 genes were reported as AD-related genes after mapping the identified SNPs onto the genome. Further gene set enrichment analyses showed that the significant pathway enrichments are associated with AD. These findings are in line with AD pathology and maybe contributes to partially explain the “missing heritability” of AD. Importantly, our results overlapped 11 pairs of gene–gene interaction with the PPI network, which are CSNK1A1–PTK7, HNRNPU–NEDD4, CD80–PLCG2, IGFBP7–VWF, ERBB4–ETV6, DNM3–USP25, MYT1L–NYAP2, TESPA1–OPCML, SCD5–SORBS1, LBR–PRPF4B, and PKP1–SPOCK1, as shown in Fig. 4. Further analysis of the biological significance was as follows.
Casein kinase I isoform alpha (CSNK1A1) can phosphorylate many proteins and participates in Wnt signaling. PTK7 is a versatile receptor and interact with Wnt signaling pathways. To our knowledge, synapse loss is the prominent event in early stages of AD, and the pathology of AD is amyloid- (A), and total and phosphorylated tau (T-tau and P-tau). Intriguingly, disrupted Wnt signaling may be a direct link between A-toxicity and tau hyperphosphorylation. In parallel, A may trigger the deregulation of Wnt signaling, which could contribute to impaired synaptic plasticity and synapse dysfunction. It suggests that the deregulation of Wnt pathway is associated with AD, and a sustained loss of Wnt signaling function might control AD pathogenesis. Therefore, the CSNK1A1–PTK7 interaction may be related to Wnt signaling pathways and synaptic pathology in AD, ultimately promoting the progression of AD42–46.
As the largest among the HNRNP proteins, HNRNPU has been identified as an RNA binding protein to promote RNA stability and regulate gene transcription. Researchers have noted that MiR-132-3p may alleviates the impairment of learning and memory abilities of AD patients by negatively regulating HNRNPU stabilized BACE147,48.
LncRNA nuclear enriched abundant transcript 1 (NEAT1) interacts with NEDD4L and promote PTEN-induced putative kinase 1 (PINK1)’s degradation, thereby the interaction between them is assumed to be responsible for mitochondria dysfunction and cognitive impairment. Furthermore, mitochondrial dysfunctions are extremely important for the onset and development of AD pathology49. Therefore, HNRNPU–NEDD4L interaction might has effects on the impairment of cognitive and memory abilities in AD.
Although the role of phospholipase C-gamma-2 (PLCG2) in the pathogenesis of AD is only poorly understood, many genetic studies have shown PLCG2 plays an important role in late-onset AD pathophysiology and in the neural immune response. An increasing body of evidence suggest that due to the driving effect of microglia, PLCG2 has higher expression levels in brain regions of late-onset AD patients, and is expressed in human and mouse brain microglia. Moreover, the expression of PLCG2 is maintained in microglia near plaques in the cerebral tissue of an APP mouse model, and is significantly positively correlated with the density of amyloid plaques. These findings suggest that PLCG2 is associated with the inflammatory response and is induced by amyloid plaques in AD50–52. Human immunomodulatory ligand B7-1 (CD80) is a canonical costimulatory molecule, and might mediates synaptic remodeling. Microglia or macrophages lead to the upregulation of CD80, which is related to aging, central nervous system inflammation, injury, and neurodegenerative diseases53. Therefore, the interaction between CD80 and PLCG2 maybe associated with both microglia and inflammatory.
USP25 gene is a critical regulator of AD pathology, and genetic deletion of USP25 reduced amyloid deposition54. DNM3 is in neuronal tissues, and mostly expressed in the brain. DNM3 gene has been previously associated with AD pathology based on an independent proteomic study55,56. Therefore, the interaction between DNM3 and USP25 maybe associated with AD pathology indirectly.
