Abstract
A Coxiella burnetii Hsp70 homologue was identified by using an acid activation in vitro system in which protein synthesis has been followed by [35S]methionine labeling, autoradiography, and immunoblotting. The protein was one of those predominantly labeled, and the immunoblots revealed that it was recognized by anti-DnaK antibodies. The corresponding gene was isolated, and its nucleotide sequence was determined and analyzed. A single open reading frame (ORF) with a size of 1,968 bp was identified. The ORF encodes a protein containing 656 residues and having a molecular weight of 70,800. The −10 promoter sequence was shown to be identical with the consensus heat shock ς32 promoter sequence. The base composition at the presumed −35 region revealed an EcoRI site in the expected region, which is assumed to be located at the border of the cloned fragment. The gene was expressed in Escherichia coli as an intact protein. The C. burnetii 71-kDa protein sequence has a high degree of homology to sequences of the Hsp70 family. A comparison of sequences revealed that the similarity with Hsp70s from other intracellular bacteria, e.g., Legionella pneumophila and Francisella tularensis, as well as E. coli DnaK, is more than 80%. The homologous regions are found in the N-terminal and central parts of the protein sequence, and they include the signature patterns of the Hsp70 family of proteins. The presence of the 71-kDa protein in association with the cell wall as well as in the cytoplasm was demonstrated by the use of immunoelectron microscopy. The dual localization was verified by Western blot analysis of proteins in C. burnetii cell fractions, using purified antibodies directed to the 71-kDa protein.
Information is accumulating on heat shock proteins (Hsp) in pathogenic bacteria (9, 14, 29, 39, 41, 42, 47, 48). Two dominant families of heat shock proteins, Hsp60 and Hsp70, have been described. Hsp have also been extensively studied at the immunological level and are reported to be major targets of both humoral and cell-mediated immune responses (24, 30, 49). Expression of these proteins might result from stress the bacteria encounter during the infectious process, and these proteins have also been suggested to be virulence determinants (7, 23, 28, 30). The proteins have basic functions in the acquisition of the native structure of proteins in the bacterial cell and in the assembly and membrane translocation of proteins (27).
The causative agent of Q fever, Coxiella burnetii, is an obligate intracellular bacterium which replicates in the acidic phagolysosome of the host cell (3). C. burnetii causes a variety of symptoms in its human host, including influenza-like acute fever and chronic infections in the heart and liver (1). Biochemical studies have shown that bacterial metabolic activity at neutral pH is minimal, thus suggesting an in vivo requirement for the acidic conditions of the phagolysosome (20).
It has been shown that naturally released C. burnetii organisms, which were incubated in an acidic in vitro system, were extremely active in protein biosynthesis (41, 51). The acidic in vitro system is thus useful for studies of proteins induced during phagolysosome-like conditions.
C. burnetii has been shown to respond to increased temperature by producing a set of proteins (41). One of these proteins, a 62-kDa common antigen, has previously been described to be a homologue of the GroEL protein of Escherichia coli, which belongs to the Hsp60 family (46). In the present study, we focused on the initially observed, acid-activated C. burnetii proteins. A 71-kDa protein, one among the predominantly labeled proteins, was shown to be a new member of the Hsp70 family. The C. burnetii Hsp71 gene has been isolated and analyzed, and the localization of the protein in the bacterial cell is discussed.
MATERIALS AND METHODS
Strains and growth conditions.
The C. burnetii Nine Mile RSA 493 phase I strain (American isolate, kindly provided by L. Mallavia, Washington State University, Pullman, Wash.) was used in the study. The bacteria were grown in the BGM cell line (Flow Laboratories). The cell growth medium was Eagle’s minimal essential medium (MEM) supplemented with Earle’s salts, 2 mM l-glutamine, 0.2% NaHCO3 (Nordcell, Bromma, Sweden), 5% calf serum, and 1% nonessential amino acids (Sigma). Confluent cell layers were infected with the bacteria and incubated at 37°C. Fresh medium was added after 20 to 24 h. C. burnetii was collected from the media of actively growing cultures after 7 or 8 days by a differential centrifugation method. An initial centrifugation at 4,000 × g for 8 min at 4°C removed cell debris, and a second centrifugation at 25,000 × g for 20 min at 4°C collected the bacteria. The number of bacteria was initially estimated by measuring A565, and the concentration was verified by plaque counting (43).
E. coli XL-Blue MFR′ (Stratagene) was used as the host strain for recombinant plasmids and bacteriophage Lambda ZAP II. E. coli cells were grown in Luria broth with 12.5 μg of tetracycline/ml.
In vitro acid activation and metabolic labeling.
C. burnetii cells were isolated as described above, washed once, and resuspended in buffer A (pH 6.8) (10 mM NaCl, 20 mM KH2PO4, 100 mM KCl, 60 mM glycine, and 250 mM sucrose). The bacteria were preincubated in this buffer for 3 h at room temperature. An acidic milieu was created by adding components similar to those described by Thompson et al. (41). The composition of buffer B (pH 4.5) was 48 mM KH2PO4, 240 mM KCl, 24 mM NaCl, and 48 mM MgCl2, and that of buffer C was 550 mM sucrose, 24 mM glucose, 24 mM glutamate, 176 mM glycine, and 0.21 mM each of the other amino acids. The final reaction mixture was obtained by mixing 100 μl of buffer A with 450 μl of each of buffer B and buffer C. To the reaction mixture, which contained 1.5 × 109 bacteria/ml, 250 μCi of l-[35S]methionine/ml (specific activity, 1,000 Ci/mmol) was added. Samples (100 μl) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were withdrawn at various intervals after a shift to the acidic buffer and/or increased temperature, and the bacteria were harvested by centrifugation (12,000 × g for 45 min at 4°C) and washed once in 1.5 ml of 10 mM Tris-HCl–5 mM MgCl2 (pH 8). The pellets were dissolved in sample buffer (125 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 25% [vol/vol] glycerol, 5% [vol/vol] 2-β-mercaptoethanol), sonicated four times for 15 s each time on ice, and boiled for 5 min.
