Abstract
Genetic testing has undergone a revolution in the last decade, particularly with the advent of next-generation sequencing and its associated reductions in costs and increases in efficiencies. The Undiagnosed Diseases Network (UDN) has been a leader in the application of such genomic testing for rare disease diagnosis. This review discusses the current state of genomic testing performed within the UDN, with a focus on the strengths and limitations of whole-exome and whole-genome sequencing in clinical diagnostics and the importance of ongoing data reanalysis. The role of emerging technologies such as RNA and long-read sequencing to further improve diagnostic rates in the UDN is also described. This review concludes with a discussion of the challenges faced in insurance coverage of comprehensive genomic testing as well as the opportunities for a larger role of testing in clinical medicine.
Keywords: genetic testing, whole-exome sequencing, whole-genome sequencing, RNA sequencing, long-read sequencing, insurance coverage
INTRODUCTION
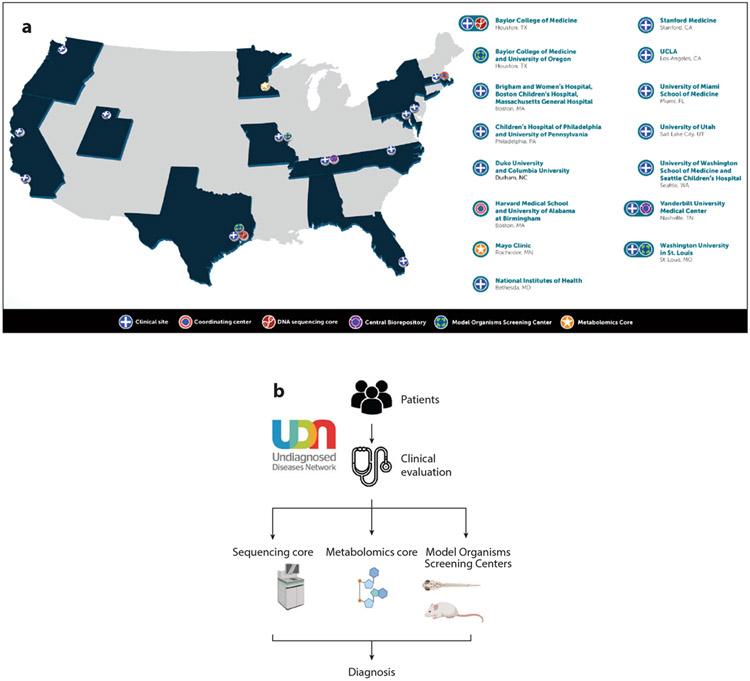
Established in 2014, the Undiagnosed Diseases Network (UDN), funded by the National Institutes of Health (NIH) Common Fund, is a multicenter collaborative effort that takes a multi-omic approach to inform deep clinical phenotyping to solve complex medical cases (1). The UDN was an extension of the Undiagnosed Diseases Program established in 2008 within the Intramural Research Program of the NIH (2, 3). The UDN incorporates state-of-the-art testing, including whole-exome sequencing (WES), chromosomal microarray (CMA), metabolomics, and model organism screening, as part of clinical research evaluations to arrive at a diagnosis (Figure 1). The overall diagnostic rate within the UDN is ~35% (4), which is notable since many patients have already undergone extensive negative diagnostic workups, including with WES and CMA. The UDN has also led the way in implementing emerging technologies such as whole-genome sequencing (WGS), RNA sequencing (RNA-seq), and long-read sequencing to improve diagnostic rates of unsolved cases.
Figure 1.
(a) Undiagnosed Diseases Network (UDN) composition of clinical sites and cores performing sequencing, metabolomics, and model organism screening. (b) Overall UDN workflow from patient enrollment to clinical evaluation through analysis of sequencing and other data to arrive at a diagnosis. Panel (a) taken as a screen shot from https://undiagnosed.hms.harvard.edu/udn-sites/.