MYT1L is a critical mediator of induced neuron cell reprogramming, which can convert into cholinergic neuronal cells. The cholinergic deficit in the brain is involved in the pathological process of cognitive dysfunction, and may be related to cognitive decline in AD57. As a phosphoprotein family, NYAP is composed of NYAP1, NYAP2, and Myosin16/NYAP3. The NYAPs are expressed predominantly in developing neurons, and controlled remodeling of the actin cytoskeleton58. Previous reports have shown an impairment in actin cytoskeleton dynamics can contribute to AD pathology and underlie the synaptic failure in AD. Indeed, synapse loss is an early event in the process of AD and is associated with cognitive decline59. Therefore, the MYT1L–NYAP2 interaction maybe related to the pathogenesis of AD indirectly, and might associate with both neuronal cells and cognitive decline in AD.
IGFBP7 is a critical regulator of AD-like memory consolidation60. Von Willebrand antigen 2 (VWF) is the von Willebrand factor. It suggests that VWF may contribute to A cerebrovascular deposition, thus associated with pathogenesis of AD indirectly61. Therefore, both IGFBP7 and VWF genes have been implicated in AD.
ERBB4 gene mediates A-induced neuropathology through pathway associated with tau62. ETV6 gene is a regulator of AD, which maybe involved in cellular senescence63.
The stearoylCoA desaturase-5 (SCD5) gene participates in the regulation of neuronal cell growth and differentiation, and may be a molecular link signaling and those neurodegenerative conditions64. The SORBS1 gene is a common gene for cognition changing and has potential function in cognitive impairment. SORBS1 expression has been found to be upregulated in the hippocampus of AD patients65.
OPCML gene associated with specific hippocampal cell types, which including subtypes of neurons and glial cells, and plays an important role in the development of AD-related pathology66. TESPA1 is involved in the Ca2+ transfer and genetic variation in TESPA1 might influence the aging process. Mounting evidence has shown that the accumulation of mitochondrial Ca2+ is significantly related to neuronal apoptosis and other pathways, and enhanced neuronal apoptosis can lead to the neurodegeneration in AD67,68.
The oligomeric tau (oTau) is the key toxic species in tauopathy and can elicit disruption of the nucleocytoplasmic interface which is greatly disrupted in AD. Noteworthy, LaminB2 and lamin B Receptor (LBR) is co-localized with oTau. Thus, it suggests that LBR gene may be related to AD69. The pre-mRNA processing factor 4 homolog B (PRPF4B) is a kinase involved in mRNA splicing. The current findings suggest that Loss of function of homeodomain-interacting protein kinase 2 (HIPK2) has been linked to the development of Alzheimer’s disease, and the homeodomain-interacting protein kinases (HIPKs) reside on a branch together with PRPF4B. However, PRPF4B has not yet been reported as AD-related gene70.
The Kazal-like domains proteoglycan 1 (SPOCK1) endocytic pathway modulates by A precursor protein APP may be related to the accumulation of A and A in the cerebrospinal fluid and brain tissue in patients with AD71. Mitochondrial autophagy or mitophagy is a quality control mechanism that culls defective mitochondria to prevent cellular damage and dysfunction. Mitochondrial dysfunction underlies many age-related human pathologies, such as AD, Parkinson’s disease (PD) and type II diabetes. Although the knockout of both mitochondrial kinases PKP1 gene and PKP2 gene (encoding plakophilin 1 and 2) were found to be related with the mitophagy trafficking, the correlation between PKP1 and AD has not yet been reported72.
Taken together, five pairs of gene–gene interactions showed strong association with AD: CSNK1A1–PTK7, HNRNPU–NEDD4, CD80–PLCG2, IGFBP7–VWF, and ERBB4–ETV6. Among them the CSNK1A1–PTK7 interaction maybe related to the synapse dysfunction and Wnt signaling pathways, the HNRNPU–NEDD4L interaction might has an impact on the impairment of cognitive and memory abilities in AD, and the CD80–PLCG2 interaction maybe associated with both microglia and inflammatory. Two pairs of gene–gene interactions maybe related to the pathogenesis of AD indirectly: DNM3–USP25 and MYT1L–NYAP2. And the other two pairs of gene–gene interactions may be related to hippocampus, neuronal cells and cognitive impairment: TESPA1–OPCML and SCD5–SORBS1. Noteworthy, the possible mechanisms behind LBR–PRPF4B and PKP1–SPOCK1 interactions warrant further investigation.