SDS-PAGE.
Denaturing SDS-PAGE was carried out in a modified Laemmli buffer system (26). The samples were prepared as described above and separated on a uniform 12.5% acrylamide–N-bisacrylamide (37.5:1) gel. The electrophoresis was run at 0.02 mA/cm2 and room temperature for 16 h by using a dual vertical slab electrophoresis cell (Protean II; Bio-Rad). The molecular weight protein standards were Rainbow or Rainbow 14C-methylated standards (Amersham International). The polyacrylamide gel was stained in 0.1% Coomassie brilliant blue R in 45% methanol and 9% acetic acid at 37°C for 30 min and destained for 3 h in 7% methanol and 5% acetic acid.
Bacterial cell fractionation.
Bacterial cells were fractionated by a modification of the method of Osborn et al. (35). The bacteria (approximately 2.5 × 1010) were collected by centrifugation at 25,000 × g for 20 min at 4°C, and the pellet was washed once in phosphate-buffered saline (PBS), resuspended in 100 mM Tris-HCl (pH 8.0)–10 mM EDTA containing 50 μg of lysozyme/ml and incubated for 2 h at 25°C. After sonication on ice 10 times for 15 s each time, the sample was centrifuged at 4,000 × g for 20 min. The supernatant, containing soluble cytoplasmic proteins as well as membrane proteins, was subjected to a centrifugation at 360,000 × g for 2 h at 4°C. The soluble cytoplasmic proteins in the supernatant were precipitated with a 1/10 volume of a saturated solution of trichloroacetic acid on ice for 16 h, collected by centrifugation at 4,000 × g for 20 min, and resuspended in 2% SDS. The membrane protein pellet was washed once in 0.25 M sucrose–3.3 mM Tris-HCl (pH 8)–1 mM EDTA and recentrifuged for another hour at 360,000 × g. This final pellet was dissolved in 0.25% sucrose–5 mM EDTA (pH 7.5) and subjected to a density gradient centrifugation at 200,000 × g for 14 h. The gradient consisted of blocks with 30, 35, 40, 45, and 50% sucrose in 5 mM EDTA. The gradient was fractionated from the bottom, and the A280 of each of the fractions was measured (35).
Two-dimensional (2-D) gel electrophoresis.
Preparation of proteins was as described above for SDS-PAGE. The first electrophoresis (in one dimension) of acid-activated C. burnetii proteins (at 42°C and pH 5.2 for 4 h) and the second electrophoresis were performed as described by Ericsson et al. (15).
Immunoblotting and autoradiography.
The antigens were separated on polyacrylamide gel and transferred to nitrocellulose (NC) membranes in a semidry electroblotter according to the instructions of the manufacturer (Ancos Aps, Stykke, Denmark) but with the Laemmli buffer (26). To reduce nonspecific binding, the membrane was preincubated with 5% (wt/vol) defatted milk in TBS (0.5 M NaCl, 0.01 M Tris-HCl [pH 8.2]) for 2 h. The NC membrane was incubated with serum diluted in TBS containing 2.5% defatted milk. After 2 h at room temperature the membrane was washed five times for 5 min each time in TBS containing 0.5% Tween 20 (vol/vol) and incubated for 1 h with conjugated antibodies (alkaline phosphatase-labeled swine anti-rabbit immunoglobulin antibodies [Dakopatts, Gløstrup, Denmark]) in TBS containing 2.5% defatted milk. After they were washed five times for 5 min each time in TBS containing 0.5% Tween 20 (vol/vol) and three times in rinsing buffer (50 mM Tris-HCl [pH 9.8], 3 mM MgCl2), the bands were visualized by the addition of nitroblue tetrazolium (0.1 mg/ml) and 5-bromo-4-chloro-3-indolylphosphate (0.05 mg/ml) to the rinsing buffer.
Autoradiography was performed by exposure of Enhance-treated, dried polyacrylamide gels to Hyperfilm-MP film (Amersham International) for 2 to 5 days at −80°C.
Immune sera.
Anti-71-kDa-protein-specific antiserum was raised by immunizing rabbits with 100 μg of protein, which was electroeluted from polyacrylamide gel slices and dialyzed. The rabbits were boostered twice with homogenized and dried polyacrylamide gel slices, containing about the same amount of protein, dissolved in 0.9% NaCl.
Anti-71-kDa-protein antibodies were purified from the polyclonal serum by immunoadsorption by using NC membrane spots containing the 71-kDa protein. C. burnetii total proteins were separated by 2-D PAGE and blotted onto an NC membrane, which was stained with Coomassie brilliant blue R. The 71-kDa protein spot was cut out, incubated in a blocking buffer (50 mM Tris [pH 7.0], 0.15 M NaCl, 2.5 mM EDTA, 20% fetal calf serum) for 1 h at room temperature, and then incubated overnight at 4°C in rabbit antiserum diluted 1:10 in the blocking buffer. After being washed twice for 15 s each time in PBS, twice in PBS with 0.1% Triton X-100, three times in PBS, and finally once with distilled water by intensive shaking, the antibodies were eluted with 0.1 M glycine-HCl (pH 2.5) for 2 min. The eluate was neutralized by addition of a 1/10 volume of 1 M Tris-HCl (pH 8.0) and diluted 1:1 in the blocking buffer.
E. coli DnaK-specific rabbit antiserum was kindly provided by M. Zylicz, University of Gdansk, Gdansk, Poland, and rabbit GroEL-specific anti-Legionella micdadei antiserum (5) was kindly provided by P. Hindersson, Statens Seruminstitut, Copenhagen, Denmark.
Amino acid sequencing.
Unlabeled proteins, isolated from stress-exposed bacteria (at 42°C and pH 5.2 for 4 h) were separated by 2-D gel electrophoresis and transferred to a polyvinylidene difluoride transfer membrane (Millipore Corp.). The membrane was stained with Coomassie brilliant blue R, and the spot corresponding to the anti-DnaK-serum-binding protein was excised. The protein was subjected to N-terminal sequencing by the Edman degradation technique (Bo Ek, Biomedical Center, University of Uppsala, Sweden). The sequence was analyzed for homology to known protein sequences in the Swiss protein database (version 32).