Clinical genetic testing has evolved rapidly from Sanger-based single-gene tests to multiple-gene panels to WGS. A major revolution in genetic testing occurred around 2008 with the advent of next-generation sequencing (NGS) that dramatically decreased the cost of such testing and revolutionized the field of molecular genetics (5). The primary provider of this technology is currently Illumina, and the approach of NGS is to generate millions of short reads of DNA, typically around 100–200 base pairs in length, that are then aligned to the human reference sequence. Before NGS, most genetic testing was based on Sanger sequencing, which was more expensive and slower than NGS and not suited for high-throughput applications (6). The Human Genome Project, for example, used Sanger sequencing to generate the human reference genome at a cost of $3 billion over 13 years (7). In contrast, the cost (excluding interpretation and computational processing) of today’s sequencing per human genome is less than $1,000 and can be achieved in less than a day (8). However, it is important to note that NGS would not be possible today without the efforts of the Human Genome Project that generated the original human reference sequence.
The application and utility of genetic testing are well established in pediatrics and continue to expand in adults as precision medicine efforts aim to shift care from a reactive to a preventative model (9). For example, the American College of Medical Genetics and Genomics (ACMG) has published evidence of improved outcomes from genomic testing in pediatric patients with congenital anomalies or intellectual disability (10). Similarly, in 2018, the Heart Failure Society of America and ACMG published guidelines recommending genetic testing for adult patients with cardiomyopathies to assist in management (11). Large-scale NIH-funded programs such as Electronic Medical Records and Genomics (eMERGE) and All of Us aim in part to return actionable clinical genetic findings to improve health outcomes (12, 13). Our experience in the UDN has provided important lessons about the roles and applications of genomic testing in clinical medicine in both children and adults.
LIMITATIONS OF WHOLE-EXOME SEQUENCING AND ADVANTAGES OF WHOLE-GENOME SEQUENCING
Widespread adoption of WES in clinical practice has improved diagnosis for rare Mendelian conditions; however, studies generally report diagnostic rates of 25–30% using WES alone (14-16). The UDN has been a leader in implementing WGS for rare disease diagnosis. WGS is becoming a first-tier diagnostic test for individuals with suspected genetic conditions because it can detect most types of variants in a single test (Table 1), enabling more rapid and less costly diagnoses (17-20). It can detect single-nucleotide variants, small insertion/deletions, copy-number variants, balanced and complex structural variation, many repeat expansions, and mitochondrial variants (17, 21, 22). Also, WGS is a genome-wide screen covering all coding and noncoding regions, whereas WES targets only the 1% coding region of the genome and NGS gene panels test specific genes. As such, WGS probes not only exonic regions but also intronic, intergenic, and regulatory regions that have been shown to cause disease and are frequently missed with other testing modalities (23). One limitation is of course our ability to interpret the pathogenicity of this abundance of data.
Table 1.
Comparison of reportable variant types detected by different genetic testing assays
| Genetic test |
SNVs/Indels | CNVs | Mosaic variants |
Repeat expansions |
Balanced and complex SVs |
mtDNA | Genes tested |
VUS potentiala |
Costa |
|---|---|---|---|---|---|---|---|---|---|
| Sanger | Yes | No | Limited | No | No | If included | Single–few | + | $$ |
| CMA | No | Yes | Yes (CNV only) | No | No | No | Up to ~20,000 | ++ | $$ |
| NGS panel | Yes | Yes | Yes | No | No | If included | Few–hundreds | ++ | $ |
| WES | Yes | Limited | Limited (depends on coverage) | No | No | If included | ~20,000 (coding regions only) | +++ | $$$ |
| WGS | Yes | Yes | Limited (depends on coverage) | Yes | Yes | Yes | ~20,000 (coding and noncoding regions) | ++++ | $$$$ |
VUS potential and cost are compared on a scale from one symbol (lowest) to four (highest).
Abbreviations: CMA, chromosomal microarray; CNV, copy-number variation; Indel, small (<150 base pair) insertion/deletion; mtDNA, mitochondrial DNA; NGS, next-generation sequencing; SNV, single-nucleotide variant; SV, structural variant; VUS, variant of uncertain significance; WES, whole-exome sequencing; WGS, whole-genome sequencing.