Materials and methods
Subjects
The phenotyping data and genotyping data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The genotyping data were collected using two arrays of the Illumina 2.5M array and the Illumina OmniQuad array from the ADNI-1, ADNI-GO, and ADNI-2 cohorts. It should be noted that ADNI-1, ADNI-GO, and ADNI-2 are the first three of the four stages contained in ADNI, and the last stage is ADNI-3. The subjects included 1178 individuals, including 639 males and 539 females, and obtained 687,414 SNP after genotyping. In this study, stringent Quality control (QC) was performed using the PLINK v.1.9 software, and SNPs that do not satisfy the criteria of QC will be removed. The QC procedures for genotyping data is mainly composed of eight steps, including call rate check per SNPs, call rate check per subject, the minimum allele frequencies, gender check, pedigree samples identification, Hardy-Weinberg equilibrium test, population stratification, and chromosome removal. Therefore, strict QC procedures were followed:
call rate per SNP ;
call rate check per subject ;
minor allele frequency ;
gender check followed the F values: if the F value is less than 0.2, the sample is judged to be female, and if greater than 0.8, it is predicted to be male;
individuals from the same descent were removed;
Hardy–Weinberg equilibrium test of p ;
population stratification analysis: excluded 89 subjects from the analysis that were non-Hispanic Caucasians;
SNPs on chromosome 1–22.
After QC, 858 valid subjects of CSF P-tau/A ratio remained, and 563,980 SNPs qualified for subsequent genome-wide SNP-SNP interaction analyses. The study cohort comprised 284 normal controls (NC), 459 individuals with mild cognitive impairment (MCI), and 115 AD participants. Table 2 presents the demographic and clinical characteristics of the selected participants at baseline, with detailed data provided in Supplementary Table S2 online. The distribution of the P-tau/A ratio within our study population is shown in Fig. 5.
Table 2.
Demographic and clinical characteristics of 858 ADNI participants at baseline analyzed in this study.
| Diagnosis | NC (N = 284, 33.1%) | MCI (N = 459, 53.4%) | AD (N = 115, 13.4%) |
|---|---|---|---|
| Age (years) | 73.71 (5.80) | 71.85 (7.40) | 74.92 (8.30) |
| Women | 145 (51.0%) | 189 (41.1%) | 43 (37.3%) |
| Education (years) | 16.52 (2.61) | 16.22 (2.76) | 15.70 (2.72) |
| CDR-SOB | 0.05 (0.21) | 1.32 (1.00) | 3.90 (2.14) |
| MMSE | 27.07 (7.35) | 24.88 (8.89) | 20.23 (8.11) |
| CSF A | 199.42 (51.75) | 173.82 (52.59) | 137.27 (35.72) |
| CSF P-tau | 32.8 (16.56) | 39.69 (20.86) | 53.03 (22.97) |
| P-tau/A ratio | 0.19 (0.13) | 0.27 (0.19) | 0.41 (0.21) |
Data are number (%) or mean (s.d.).
AD Alzheimer’s disease, ADNI Alzheimer’s Disease Neuroimaging Initiative, CDR-SOB clinical dementia rating-sum of boxes, CSF cerebrospinal fluid, NC normal controls, MCI mild cognitive impairment, MMSE Mini-Mental Status Examination.
Figure 5.