Preparation of chromosomal DNA from C. burnetii.
The bacteria were collected as described above, resuspended in a solution containing 50 mM Tris-HCl (pH 7.8), 10 mM MgSO4, and 20 μg of DNase/ml and incubated at 37°C for 30 min. Proteinase K (50 μg/ml) and SDS (0.5%) were added, and the incubation was continued at 56°C for 1 h. Preparation of chromosomal DNA was performed by a modification of the procedure described by Vodkin et al. (44). After the addition of 15% sucrose, 1 mg of lysozyme/ml, 100 mM Tris-HCl (pH 7.8), and 1 mM EDTA, the incubation was continued at 37°C for 16 h. Finally, 100 mM Tris-HCl (pH 12.0), 1 mM EDTA, and 5% SDS were added, and the lysate was incubated for 1 h at 56°C. The lysate was treated once with 1 volume of phenol and once with 1 volume of phenol-chloroform (1:1, vol/vol), and finally the nucleic acid was precipitated with 3 volumes of ice-cold 99.5% ethanol. After 30 min at −20°C, nucleic acid was collected by centrifugation at 16,000 × g for 30 min. The pellet was resolved in TE buffer (10 mM Tris-HCl [pH 7.2], 1 mM EDTA) and treated with 10 μg of RNase/ml at 37°C for 1 h. One volume of phenol was added, and after that the preparation was treated twice with phenol-chloroform (1:1, vol/vol). The DNA was reprecipitated by the addition of a 1/10 volume of 3 M Na-acetate and 3 volumes of 99.5% ethanol, washed twice with 80% ethanol, and finally air dried before it was resolved in TE buffer.
Cloning of the 71-kDa-protein gene.
Random cutting of the C. burnetii chromosomal DNA was achieved with the aid of EcoRI* activity, which was generated by digestion of 2 μg of total DNA with EcoRI (40 U) in a low-ion-concentration reaction buffer (2.5 mM MnCl2, 2.5 mM MgCl2, 10 mM NaCl, 10% glycerol [pH 8.5]) at 37°C. The digestion reactions were terminated after 6, 8, and 16 h, and all three mixtures were pooled. The digested fragments were separated by electrophoresis on a 0.8% agarose gel (SeaKem GTG agarose; FMC Bio Products), and fragments between 2 and 9 kb in length were excised. The purified fragments were ligated to the EcoRI-digested Lambda ZAP II vector and packed by using the Gigapack II extract according to the protocol provided by the manufacturer (Stratagene). Approximately 6,000 individual phage particles of the C. burnetii library were plated. Detection of positive clones with antibodies directed to the 71-kDa protein was performed according to the method described by Mierendorf et al. (32). In vivo excision of the pBluescript vector along with the inserted DNA of each positive clone was done according to the protocol of the supplier of the Lambda ZAP II cloning system (Stratagene).
DNA sequencing.
The nucleotide sequence was determined by the dideoxynucleotide chain-termination method with the Thermo Sequenase-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham International). Universal primers and primers based on the sequenced DNA were synthesized and infrared dye labeled by MWG-Biotech, Ebersberg, Germany (Table 1). The software package GCG was used to assemble and analyze the sequences, and homologies to other proteins or genes were analyzed by using the FASTA and Gap methods of the GCG Wisconsin Sequence Analysis Package, version 8.0, and the GenBank or Swiss protein database.
TABLE 1.
Primers used for sequencing of the C. burnetii 71-kDa-protein gene
| Specificity | Positions | Sequencea |
|---|---|---|
| C. burnetii | 1874–1856 | GAG GAA TGC TCC GTT AGC G |
| 1080–1070 | CGA TTT CAG AGA CTT TAA GAC C | |
| 1271–1252 | GTA ACG TCC AAC AGC AAC AC | |
| 636–619 | CTT GTC CAT CCC GTA AGC | |
| 1435–1419 | CGG AAG CCA TTT CAC GT | |
| 257–275 | CGG ACA GAA CCT TGT ACG C | |
| 619–635 | GCT TAC GGG ATG GAC AA | |
| 1252–1271 | GTG TTG CTG TTG GAC GTT AC | |
| 1539–1556 | GCA CGT ATC GGC GAA AGA | |
| E. coli | 605–622 | (ACGT)GG(ACGT)AC(ACGT)GT(ACGT) AT(ACGT)AC(ACGT)GC |
The direction of the sequences is 5′ to 3′.
Expression of the C. burnetii 71-kDa-protein gene in E. coli.
E. coli cells harboring a plasmid with the Hsp71 gene cloned downstream of the lac promoter (clone 71) were induced by isopropyl-β-d-thiogalactopyranoside (IPTG) as follows. Clone 71 was grown in Luria broth supplemented with 12.5 μg of tetracycline/ml and 50 μg of carbenicillin/ml at 37°C overnight. The culture was diluted 1/10 in the same medium and incubated at 37°C for 2 h. After addition of 200 μM IPTG the incubation was continued for another 3 h. The bacteria were collected by centrifugation at 10,000 × g, resuspended in water, diluted in sample buffer, and subjected to a 12.5% PAGE.
Immunoelectron microscopy.
Bacteria were isolated from continuously infected cultures, washed twice in PBS, fixed for 1 h in 3% formaldehyde in PBS at 4°C, and infused in 20% polyvinylpyrrolidone with 2.3 M sucrose for 3 h. The samples were frozen in liquid N2 and cryosectioned. The sections were placed on a drop of 2.3 M sucrose in PBS and mounted on Formvar-coated nickel grids. The grids were incubated in PBS containing 0.01% blocking reagent (Boehringer Mannheim Biochemica, Mannheim, Germany) for 30 to 60 min, rinsed in the same buffer for 30 min, and incubated for 1 h with the 1:10-diluted primary antibodies, which had been purified from a polyclonal rabbit anti-71-kDa-protein antiserum. The secondary antibodies were collodial gold-conjugated goat anti-rabbit immunoglobulin G antibodies, diluted 1:10. After 1 h of incubation the grids were washed for 15 min in PBS and 5 min in distilled water and embedded in methylcellulose. The immunolabeled sections were examined in a JEOL 100-CX electron microscope.