One UDN clinical site recently reported that 28% of its diagnoses required testing beyond WES. The vast majority had a causative noncoding variant or copy number variant that had been missed on prior testing (24). In fact, of new diagnoses from July 2015 to September 2019 across four clinical UDN sites, WGS alone was responsible for 45% of the diagnoses in cases with prior nondiagnostic WES (25). One downside of WGS is its increased cost compared to multigene panels and WES, although this is already rapidly decreasing with new technologies. However, because WGS can effectively replace serial genetic testing with a single assay, it has been shown to lead to faster diagnoses, improved outcomes, and decreased cost of care (19, 26). Another drawback of WGS is the increased number of variants of uncertain significance it identifies. This is because of our limited ability to interpret noncoding variation with current ACMG guidelines that focus primarily on coding changes (23, 27). However, techniques such as RNA-seq and high-throughput functional assays may help overcome this issue and bring WGS into mainstream practice. With the continued fall of sequencing costs, clinical WGS will likely replace WES and negate the need for multiple testing modalities such as CMA and mitochondrial DNA sequencing in the coming years.
ROLE OF RNA SEQUENCING IN RARE DISEASE DIAGNOSIS
RNA or transcriptome sequencing (RNA-seq) is emerging as another tool in the genetic diagnostic toolbox. Though RNA-seq is available in a limited manner for targeted clinical applications (28) and in the analysis of somatic fusion events in diagnosing cancer genomic changes, the UDN has pioneered its use for rare disease diagnosis across varied phenotypes and indications (23, 29, 30). In contrast to DNA sequencing, RNA-seq can provide evidence of the functional impact of noncoding variants on splicing, decreased gene expression via nonsense-mediated decay, allele-specific expression, and other findings (31). In the case of splice-altering variants, for example, bioinformatic predictions have mixed effectiveness (32), but RNA-seq can be used to support or refute the effects of a variant on gene splicing (23, 33). Overall, RNA-seq has led to a 7.5–36% improvement in diagnostic rates depending on the sampled tissue and clinical phenotype (23, 29-31, 34, 35) (Table 2) and aids in the resolution of variants of uncertain significance (28, 33). Among four clinical UDN sites specifically, RNA-seq contributed to a diagnosis in 16% of newly diagnosed cases (25).
Table 2.
Comparison of published RNA-seq studies and their contrasting approaches and results
| Citation | UDN study | Phenotypes | Tissue | Events detected |
Analysis methoda |
RNA-seq diagnostic rate |
|---|---|---|---|---|---|---|
| 35 | No | Neuromuscular | Muscle | Splicing | Candidate + Outlier | 35% |
| 34 | No | Neuromuscular | Muscle, fibroblasts, fibroblast-derived myotubes | Expression, splicing, ASE | Outlier | 36% |
| 29 | Yes | Multiple (neurologic most common) | Whole blood | Expression, splicing, ASE | Outlier | 7.5% |
| 30 | Yes | Multiple (neurologic most common) | Whole blood, fibroblasts, muscle, bone marrow | Expression, splicing, ASE | Candidate | 18% |
| 23 | Yes | Multiple (neurologic most common) | Whole blood, fibroblasts | Expression, splicing, ASE | Outlier | 17% |
The candidate method relies on variants first identified on primary WES/WGS to direct RNA-seq analysis. In contrast, the outlier technique identifies novel abnormalities in expression and splicing in a large cohort to direct genomic analysis.
Abbreviation: ASE, allele-specific expression.