The distribution plot of P-tau/A ratio after quality control. (a) Quantile–quantile (Q–Q) plot of quantitative trait P-tau/A ratio. (b) The bar plot displays the statistical distribution of the quantitative trait P-tau/A ratio across the number of individuals. (c) The scatter plot shows the distribution of the quantitative trait P-tau/A ratio across the ID of individuals. The red spots represent the ID of Alzheimer’s disease (AD). The blue spots represent the ID of mild cognitive impairment (MCI). The green spots represent the ID of normal controls (NC).
Linear detection model of SNP–SNP interaction
This article uses the multiple linear regression model, a statistical model, for interaction detection. Multiple linear regression is a classical statistical learning method widely applied in modeling and prediction of various real-world problems. The model establishes a linear relationship between multiple independent variables and a dependent variable, and estimates the model parameters using the least squares method (LSM). The multiple linear regression model has strong interpretability and predictive performance. Using a linear model to detect SNP interactions allows for intuitive interpretation of the impact of interactions on phenotypes, providing relevant statistics and significance detection results. Additionally, the linear model can handle issues like interaction and collinearity among multiple independent variables, effectively capturing the influence of SNP–SNP interactions on phenotypes.
Moreover, the parameter estimation methods of linear models are relatively simple, using classic optimization algorithms such as LSM, which are computationally efficient and capable of handling large-scale genetic data. Furthermore, these methods facilitate the interpretation of model parameters and statistics. Taking into account the factors mentioned above, this article utilizes multiple linear regression to detect SNP–SNP interactions.
In this model, the dependent variable is predicted based on a linear combination of the independent variables, each multiplied by its respective coefficient. The data in this article consists of m subjects, each having n SNPs and one quantitative trait phenotype. Let S be the genotype matrix of the SNP array, expressed as:, where any SNP genotype vector expressed as: , where represents the genotype data of the ith individual at the jth SNP locus. Let Y be the measured quantitative trait, represented by an M1 phenotype matrix. The essence of interaction detection is to consider interaction factors based on the main effects of SNPs and calculate the p-values of these interaction factors.
It is worth noting that the SNPSNP model consisted of the same SNP and covariate terms (include age, gender, Dx) as the additive model, while with an additional multiplicative interaction term for the SNPSNP model. Finally, the p-value of the interaction is calculated according to the F statistic.
Functional enrichment analysis and functional annotation
Functional enrichment analysis is a computational method that determines if a group of genes (proteins) is associated with specific biological functions. Annotation analysis assigns functional information to genes (proteins) based on available databases. When combined, these analyses help researchers understand the biological implications of gene or protein sets by identifying related functions and providing detailed annotations. This approach is widely used in biological studies to gain insights into underlying mechanisms and pathways. In this study, genome annotations and analyses were conducted utilizing two distinct annotation tools: Ensembl (http://www.ensembl.org/) and ANNOVAR (https://annovar.openbioinformatics.org/). The results demonstrated a high level of concordance between the findings obtained from both tools, accompanied by comprehensive gene annotation information presented in Supplementary Table S3 online. For SNPs that could not be directly mapped to genes, we performed gene annotation using Homo sapiens genome assembly GRCh37 (hg19) with a distance threshold of 100 kb. This approach enabled the identification of potential regulatory elements and functional associations in close genomic proximity to the SNPs of interest, facilitating the exploration of their potential biological significance. PhenGenI Association 2021 and HDSigDB Human 2021 of Enrichr libraries were used to discuss the correlation between the identified genes and AD. Our intention was to preliminarily assess the association of two genes in a pair of gene–gene interactions with AD at the genetic level using these two databases. We achieved this by searching for whether these genes contained items related to AD. Specifically, if a gene contained items related to AD, we included it in the group of genes reported as being associated with AD. Genes that did not contain items related to AD were placed in the group of genes not yet reported as being associated with AD. In addition, pathway analyses for KEGG and GO were performed on all results with-log10 adjusted p-values. The PPI network plays a crucial role in the prediction of genotype–phenotype associations. As one key role of PPI network, functional annotation can infer the functions of genes and their associated phenotypic features. Therefore, PPI network enrichment analysis was also performed on gene–gene interaction pairs to analyze further their biological implications.