Nucleotide sequence accession number.
The nucleotide sequence encoding the C. burnetii Hsp71 was submitted to the EMBL Nucleotide Sequence Database and given the accession no. AJ005700.
RESULTS
Proteins synthesized in an acidic environment.
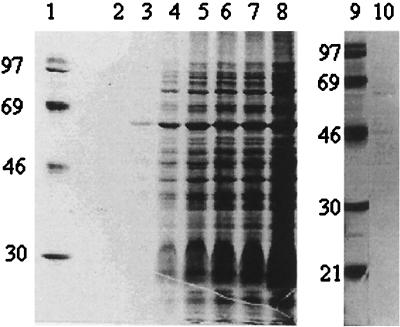
C. burnetii cells were subjected to incubations at 37 and 42°C in an acidic complex buffer (pH 5.2). The synthesis of proteins was followed by [35S]methionine labeling and separation of labeled proteins on polyacrylamide gels. The gels were dried and autoradiographed or transferred to NC membranes for Western blot analysis. During the incubation at 42°C, [35S]methionine was incorporated into about 35 proteins within a 4-h period (Fig. 1, lanes 2 to 8). The first proteins to be labeled, i.e., proteins with apparent molecular weights of approximately 60,000 and 80,000, appeared in approximately 60 min.
FIG. 1.
Autoradiogram of [35S]methionine-labeled proteins after incubation of C. burnetii at 42°C and pH 5.2 in a cell-free system. Samples were withdrawn after 30 (lane 2), 60 (lane 3), 90 (lane 4), 120 (lane 5), 150 (lane 6), 180 (lane 7), and 240 (lane 8) min. Lanes 1 and 9, molecular mass standards (with masses noted in kilodaltons); lane 10, protein of C. burnetii incubated at 42°C and pH 7 for 240 min. Each lane contains proteins corresponding to approximately 107 bacteria.
The protein profile produced by C. burnetii at 37°C and pH 5.2 was almost identical to the one obtained at 42°C but with weaker bands (data not shown). In contrast, there was only a limited protein synthesis observed at 42°C and pH 7, and the weak bands were not identical with those produced at pH 5.2 (Fig. 1, lane 10). This result is in agreement with previous observations by Thompson et al., who showed that the C. burnetii protein synthesis at low pH was optimal at or above 42°C (41).
An Hsp70 homologue.
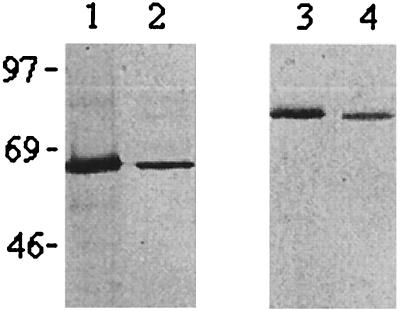
Two of the most well characterized families of Hsp are the Hsp60 and Hsp70 groups. Previously, a 62-kDa protein, a C. burnetii major protein, has been demonstrated to be a member of the Hsp60 family (46). The 60-kDa protein induced in the present experiments was shown by Western blotting with an anti-GroEL antiserum to be the Hsp60 analogue (Fig. 2, lane 2). This band was also observed in immunoblottings of proteins produced by bacteria growing in cell cultures (Fig. 2, lane 1).
FIG. 2.
Western blots of C. burnetii proteins either prepared from bacteria isolated from continuously growing cell cultures (lanes 1 and 3) or produced by bacteria in a cell-free system at 42°C and pH 5.2 for 4 h (lanes 2 and 4). Lanes 1 and 2, anti-GroEL antiserum; lanes 3 and 4, anti-DnaK antiserum. Molecular mass standards (kDa) are marked at the left side of the Western blot.
This finding raised the tempting possibility that the other predominant protein identified among those induced early, the C. burnetii protein migrating as an 80-kDa protein, was a member of the Hsp70 family. A polyclonal antiserum raised against the E. coli DnaK protein was used in immunoblot analysis of the proteins synthesized in the in vitro cell-free system. The antibodies recognized a single band, corresponding to the larger of the predominant proteins in samples from 4-h in vitro incubations (Fig. 2, lane 4). In addition, samples prepared from bacteria grown in cell cultures also contained this protein (Fig. 2, lane 3).
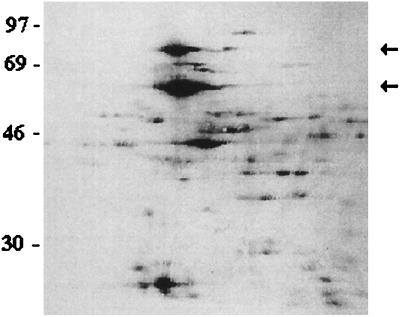
The existence of only single proteins in the predominantly labeled protein bands was verified by a 2-D gel electrophoresis of [35S]methionine-, in vitro-labeled proteins (Fig. 3). Moreover, Western blotting with the anti-DnaK serum demonstrated a single spot at the position for the Hsp70 homologue (data not shown).
FIG. 3.
Autoradiogram of a 2-D gel with [35S]methionine-labeled proteins from in vitro-induced C. burnetii (at 42°C and pH 5.2 for 4 h). The first dimension spans the pH range from 4 to 7, from left to right. The Hsp60 and Hsp70 homologues are indicated by arrows. The molecular mass standards (kDa) are marked at the left side of the autoradiogram.
The protein was excised from a membrane after blotting of a 2-D polyacrylamide gel, and the N-terminal amino acid sequence was determined to be MAEIIGIDLG. This sequence was shown to have a strong homology with the N-terminal amino acids of the E. coli DnaK protein, the most well described member of the Hsp70 family (Fig. 4).
FIG. 4.