A critical issue to consider with RNA-seq is what tissue to use, since gene expression varies widely with tissue type. While whole blood RNA is the easiest to collect, the expression of genes associated with most phenotypes is poor (23, 34), limiting the utility of RNA-seq to contribute to a diagnosis. At the Baylor College of Medicine (BCM) UDN clinical site, we perform RNA-seq on cultured fibroblasts obtained with a skin biopsy because studies have shown much higher expression of a broader number of genes in skin than in whole blood (23, 36). Using fibroblast-derived RNA-seq, we have increased diagnostic rates by 17%, in contrast with the 7.5% reported at another UDN site using blood transcriptome sequencing alone (29). Many genes are still not well expressed in clinically accessible tissues (36); however, techniques such as fibroblast trans-differentiation and/or use of induced pluripotent stem cell-derived target cells may overcome this limitation (34).
The method of RNA-seq analysis is another issue, specifically whether to pursue a traditional candidate variant approach or a transcriptome-directed strategy. The former is an extension of standard practice where candidate variants first identified on WES, for example, are manually reviewed in the transcriptome to determine any functional consequences. In contrast, a transcriptome-directed strategy uses outliers of expression or splicing in the RNA-seq itself to guide subsequent analysis of sequencing data and thereby find the underlying cause. At BCM, the RNA-seq directed method has proven to be more effective than a traditional candidate approach, revealing disease-causing variants commonly missed on WES and CMA and being a powerful tool for novel gene discovery (23). A comparison of the previously published studies, including tissues, analysis method, and diagnostic rates, is shown in Table 2.
RNA-seq shows promise to further improve diagnostic rates for rare diseases, in particular as a complement to WGS in WES/CMA-negative cases. Before RNA-seq becomes a widely accepted clinical test, standardization of techniques and interpretation criteria are needed, in the same way that the ACMG and the Association for Molecular Pathology (AMP) developed guidelines for DNA variants (27).
NEED FOR ONGOING REANALYSIS
Of the roughly 20,000 genes in the human genome, only ~4,400 are currently associated with a Mendelian phenotype in the Online Mendelian Inheritance in Man (OMIM) database (37). However, gene discovery continues rapidly, with ~250 new gene–disease associations entered annually in OMIM (38, 39). One major WES laboratory achieved a 22% improvement in diagnostic rate by reanalysis of existing sequencing data that was overwhelmingly driven by newly implicated disease genes (40). Bioinformatic tools continue to evolve, allowing for improved identification of pathogenic copy-number changes from NGS data (41). Adaptations to the 2015 ACMG and AMP guidelines for variant interpretation (27) can also yield improvement in variant classification over time. For example, recent gene-specific interpretation guidelines permit upgraded variant interpretation for genes like MYH7 (42). Reanalysis of data with consideration of alternative transcripts has also been shown to increase diagnostic rates and resolve uncertain results (43). For these reasons, most diagnostic laboratories now offer periodic WES reanalysis, a practice endorsed by the ACMG (44). One UDN site recently reported 23% of individuals receiving a diagnosis after reanalysis of nondiagnostic WES done before UDN enrollment (45). At our own BCM clinical UDN site, we perform such reevaluation of pre-UDN WES data on a routine basis for these reasons before considering alternative testing, such as WGS or RNA-seq, with similar yields.
EMERGING TECHNOLOGIES: LONG-READ SEQUENCING
Sequencing technology continues to evolve, offering hope for further improvement in diagnostic rates and our understanding of genetic diseases. As impactful as NGS has been, it has several limitations resulting from the short (100–200 base pair) reads upon which it is based. For example, NGS has difficulty in genomic regions with high homology (e.g., pseudogenes), repetitive sequences, and high GC content (46, 47). Complex structural rearrangements are also challenging to detect and characterize with NGS; it has been shown to have low sensitivity (30–70%) and a high false discovery rate (as high as 85%) for these variant types (48). Particularly important for autosomal recessive conditions, phasing of heterozygous variants (cis or trans) is generally not possible with NGS without parental information (49).