Conclusion
In this paper, we conducted a genome-wide SNP–SNP interaction analysis based on P-tau/A ratio. We used a linear regression model with age, gender, and Dx as covariates. To expedite the search speed for SNP–SNP interactions at the whole-genome level, we employed a parallel computing approach based on multiple GPUs. By using P-tau/A ratio as QT, we increased statistical power and facilitated the identification of SNP–SNP interactions with marginal main effects. We identified 961 SNP–SNP interaction pairs with marginal main effects that contribute to explained a relatively high-level variance of P-tau/A ratio. Notably, a total of 432 previously reported AD-related genes were replicated, and 11 gene–gene interaction pairs were found to overlap with the PPI network. Nine of these gene–gene interaction pairs were potentially related to synaptic dysfunction, Wnt signaling pathway, oligodendrocytes, inflammation, hippocampus, and neuronal cells, which may ultimately lead to cognitive decline in AD. The potential mechanisms underlying the interactions of LBR-PRPF4B and PKP1-SPOCK1 warrant further investigation. Our findings contribute to the partial elucidation of the “missing heritability” of AD. Future research should involve various methods and datasets to validate and further elucidate the biological significance of our results.
Supplementary Information
Acknowledgements
The authors acknowledge the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database.
Author contributions
Q.Z.: Conceptualization, Writing-Original Draf, Formal analysis. J.L.: Software, Methodology. H.L.: Validation, Software. L.A.: Validation. Y.X.: Validation. D.C.: Writing-Review & Editing. All authors revised the manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (grant number: 62206044).
Data availability
The data used in this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI). The database is the (http://adni.loni.usc.edu/, accessed on 1 September 2023) of the Alzheimer’s disease neuroimaging database.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-54541-8.
References
- 1.Pang, C. et al. Identification and analysis of Alzheimer’s candidate genes by an amplitude deviation algorithm. J. Alzheimer’s Dis. Parkinson.9 (2019). [DOI] [PMC free article] [PubMed]
- 2.Abdullah MN, Wah YB, Majeed ABA, Zakaria Y, Shaadan N. Identification of blood-based transcriptomics biomarkers for Alzheimer’s disease using statistical and machine learning classifier. Inform. Med. Unlock. 2022;33:101083. doi: 10.1016/j.imu.2022.101083. [DOI] [Google Scholar]
- 3.Hardy JA, Higgins GA. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe, D. J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. (2001). [DOI] [PubMed]
- 5.Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasqualetti, G., Thayanandan, T. & Edison, P. Influence of genetic and cardiometabolic risk factors in Alzheimer’s disease. Ageing Res. Rev. 101723 (2022). [DOI] [PubMed]
- 8.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordell HJ. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 12.Phillips PC. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann. Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- 14.Moore JH. A global view of epistasis. Nat. Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- 15.Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Zhang Q, Meng F, Li B. Objective-hierarchy based large-scale evolutionary algorithm for improving joint sparsity-compression of neural network. Inf. Sci. 2023;640:119095. doi: 10.1016/j.ins.2023.119095. [DOI] [Google Scholar]
- 17.Abd El Hamid, M. M., Shaheen, M., Omar, Y. M. & Mabrouk, M. S. Discovering epistasis interactions in Alzheimer’s disease using integrated framework of ensemble learning and multifactor dimensionality reduction (MDR). Ain Shams Eng. J.14, 101986 (2023).