Alignment of the deduced amino acid sequence of the C. burnetii Hsp71 gene with four members of the Hsp70 family of proteins. Cb, C. burnetii; Lp, L. pneumophila; Ft, F. tularensis; Ec, E. coli; Cp, C. pneumoniae. Bold letters denote the N-terminal amino acid residues determined for the purified protein. Asterisks denote amino acid residues identical to residues found in the C. burnetii sequence. Shaded areas denote the three PROSITE Hsp70 family signatures.
DNA sequencing and characterization of the Hsp70 homologue gene.
A gene library was constructed using the Lambda ZAP II vector and EcoRI*-digested C. burnetii DNA. Two positive clones were identified in the C. burnetii gene library by using antibodies directed to the purified protein. Phagemids were isolated from these clones. One of the clones was shown to contain the 5′ region of the gene by PCR amplification. The members of the primer pair used were based on the N-terminal amino acid sequence obtained for the C. burnetii protein and the E. coli DnaK sequence positions 605 to 622. The clone was later shown to contain the entire gene on a 2.1-kb insert.
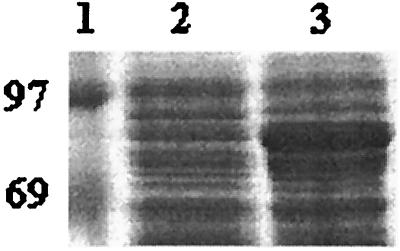
The sequence strategy included the use of an Hsp70 N-terminal primer designed from the determined C. burnetii amino acid sequence, the forward and reverse primers of M13, and sequence primer walking. Sequence analysis identified a single open reading frame with a size of 1,968 bp. The deduced amino acid sequence contains 656 residues, which corresponds to a protein with a molecular weight of 70,800. We will herein refer to this gene as the C. burnetii Hsp71 gene. The presence of initiation and stop codons at positions 70 and 2,037, respectively, together with results for the alignment with Hsp70 sequences, indicated that the cloned fragment encompasses the full-length Hsp71 gene (Fig. 4 and Table 2). Furthermore, the expression of the clone in E. coli revealed the production of a protein with the expected size, which migrated as an 80-kDa protein, as disclosed by SDS-PAGE (Fig. 5).
TABLE 2.
Alignment of heat shock ς32 promoters
| Hsp gene | −35 region | Spacing (bp) | −10 region | Refer-ence |
|---|---|---|---|---|
| Consensusa | CTTGAAA | 12–16 | CCCCAT-T | 18 |
| DnaK, E. coli P1 | CTTGATG | 13 | CCCCATTT | 18 |
| DnaK, E. coli P2 | GTTGAAA | 13 | CCCTATTA | 18 |
| Hsp70, L. pneumophila | CTTGAAA | 13 | CCCCATTT | 2 |
| Hsp70, F. tularensis | CTTGAAA | 13 | GCCCATCT | 50 |
| Hsp70, C. burnetii | ---GAAT | 12 | CCCCATGT | This paper |
| Hsp60, C. burnetii | CTTGAAT | 13 | CCTTATAT | 46 |
The consensus sequence is derived from an alignment of E. coli heat shock promoters.
FIG. 5.
Expression of the C. burnetii 71-kDa-protein gene in E. coli. Lane 1, molecular mass standards (kDa); lane 2, clone 71; and lane 3, IPTG-induced clone 71.
A Shine-Dalgarno sequence was located at position 57, and a −10 promoter sequence identical with the E. coli heat shock ς32 promoter sequence was identified (Table 2). We were not able to find a complete −35 region. However, an alignment of the postulated promoter region with the consensus sequence and postulated promoter regions of DnaK genes from other bacteria indicates that an EcoRI site is located in the −35 region. The spacing between the −10 and −35 regions in E. coli ς32 promoters ranges from 12 to 16 bp and is 13 bp for the DnaK genes of Legionella pneumophila and Francisella tularensis (Table 2). The sequence GAAT, at the 5′ end of the EcoRI fragment, is found 12 bp upstream from the postulated −10 region of the C. burnetii Hsp71 gene and probably constitutes the second half of the −35 region. Thus, the cloned fragment includes only part of this region. The gene for the Hsp60 homologue of C. burnetii has also been shown to contain a heat shock promoter with strong sequence homology to the E. coli consensus promoter (Table 2).
Sequence homologies.
The deduced amino acid sequence of the C. burnetii Hsp71 gene was compared with other Hsp70 sequences. This comparison revealed extensive homologies (Fig. 4). The homologies are found in the N-terminal and central parts of the sequence. The similarities at the amino acid level to the Hsp70s of L. pneumophila, F. tularensis, and E. coli are 86%, 83%, and 80%, respectively. Moreover, the three PROSITE signature patterns (4) for the Hsp70 family of proteins are present in the C. burnetii Hsp71 amino acid sequence (Fig. 4).
Cellular localization.
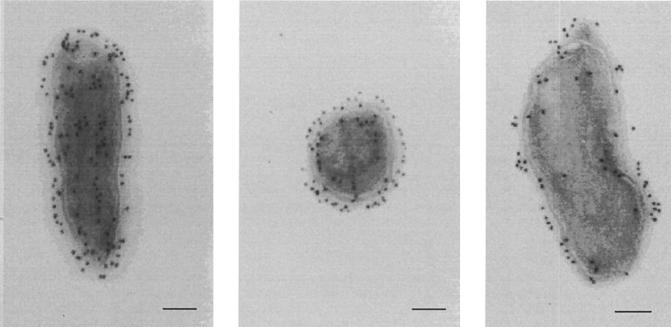
Anti-71-kDa-protein antibodies were used as primary antibodies for immunolabeling of the protein in ultrathin sections of C. burnetii. The gold particles were found inside the bacterial cell as well as in association with the cell wall (Fig. 6). Many gold particles were located in the outermost part of the cell wall. Counting of the gold particles revealed that on average 53.3% (standard deviation, 1.3%) were located in the cytoplasm and 47.3% (standard deviation, 1.6%) were found in association with the cell wall. About 2% of the gold particles were within a comparable area of background.