In contrast, long-read sequencing from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (Nanopore) generates reads from 10 kb to >1Mb in length (48), overcoming many of the shortcomings inherent to NGS. Long-read sequencing has already been applied to rare disease research, where it has been particularly adept with complex structural rearrangements (46, 48). For example, long reads were recently used to discover a pathogenic GGC repeat expansion missed by NGS in the NOTCH2NLC gene associated with neuronal intranuclear inclusion disease (50). Unlike short reads, long reads can span the entire expanded repeat, making detection more straightforward (46). We have begun using Nanopore sequencing at our UDN clinical site, in one case diagnosing a patient with NEMO deleted exon 5 autoinflammatory syndrome (NEMO-NDAS) (51) by identifying a pathogenic Inhibitor Of Nuclear Factor Kappa-B Kinase, Regulatory Subunit Gamma (IKBKG) variant missed on WES due to a nearby pseudogene. Despite the advantages of long-read sequencing compared to NGS, its higher cost and base calling error rates (46, 52) will need to improve before it is routinely used in the clinical space.
INSURANCE COVERAGE FOR COMPREHENSIVE GENETIC TESTING
Despite the well-documented utility and cost-effectiveness of genetic testing in the diagnosis and management of patients with rare diseases (53-55), insurance coverage for comprehensive molecular tests remains an obstacle. A retrospective analysis of patients enrolled in the UDN revealed that 40% of patients undergoing WES through the UDN had previously faced insurance coverage barriers to testing (56). Among those previously denied, WES yielded a diagnosis in 35% of these patients, resulting in management changes in a majority of them (61%) (56). Nevertheless, a frequent reason for insurance denial is that such testing is considered experimental or investigational and thus is not accepted practice (56, 57). Coverage for WES in adults is particularly low, despite the high diagnostic yield in patients with moderate–severe intellectual disability (58).
Even fewer insurers cover WGS at this time, with many considering it investigational. Payer opinions may evolve as studies demonstrate not only improved diagnostic rates with WGS but also reduced healthcare costs (59). In a pilot study carried out in California from 2018 to 2020, rapid WGS in 178 critically ill babies led to a diagnosis in 43%, management changes in 31%, and $2.5 million in healthcare savings (26). Cost savings were driven by fewer days hospitalized, fewer major surgeries and invasive procedures, and avoided diagnostic tests such as CMA (26). As a result of these findings, Blue Shield of California now covers the cost of rapid WGS in critically ill children (60). Similar pilot studies in other states are currently under way or are in development.
Nevertheless, a more rigorous economic analysis of genomic sequencing is needed to direct reimbursement policy and measure the cost-effectiveness of such testing. Such economic studies have been lacking and are challenging to do (61, 62). In a recent review of 171 articles describing the use of comprehensive genomic sequencing in infants and children, only 4 had a primary economic evaluation aim (55). One report on infants in neonatal intensive care units who were suspected to have a genetic diagnosis surprisingly found higher costs for patients who underwent WES than for those who did not when a detailed economic analysis was done (63). In addition, the perceived value of these comprehensive assays for clinicians and families will be important to measure (63-65). Collectively, such analyses will establish a framework from which to demonstrate clinical utility and cost-effectiveness of genomic sequencing. With the development of much less costly sequencing platforms, the fundamental determinants of cost-effectiveness will further help to overcome payer resistance for covering WGS.
CONTINUED CLINICAL PHENOTYPING
Detailed clinical phenotyping remains an integral component of the diagnostic process. In some cases, a molecular diagnosis is not needed if a patient is found to meet clinical diagnostic criteria for a specific disorder after deep phenotyping. Within the UDN, Shashi et al. recently diagnosed individuals with oral-facial-digital syndrome and multiple pterygium syndrome based on clinical features in the absence of a molecular diagnosis (45). Deep phenotyping may also suggest performing an alternative test if no causative variants are found on the initial assay. One UDN clinical site recently reported three such patients who were negative on WES but were diagnosed after targeted Sanger sequencing and CNV analysis revealed the causative variants (66). In those cases, detailed phenotyping prompted the sequencing of specific genes that were felt to be a strong fit for the clinical features. The disease-causing variants were missed on WES due to the inherent limitations of the technology. Phenotyping can also guide diagnosis by leading to the reinterpretation of variants of uncertain significance (45). Automated filtering algorithms may exclude or deprioritize certain variants from further analysis if they miss specific thresholds (e.g., in silico predictions, population frequency). Detailed phenotyping and the subsequent generation of an accurate differential diagnosis may rescue such uncertain variants and trigger additional investigations. Ultimately, phenotyping informs the generation of candidate gene lists that may lead to downstream functional studies, gene matching, and other studies that may enable the conversion of a variant of uncertain significance to a pathogenic variant. We have also previously demonstrated that nearly 5% of cases carry multiple molecular diagnoses (67). Broad phenotyping in such cases is critical in identifying features of each diagnosis that may erroneously be attributed to a single cause and lead to an incomplete molecular diagnosis.