- 18.Mackay TF. Epistasis and quantitative traits: Using model organisms to study gene–gene interactions. Nat. Rev. Genet. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, et al. Genome-wide association and interaction studies of CSF T-tau/A42 ratio in ADNI cohort. Neurobiol. Aging. 2017;57:247e1. doi: 10.1016/j.neurobiolaging.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J. et al. Genetic interactions explain variance in cingulate amyloid burden: An AV-45 pet genome-wide association and interaction study in the ADNI cohort. BioMed Res. Int.2015 (2015). [DOI] [PMC free article] [PubMed]
- 21.Visscher PM, et al. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray NR, et al. Research review: Polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- 23.Clark CM, et al. Cerebrospinal fluid tau and -amyloid: How well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch. Neurol. 2003;60:1696–1702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- 24.Kang J-H, et al. Association of cerebrospinal fluid -amyloid 1–42, t-tau, p-tau181, and -synuclein levels with clinical features of drug-naive patients with early parkinson disease. JAMA Neurol. 2013;70:1277–1287. doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korolev IO, Symonds LL, Bozoki AC, Initiative ADN. Predicting progression from mild cognitive impairment to Alzheimer’s dementia using clinical, MRI, and plasma biomarkers via probabilistic pattern classification. PloS one. 2016;11:e0138866. doi: 10.1371/journal.pone.0138866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman M, et al. Cerebrospinal fluid profile in frontotemporal dementia and Alzheimer’s disease. Ann. Neurol. 2005;57:721–729. doi: 10.1002/ana.20477. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Q., Liu, H., Ao, L., Liang, H. & Chen, D. A GPU-based approach for detecting genome-wide SNP–SNP interactions of quantitative trait in ADNI cohorts. In 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2564–2570 (IEEE, 2022).
- 28.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–D592. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, et al. Cell adhesion molecules contribute to Alzheimer’s disease: Multiple pathway analyses of two genome-wide association studies. J. Neurochem. 2012;120:190–198. doi: 10.1111/j.1471-4159.2011.07547.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, et al. Roles and mechanisms of axon-guidance molecules in Alzheimer’s disease. Mol. Neurobiol. 2021;58:3290–3307. doi: 10.1007/s12035-021-02311-2. [DOI] [PubMed] [Google Scholar]
- 33.Inestrosa NC, Toledo EM. The role of wnt signaling in neuronal dysfunction in Alzheimer’s disease. Mol. Neurodegener. 2008;3:1–13. doi: 10.1186/1750-1326-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popugaeva E, Pchitskaya E, Bezprozvanny I. Dysregulation of intracellular calcium signaling in Alzheimer’s disease. Antioxid. Redox Signal. 2018;29:1176–1188. doi: 10.1089/ars.2018.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caltagarone, J., Jing, Z. & Bowser, R. Focal adhesions regulate a signaling and cell death in Alzheimer’s disease. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis.1772, 438–445 (2007). [DOI] [PMC free article] [PubMed]
- 36.Dourlen P, Kilinc D, Malmanche N, Chapuis J, Lambert J-C. The new genetic landscape of Alzheimer’s disease: From amyloid cascade to genetically driven synaptic failure hypothesis? Acta Neuropathol. 2019;138:221–236. doi: 10.1007/s00401-019-02004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, P. J., Ren, Y. & Yan, Z. Epigenomic analysis of Alzheimer’s disease brains reveals diminished CTCF binding on genes involved in synaptic organization. Neurobiol. Dis. 106192 (2023). [DOI] [PMC free article] [PubMed]