FIG. 6.
Immunogold staining of C. burnetii cells with primary antibodies directed to the 71-kDa protein and secondary antibodies conjugated to collodial gold. Bars represent 0.1 μm.
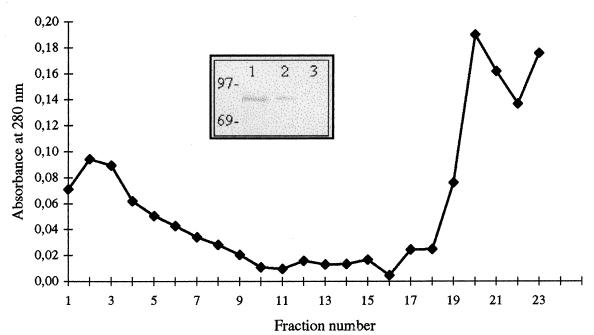
A separation of the membrane proteins was performed by the procedure of Osborn et al. (35) (Fig. 7). Immunoblot analysis revealed that rabbit antibodies recognized a single band in both membrane fractions as well as a faint band in the cytoplasmic fraction (Fig. 7). These results strongly supported a cell wall association in addition to the localization in the cytoplasmic compartment for the C. burnetii 71-kDa Hsp70 homologue.
FIG. 7.
Osborne gradient of C. burnetii membranes and (inset) Western blot of C. burnetii cell fractions with anti-DnaK antibodies. Lane 1, membrane fraction 1 (gradient fractions 1 to 5); lane 2, membrane fraction 2 (gradient fractions 19 to 22); lane 3, cytoplasmic fraction (TCA-precipitated supernatant proteins). The molecular mass standards (kDa) are marked at the left side of the Western blot.
DISCUSSION
We showed that in an acidic environment in vitro—at 37°C or 42°C—C. burnetii induced the expression of about 35 proteins. The smaller of the two predominant proteins was shown to be the 62-kDa Hsp60 homologue previously described by Vodkin and Williams (46). In this study we present evidence that the larger of the predominantly labeled C. burnetii proteins is a new member of the Hsp70 family.
The heat shock response is universal, and the stress proteins are among the most highly conserved genetic elements known. Comparison of the entire Hsp70 sequences of several pathogenic species, such as Borrelia burgdorferi, Brucella ovis, Chlamydia pneumoniae, F. tularensis, and Mycobacterium tuberculosis, have demonstrated an extreme conservation (9, 11, 14, 25, 47). Proteins from the most distantly related species have at least 45% identity, and among all proteins analyzed more than one-sixth of the amino acids are identical in the aligned region (6). In fact, Hsp70 is the most conserved protein known to date and is found in all flora and fauna (19).
Alignment of the deduced amino acid sequence of C. burnetii Hsp71 with published Hsp70 sequences revealed that the protein has a high degree of similarity to members of the Hsp70 family. The homologous regions are found in the N-terminal and central parts of the sequence. Also, the signature patterns of this family of proteins are found in the deduced amino acid sequence. Mutational analysis and proteolysis experiments have shown that bacterial and eucaryotic Hsp70s can be divided into two main functional domains (10, 33). The highly conserved amino-terminal domain is required for ATP binding and ATPase activity, while the more variable C-terminal domain is involved in peptide binding. The C-terminal region is also thought to specify the compartmentalization of the protein in the cell.
Hsp60 and Hsp70 are generally considered to be cytoplasmic proteins in most procaryotes, but there have been reports suggesting that they are membrane associated in some organisms or secreted from the cells (8, 12, 16, 21, 22, 40). The results obtained from immunogold labeling and immunoblot analysis of cell fractions strongly suggest a cell wall association of the C. burnetii Hsp71 in addition to the cytoplasmic localization. Another example of an Hsp with these dual localizations is the C. burnetii Hsp60 homologue (45). Furthermore, the proteins of the Hsp70 family, like the E. coli DnaK protein and the B. burgdorferi Hsp70 protein, are predominantly found in the cytoplasm and have a possible association with the inner membrane. It is, however, possible that these proteins may be associated with the membrane artifactually because of their chaperone functions (8). The hydrophobic binding site of the protein, which facilitates its interaction with hydrophobic regions of other proteins, might also permit it to interact with the hydrophobic regions of molecules such as phospholipids or lipopolysaccharides (38).
The C. burnetii Hsp71 is one of the immunodominant proteins identified in Western blottings with sera from humans naturally infected or vaccinated with C. burnetii (data not shown). This finding is in agreement with various publications, where antibodies to Hsp of several pathogenic species have been reported to be major targets of antibodies (24, 49).
In addition to their immunogenic roles, a multitude of biologic functions have been ascribed to the proteins of the Hsp60 and Hsp70 families. There is, however, limited information concerning the direct interaction between Hsp and the host or about the function of stress proteins in disease pathogenicity. The data presented show that the C. burnetii Hsp71 is one of the proteins induced under early acid-activation protein synthesis, and this may indicate a role in the natural infection. Generally, the Hsp might be expected to be important in the different stages of the infectious process. For example, it has been suggested that Chlamydiaceae DnaK and GroEL are important in the early phases of infection. During the transformation of Chlamydiaceae elementary bodies to reticulate bodies, there is an early and extensive expression of DnaK and GroEL (29). This is an interesting finding given the similarities of the developmental cycles of Chlamydiaceae and C. burnetii, including the conversion from an infectious extracellular form to a metabolically active reproductive form (31, 34).
An example of an Hsp with a defined biological function is a 66-kDa Hsp60 homologue of Salmonella typhimurium, which was shown to be responsible for binding of the bacterium to intestinal mucus (13). In this case the protein is secreted into the supernatant, from which it can be isolated in both dimeric and polymeric forms. Furthermore, Haemophilus ducreyi produces a GroEL Hsp which is associated with the cell surface and has the capacity to bind to eucaryotic cells (17). There are also other examples demonstrating specific effects of bacterial Hsp70s on host cells; purified Hsp from L. pneumophila, E. coli, M. tuberculosis, Mycobacterium leprae, and Mycobacterium bovis were demonstrated to directly induce the production of cytokines in macrophages (36, 37).