CONCLUSION
Genetic testing technology continues to evolve with the goal of improving diagnostic rates for rare diseases. As sequencing costs fall and our understanding of the entire genome increases, the role of such testing in clinical medicine will continue to grow. The UDN has led the field in its application of current and emerging technologies for diagnostic purposes and a better understanding of disease mechanisms. Increasing access to comprehensive genomic testing outside a research setting such as the UDN will be an important step in integrating genomics into routine health care. While this will increasingly drive a genotype-first approach to clinical medicine, the continued need for deep and broad clinical phenotyping will remain, especially as increasingly numerous rare variants are correlated with unique presentations or combinations of presentations that require old-fashioned hypotheses testing with downstream studies.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health grant U01 HG007709 (B.L.), U54 HD083092.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. The Dept. of Molecular and Human Genetics received funding from Baylor Genetics, a joint venture diagnostic laboratory of Baylor College of Medicine and H.U. Holdings. None of the authors received direct compensation from Baylor Genetics diagnostic laboratory.
LITERATURE CITED
- 1.Gahl WA, Wise AL, Ashley EA. 2015. The Undiagnosed Diseases Network of the National Institutes of Health: a national extension. JAMA 314(17):1797–98 [DOI] [PubMed] [Google Scholar]
- 2.Tifft CJ, Adams DR. 2014. The National Institutes of Health Undiagnosed Diseases Program. Curr. Opin. Pediatr 26(6):626–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gahl WA, Tifft CJ. 2011. The NIH Undiagnosed Diseases Program: lessons learned. JAMA 305(18):1904–5 [DOI] [PubMed] [Google Scholar]
- 4.Splinter K, Adams DR, Bacino CA, et al. 2018. Effect of genetic diagnosis on patients with previously undiagnosed disease. N. Engl. J. Med 379(22):2131–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker ML. 2010. Sequencing technologies—the next generation. Nat. Rev. Genet 11(1):31–46 [DOI] [PubMed] [Google Scholar]
- 6.Slatko BE, Gardner AF, Ausubel FM. 2018. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol 122(1):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431(7011):931–45 [DOI] [PubMed] [Google Scholar]
- 8.Natl. Hum. Genome Res. Inst. DNA Sequencing Costs: Data. NHGRI, Bethesda, MD. https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data [Google Scholar]
- 9.Collins FS, Varmus H. 2015. A new initiative on precision medicine. N. Engl. J. Med 372(9):793–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malinowski J, Miller DT, Demmer L, et al. 2020. Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability. Genet. Med 22(6):986–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershberger RE, Givertz MM, Ho CY, et al. 2018. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med 20(9):899–909 [DOI] [PubMed] [Google Scholar]
- 12.eMERGE Consortium. 2019. Harmonizing clinical sequencing and interpretation for the eMERGE III Network. Am. J. Hum. Genet 105(3):588–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, Rutter JL, Goldstein DB, et al. 2019. The “All of Us” research program. N. Engl. J. Med 381(7):668–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posey JE, Rosenfeld JA, James RA, et al. 2016. Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet. Med 18(7):678–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Muzny DM, Xia F, et al. 2014. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312(18):1870–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark MM, Stark Z, Farnaes L, et al. 2018. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom. Med 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lionel AC, Costain G, Monfared N, et al. 2018. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med 20(4):435–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilissen C, Hehir-Kwa JY, Thung DT, et al. 2014. Genome sequencing identifies major causes of severe intellectual disability. Nature 511(7509):344–47 [DOI] [PubMed] [Google Scholar]