- 38.Doglio LE, et al. Retracted: -cleavage-independent functions of presenilin, nicastrin, and aph-1 regulate cell-junction organization and prevent tau toxicity in vivo. Neuron. 2006;50:359–375. doi: 10.1016/j.neuron.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 39.Lee AK, Khaled H, Chofflet N, Takahashi H. Synaptic organizers in Alzheimer’s disease: A classification based on amyloid- sensitivity. Front. Cell. Neurosci. 2020;14:281. doi: 10.3389/fncel.2020.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol D, Maloney B, Long J, Ray B, Lahiri D. Autism, Alzheimer disease, and fragile x: app, fmrp, and mglur5 are molecular links. Neurology. 2011;76:1344–1352. doi: 10.1212/WNL.0b013e3182166dc7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soba P, et al. Homo-and heterodimerization of app family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng Y, et al. Potential biomarkers to detect traumatic brain injury by the profiling of salivary extracellular vesicles. J. Cell. Physiol. 2019;234:14377–14388. doi: 10.1002/jcp.28139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger H, Wodarz A, Borchers A. Ptk7 faces the wnt in development and disease. Front. Cell Dev. Biol. 2017;5:31. doi: 10.3389/fcell.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palomer E, Buechler J, Salinas PC. Wnt signaling deregulation in the aging and Alzheimer’s brain. Front. Cell. Neurosci. 2019;13:227. doi: 10.3389/fncel.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inestrosa NC, Varela-Nallar L. Wnt signaling in the nervous system and in Alzheimer’s disease. J. Mol. Cell Biol. 2014;6:64–74. doi: 10.1093/jmcb/mjt051. [DOI] [PubMed] [Google Scholar]
- 46.Inestrosa NC, Montecinos-Oliva C, Fuenzalida M. Wnt signaling: Role in Alzheimer disease and schizophrenia. J. Neuroimmune Pharmacol. 2012;7:788–807. doi: 10.1007/s11481-012-9417-5. [DOI] [PubMed] [Google Scholar]
- 47.Rooney K, et al. DNA methylation episignature and comparative epigenomic profiling of HNRNPU-related neurodevelopmental disorder. Genet. Med. 2023;25:100871. doi: 10.1016/j.gim.2023.100871. [DOI] [PubMed] [Google Scholar]
- 48.Qu J, et al. MicroRNA-132-3p alleviates neuron apoptosis and impairments of learning and memory abilities in Alzheimer’s disease by downregulation of hnrnpu stabilized bace1. Cell Cycle. 2021;20:2309–2320. doi: 10.1080/15384101.2021.1982507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Zhao J, Wang W, Zhou J, Zhang J. Depletion of lncRNA neat1 rescues mitochondrial dysfunction through nedd4l-dependent pink1 degradation in animal models of Alzheimer’s disease. Front. Cell. Neurosci. 2020;14:28. doi: 10.3389/fncel.2020.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magno L, et al. Alzheimer’s disease phospholipase c-gamma-2 (plcg2) protective variant is a functional hypermorph. Alzheimer’s Res. Ther. 2019;11:1–11. doi: 10.1186/s13195-019-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Der Lee SJ, et al. A nonsynonymous mutation in plcg2 reduces the risk of Alzheimer’s disease, dementia with Lewy bodies and frontotemporal dementia, and increases the likelihood of longevity. Acta Neuropathol. 2019;138:237–250. doi: 10.1007/s00401-019-02026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai AP, et al. Plcg2 is associated with the inflammatory response and is induced by amyloid plaques in Alzheimer’s disease. Genome Med. 2022;14:17. doi: 10.1186/s13073-022-01022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morano, N. C. et al. Human immunomodulatory ligand b7-1 mediates synaptic remodeling via the p75 neurotrophin receptor. J. Clin. Invest.132 (2022). [DOI] [PMC free article] [PubMed]
- 54.Zheng, Q. et al. Usp25 inhibition ameliorates Alzheimer’s pathology through the regulation of app processing and a generation. J. Clin. Invest.132 (2022). [DOI] [PMC free article] [PubMed]