In conclusion, we have identified a new member of the Hsp70 family, C. burnetii Hsp71. The protein has a deduced amino acid sequence with strong homology with the Hsp70 family of proteins. The C. burnetii 71-kDa protein is demonstrated to be located in the cytoplasmic compartment as well as in association with the cell wall. The protein is one of the two initially synthesized in bacterial cells in an acidic environment at 37°C, similar to phagolysosomes. This might indicate a role in the infectious process.
ACKNOWLEDGMENTS
We thank B. Ek, BMC, Uppsala, Sweden, who kindly performed the N-terminal amino acid sequencing, and M. Zylicz, University of Gdansk, Gdansk, Poland, and P. Hindersson, Statens Seruminstitut, Copenhagen, Denmark, for providing antisera. We are grateful to A. Norqvist for critically reading the manuscript and to L. Johansson for performing the electron microscopy work.
REFERENCES
- 1.Aitken I D, Bögel K, Cracea E, Edlinger E, Houwers D, Krauss H, Rády M, Rehácek J, Schiefer H G, Schmeer N, Tarasevich I V, Tringali G. Q fever in Europe: current aspects of aetiology, epidemiology, human infection, diagnosis and therapy. Infection. 1987;15:323–327. doi: 10.1007/BF01647731. [DOI] [PubMed] [Google Scholar]
- 2.Amemura-Maekawa J, Watanabe H. Cloning and sequencing of the dnaK and grpE genes of Legionella pneumophila. Gene. 1997;197:165–168. doi: 10.1016/s0378-1119(97)00257-6. [DOI] [PubMed] [Google Scholar]
- 3.Baca O G, Paretsky D. Q fever and Coxiella burnetii: a model for host-parasite interactions. Microbiol Rev. 1983;47:127–149. doi: 10.1128/mr.47.2.127-149.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangsborg J M, Collins M T, Høiby N, Hindersson P. Cloning and expression of the Legionella micdadei “common antigen” in Escherichia coli. APMIS. 1989;97:14–22. [PubMed] [Google Scholar]
- 6.Boorstein W R, Ziegelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 7.Buchmeier N A, Heffron F. Induction of Salmonella stress proteins upon infection of macrophages. Science. 1990;248:730–732. doi: 10.1126/science.1970672. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B, Reilly P, McCarty J, Walker G C. Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J Gen Microbiol. 1993;139:95–99. doi: 10.1099/00221287-139-1-95. [DOI] [PubMed] [Google Scholar]
- 9.Carreiro M M, Laux D C, Nelson D R. Characterization of the heat shock response and identification of heat shock protein antigens of Borrelia burgdorferi. Infect Immun. 1990;58:2186–2191. doi: 10.1128/iai.58.7.2186-2191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cegielska A, Georgopoulos C. Functional domains of the Escherichia coli DnaK heat shock proteins as revealed by mutational analysis. J Biol Chem. 1989;264:21122–21130. [PubMed] [Google Scholar]
- 11.Cellier M F M, Teyssier J, Nicolas M, Liautard J P, Marti J, Sri Widada S. Cloning and characterization of the Brucella ovis heat shock protein DnaK functionally expressed in Escherichia coli. J Bacteriol. 1992;174:8036–8042. doi: 10.1128/jb.174.24.8036-8042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Yaagoubi A, Kohiyama M, Richarme G. Localization of DnaK (chaperone 70) from Escherichia coli in an osmotic-shock-sensitive compartment of the cytoplasm. J Bacteriol. 1994;176:7074–7078. doi: 10.1128/jb.176.22.7074-7078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensgraber M, Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacteria to intestinal mucus. Infect Immun. 1992;60:3072–3078. doi: 10.1128/iai.60.8.3072-3078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ericsson M, Tärnvik A, Kouppa K, Sandström G, Sjöstedt A. Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect Immun. 1994;62:178–183. doi: 10.1128/iai.62.1.178-183.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericsson M, Golovliov I, Sandström G, Tärnvik A, Sjöstedt A. Characterization of the nucleotide sequence of the groE operon encoding heat shock proteins chaperone-60 and -10 of Francisella tularensis and determination of the T-cell response to the proteins in individuals vaccinated with F. tularensis. Infect Immun. 1997;65:1824–1829. doi: 10.1128/iai.65.5.1824-1829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans D J, Jr, Evans D G, Engstrand L, Graham D Y. Urease-associated heat shock protein of Helicobacter pylori. Infect Immun. 1992;60:2125–2127. doi: 10.1128/iai.60.5.2125-2127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisk A, Ison C A, Lagergård T. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect Immun. 1998;66:1252–1257. doi: 10.1128/iai.66.3.1252-1257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 19.Gupta R S, Golding G B. Evolution of hsp70 gene and its implications regarding relationships between archaeobacteria, eubacteria and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 20.Hackstadt T, Williams J C. Biochemical stratagem for obligate parasitism of eucaryotic cells by Coxiella burnetii. Proc Natl Acad Sci USA. 1981;78:3240–3244. doi: 10.1073/pnas.78.5.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman P S, Houston L, Butler C A. Legionella pneumophila htpAB heat shock operon: nucleotide sequence and expression of the 60-kilodalton antigen in L. pneumophila-infected HeLa cells. Infect Immun. 1990;58:3380–3387. doi: 10.1128/iai.58.10.3380-3387.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarjour W, Tsai V, Woods V, Welch W, Pierce W, Shaw M, Mehta H, Dillman W, Zvaifler N, Winfield J. Cell surface expression of heat shock proteins. Arthritis Rheum. 1989;34:S44. [Google Scholar]