- 19.Kingsmore SF, Cakici JA, Clark MM, et al. 2019. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am. J. Hum. Genet 105(4):719–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertoli-Avella AM, Beetz C, Ameziane N, et al. 2021. Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort. Eur. J. Hum. Genet 29(1):141–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. 2017. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 27(11):1895–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Schulz-Trieglaff O, Shaw R, et al. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32(8):1220–22 [DOI] [PubMed] [Google Scholar]
- 23.Murdock DR, Dai H, Burrage LC, et al. 2021. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J. Clin. Investig 131(1):e141500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdick KJ, Cogan JD, Rives LC, et al. 2020. Limitations of exome sequencing in detecting rare and undiagnosed diseases. Am. J. Med. Genet. A 182(6):1400–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoch K, Esteves C, Bican A, et al. 2021. Clinical sites of the Undiagnosed Diseases Network: unique contributions to genomic medicine and science. Genet. Med 23(2):259–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rady Children’s Inst. Genom. Med. 2021. The evidence is in: Project Baby Bear. Rady Children’s Institute for Genomic Medicine. https://www.radygenomics.org/our-work/project-baby-bear/ [Google Scholar]
- 27.Richards S, Aziz N, Bale S, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17(5):405–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karam R, Conner B, LaDuca H, et al. 2019. Assessment of diagnostic outcomes of RNA genetic testing for hereditary cancer. JAMA Netw. Open 2(10):e1913900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frésard L, Smail C, Ferraro NM, et al. 2019. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med 25(6):911–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Huang AY, Wang L-K, et al. 2020. Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet. Med 22(3):490–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer LS, Bader DM, Mertes C, et al. 2017. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun 8:15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moles-Fernández A, Duran-Lozano L, Montalban G, et al. 2018. Computational tools for splicing defect prediction in breast/ovarian cancer genes: How efficient are they at predicting RNA alterations? Front. Genet 9:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wai HA, Lord J, Lyon M, et al. 2020. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet. Med 22(6):1005–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonorazky HD, Naumenko S, Ramani AK, et al. 2019. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. Am. J. Hum. Genet 104(5):1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummings BB, Marshall JL, Tukiainen T, et al. 2017. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med 9(386):eaal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aicher JK, Jewell P, Vaquero-Garcia J, et al. 2020. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet. Med 22(7):1181–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amberger JS, Bocchini CA, Schiettecatte F, et al. 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43(Database issue):D789–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenger AM, Guturu H, Bernstein JA, et al. 2017. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet. Med 19(2):209–14 [DOI] [PubMed] [Google Scholar]
- 39.Cope H, Spillmann R, Rosenfeld JA, et al. 2020. Missed diagnoses: clinically relevant lessons learned through medical mysteries solved by the Undiagnosed Diseases Network. Mol. Genet. Genom. Med 8(10):e1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, Meng L, Normand EA, et al. 2019. Reanalysis of clinical exome sequencing data. N. Engl. J. Med 380(25):2478–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang T, Liu X, Wu T-J, et al. 2019. Atlas-CNV: a validated approach to call single-exon CNVs in the eMERGESeq gene panel. Genet. Med 21(9):2135–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly MA, Caleshu C, Morales A, et al. 2018. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet. Med 20(3):351–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bodian DL, Kothiyal P, Hauser NS. 2019. Pitfalls of clinical exome and gene panel testing: alternative transcripts. Genet. Med 21(5):1240–45 [DOI] [PubMed] [Google Scholar]