- 55.Puri C, Rubinsztein DC. A location, location, location mutation impairs dnm2-mediated release of nascent autophagosomes from recycling endosomes. Autophagy. 2020;16:1353–1354. doi: 10.1080/15548627.2020.1764210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel H, Dobson RJ, Newhouse SJ. A meta-analysis of Alzheimer’s disease brain transcriptomic data. J. Alzheimer’s Dis. 2019;68:1635–1656. doi: 10.3233/JAD-181085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang X-G, et al. Myt1l induced direct reprogramming of pericytes into cholinergic neurons. CNS Neurosci. Ther. 2018;24:801–809. doi: 10.1111/cns.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoyama K, et al. NYAP: A phosphoprotein family that links pi3k to wave1 signalling in neurons. EMBO J. 2011;30:4739–4754. doi: 10.1038/emboj.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelucchi S, Stringhi R, Marcello E. Dendritic spines in Alzheimer’s disease: How the actin cytoskeleton contributes to synaptic failure. Int. J. Mol. Sci. 2020;21:908. doi: 10.3390/ijms21030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agbemenyah HY, Agis-Balboa RC, Burkhardt S, Delalle I, Fischer A. Insulin growth factor binding protein 7 is a novel target to treat dementia. Neurobiol. Dis. 2014;62:135–143. doi: 10.1016/j.nbd.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Ziliotto N, Bernardi F, Piazza F. Hemostasis components in cerebral amyloid angiopathy and Alzheimer’s disease. Neurol. Sci. 2021;42:3177–3188. doi: 10.1007/s10072-021-05327-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, Zhang L, Zhou D, Li H, Xu Y. Erbb4 mediates amyloid -induced neurotoxicity through jnk/tau pathway activation: Implications for alzheimer’s disease. J. Comp. Neurol. 2021;529:3497–3512. doi: 10.1002/cne.25207. [DOI] [PubMed] [Google Scholar]
- 63.Latorre E, Ostler EL, Faragher RG, Harries LW. Foxo1 and etv6 genes may represent novel regulators of splicing factor expression in cellular senescence. FASEB J. 2019;33:1086–1097. doi: 10.1096/fj.201801154R. [DOI] [PubMed] [Google Scholar]
- 64.Sinner DI, Kim GJ, Henderson GC, Igal RA. Stearoylcoa desaturase-5: A novel regulator of neuronal cell proliferation and differentiation. PloS one. 2012;7:e39787. doi: 10.1371/journal.pone.0039787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starnawska A, et al. Epigenome-wide association study of cognitive functioning in middle-aged monozygotic twins. Front. Aging Neurosci. 2017;9:413. doi: 10.3389/fnagi.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weller, A. E. et al. Single nucleus transcriptome data from Alzheimer’s disease mouse models yield new insight into pathophysiology. J. Alzheimer’s Dis. 1–15 (2022). [DOI] [PubMed]
- 67.Jung J, et al. Alcohol use disorder is associated with DNA methylation-based shortening of telomere length and regulated by tespa1: Implications for aging. Mol. Psychiatry. 2022;27:3875–3884. doi: 10.1038/s41380-022-01624-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsuzaki H, Fujimoto T, Tanaka M, Shirasawa S. Tespa1 is a novel component of mitochondria-associated endoplasmic reticulum membranes and affects mitochondrial calcium flux. Biochem. Biophys. Res. Commun. 2013;433:322–326. doi: 10.1016/j.bbrc.2013.02.099. [DOI] [PubMed] [Google Scholar]
- 69.Jiang L, Wolozin B. Oligomeric tau disrupts nuclear envelope via binding to lamin proteins and lamin b receptor. Alzheimer’s Dement. 2021;17:e054521. [Google Scholar]
- 70.Agnew C, et al. The crystal structure of the protein kinase hipk2 reveals a unique architecture of its cmgc-insert region. J. Biol. Chem. 2019;294:13545–13559. doi: 10.1074/jbc.RA119.009725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye Z, et al. Spock1: A multi-domain proteoglycan at the crossroads of extracellular matrix remodeling and cancer development. Am. J. Cancer Res. 2020;10:3127. [PMC free article] [PubMed] [Google Scholar]
- 72.Kolitsida P, et al. Phosphorylation of mitochondrial matrix proteins regulates their selective mitophagic degradation. Proc. Natl. Acad. Sci. 2019;116:20517–20527. doi: 10.1073/pnas.1901759116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI). The database is the (http://adni.loni.usc.edu/, accessed on 1 September 2023) of the Alzheimer’s disease neuroimaging database.