- 23.Johnson K, Charles I, Dougan G, Pickard D, O’Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann S H E. Heat shock proteins and the immune response. Immunol Today. 1990;11:129–136. doi: 10.1016/0167-5699(90)90050-j. [DOI] [PubMed] [Google Scholar]
- 25.Kornach J M, Kuo C, Campbell L A. Sequence analysis of the gene encoding the Chlamydia pneumoniae DnaK protein homolog. Infect Immun. 1991;59:721–725. doi: 10.1128/iai.59.2.721-725.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Langer T, Neupert W. Heat shock proteins hsp60 and hsp70: their roles in folding, assembly and membrane translocation of proteins. Curr Top Microbiol Immunol. 1991;167:3–30. doi: 10.1007/978-3-642-75875-1_1. [DOI] [PubMed] [Google Scholar]
- 28.Lathigra R B, Buthcher P D, Garbe T R, Young D B. Heat shock proteins as virulence factors of pathogens. Curr Top Microbiol Immunol. 1991;167:125–143. doi: 10.1007/978-3-642-75875-1_8. [DOI] [PubMed] [Google Scholar]
- 29.Lundemose A G, Birkelund S, Mose Larsen P, Fey S J, Christiansen G. Characterization and identification of early proteins in Chlamydia trachomatis serovar L2 by two-dimensional gel electrophoresis. Infect Immun. 1990;58:2478–2486. doi: 10.1128/iai.58.8.2478-2486.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lydyard P M, van Eden W. Heat shock proteins: immunity and immunopathology. Immunol Today. 1990;11:228–229. doi: 10.1016/0167-5699(90)90091-m. [DOI] [PubMed] [Google Scholar]
- 31.McCaul T F, Williams J C. Developmental cycle and Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J Bacteriol. 1981;147:1063–1076. doi: 10.1128/jb.147.3.1063-1076.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mierendorf R C, Percy C, Young R A. Gene isolation by screening λgt11 libraries with antibodies. Methods Enzymol. 1987;152:458–469. doi: 10.1016/0076-6879(87)52054-7. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto R I, Milarski K L. Expression and function of vertebrate Hsp70 genes. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. pp. 323–359. [Google Scholar]
- 34.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 36.Peetermans W E, Ilse Raats C J, van Furth R, Langermans J A M. Mycobacterial 65-kilodalton heat shock protein induces tumor necrosis factor alpha and interleukin 6, reactive nitrogen intermediates, and toxoplasmastatic activity in murine peritoneal macrophages. Infect Immun. 1995;63:3454–3458. doi: 10.1128/iai.63.9.3454-3458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Retzlaff C, Yamamoto Y, Hoffman P S, Friedman H, Klein T W. Bacterial heat shock proteins directly induce cytokine mRNA and interleukin-1 secretion in macrophage cultures. Infect Immun. 1994;62:5689–5693. doi: 10.1128/iai.62.12.5689-5693.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickarme G, Kohiyma M. Specificity of the E. coli chaperone DnaK for hydrophobic amino acids. J Biol Chem. 1993;268:26074–26077. [PubMed] [Google Scholar]
- 39.Schlesinger M J. Heat shock proteins. J Biol Chem. 1990;265:12111–12114. [PubMed] [Google Scholar]
- 40.Scorpio A, Johnson P, Laquerre A, Nelson D R. Subcellular localization and chaperone activities of Borrelia burgdorferi Hsp60 and Hsp70. J Bacteriol. 1994;176:6449–6456. doi: 10.1128/jb.176.21.6449-6456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson H A, Bolt C R, Hoover T, Williams J C. Induction of heat-shock proteins in Coxiella burnetii. Ann N Y Acad Sci. 1990;590:127–135. doi: 10.1111/j.1749-6632.1990.tb42215.x. [DOI] [PubMed] [Google Scholar]
- 42.Tilly K, Hansen R, Campbell J, Postheimer G J. Isolation of dnaJ, dnaK, and grpE homologues from Borrelia burgdorferi and complementation of E. coli mutants. Mol Microbiol. 1993;170:1227–1234. doi: 10.1111/j.1365-2958.1993.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 43.Tujulin E, Macellaro A, Lilliehöök B, Norlander L. Effect of endocytosis inhibitors on Coxiella burnetii interaction with host cells. Acta Virol. 1998;42:125–131. [PubMed] [Google Scholar]
- 44.Vodkin M H, Williams J C, Stephenson E H. Genetic heterogeneity among isolates of Coxiella burnetii. J Gen Microbiol. 1986;132:455–463. doi: 10.1099/00221287-132-2-455. [DOI] [PubMed] [Google Scholar]
- 45.Vodkin M H, Bolt C R, Williams J C. Cloning and expression of a gene of Coxiella burnetii encoding a major antigen homologous to a protein in Mycobacterium and Escherichia coli. In: Ginsberg H, Brown F, Lerner R A, Chanock R M, editors. Vaccines 88: new chemical and genetic approaches to vaccines. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 111–115. [Google Scholar]
- 46.Vodkin M H, Williams J C. A heat shock operon in Coxiella burnetii produces a major antigen homologous to a protein in both mycobacteria and Escherichia coli. J Bacteriol. 1988;170:1227–1234. doi: 10.1128/jb.170.3.1227-1234.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young D B, Garbe T R. Heat shock proteins and antigens of Mycobacterium tuberculosis. Infect Immun. 1991;59:3086–3093. doi: 10.1128/iai.59.9.3086-3093.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young D B, Mehlert A. Stress proteins in infectious diseases. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. pp. 131–165. [Google Scholar]
- 49.Young R A, Elliott T J. Stress proteins, infection and immune surveillance. Cell. 1989;59:5–8. doi: 10.1016/0092-8674(89)90861-1. [DOI] [PubMed] [Google Scholar]
- 50.Zuber M, Hoover T A, Dertzbaugh M T, Court D L. Analysis of the dnaK molecular chaperone system of Francisella tularensis. Gene. 1995;164:149–152. doi: 10.1016/0378-1119(95)00489-s. [DOI] [PubMed] [Google Scholar]
- 51.Zuerner R L, Thompson H A. Protein synthesis by intact Coxiella burnetii cells. J Bacteriol. 1983;156:186–191. doi: 10.1128/jb.156.1.186-191.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]