- 44.Deignan JL, Chung WK, Kearney HM, et al. 2019. Points to consider in the reevaluation and reanalysis of genomic test results: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med 21(6):1267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shashi V, Schoch K, Spillmann R, et al. 2019. A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet. Med 21(1):161–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitsuhashi S, Matsumoto N. 2020. Long-read sequencing for rare human genetic diseases. J. Hum. Genet 65(1):11–19 [DOI] [PubMed] [Google Scholar]
- 47.Mandelker D, Schmidt RJ, Ankala A, et al. 2016. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet. Med 18(12):1282–89 [DOI] [PubMed] [Google Scholar]
- 48.Sedlazeck FJ, Lee H, Darby CA, et al. 2018. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet 19(6):329–46 [DOI] [PubMed] [Google Scholar]
- 49.Laver TW, Caswell RC, Moore KA, et al. 2016. Pitfalls of haplotype phasing from amplicon-based long-read sequencing. Sci. Rep 6:21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sone J, Mitsuhashi S, Fujita A, et al. 2019. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet 51(8):1215–21 [DOI] [PubMed] [Google Scholar]
- 51.de Jesus AA, Hou Y, Brooks S, et al. 2020. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J. Clin. Investig 130(4):1669–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y-Z, Akdemir A, Tremmel G, et al. 2020. Nanopore basecalling from a perspective of instance segmentation. BMC Bioinform. 21(Suppl. 3):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark Z, Schofield D, Alam K, et al. 2017. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet. Med 19(8):867–74 [DOI] [PubMed] [Google Scholar]
- 54.Stark Z, Schofield D, Martyn M, et al. 2019. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet. Med 21(1):173–80 [DOI] [PubMed] [Google Scholar]
- 55.Smith HS, Swint JM, Lalani SR, et al. 2019. Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet. Med 21(1):3–16 [DOI] [PubMed] [Google Scholar]
- 56.Reuter CM, Kohler JN, Bonner D, et al. 2019. Yield of whole exome sequencing in undiagnosed patients facing insurance coverage barriers to genetic testing. J. Genet. Couns 28(6):1107–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deverka PA, Kaufman D, McGuire AL. 2014. Overcoming the reimbursement barriers for clinical sequencing. JAMA 312(18):1857–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabo A, Murdock D, Dugan S, et al. 2020. Community-based recruitment and exome sequencing indicates high diagnostic yield in adults with intellectual disability. Mol. Genet. Genom. Med 8(10):e1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sweeney NM, Nahas SA, Chowdhury S, et al. 2021. Rapid whole genome sequencing impacts care and resource utilization in infants with congenital heart disease. NPJ Genom. Med 6(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blue Shield Calif. 2020. Blue Shield of California becomes first health plan in U.S. to cover cost of rapid Whole Genome Sequencing for critically ill children. News Release, Mar. 9, Blue Shield Calif., Oakland, CA. https://news.blueshieldca.com/2020/03/09/RADY-genomics [Google Scholar]
- 61.Phillips KA, Deverka PA, Marshall DA, et al. 2018. Methodological issues in assessing the economic value of next-generation sequencing tests: many challenges and not enough solutions. Value Health 21(9):1033–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchanan J, Wordsworth S, Schuh A. 2013. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics 14(15):1833–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith HS, Swint JM, Lalani SR, et al. 2020. Exome sequencing compared with standard genetic tests for critically ill infants with suspected genetic conditions. Genet. Med 22(8):1303–10 [DOI] [PubMed] [Google Scholar]
- 64.Smith HS, Russell HV, Lee BH, et al. 2020. Using the Delphi method to identify clinicians’ perceived importance of pediatric exome sequencing results. Genet. Med 22:69–76 [DOI] [PubMed] [Google Scholar]
- 65.Pereira S, Robinson JO, Gutierrez AM, et al. 2019. Perceived benefits, risks, and utility of newborn genomic sequencing in the BabySeq project. Pediatrics 143(Suppl. 1):S6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pena LDM, Jiang Y-H, Schoch K, et al. 2018. Looking beyond the exome: a phenotype-first approach to molecular diagnostic resolution in rare and undiagnosed diseases. Genet. Med 20(4):464–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Posey JE, Harel T, Liu P, et al. 2017. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med 376(1):21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]