Abstract
Obesity is increasing worldwide and leads to a multitude of metabolic diseases including cardiovascular disease, type 2 diabetes, non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH). CYR61 is associated with the progression of NASH, but it has been described to have anti- as well as pro-inflammatory properties. Here, we sought to examine the role of liver CYR61 in NASH progression. CYR61 liver knockouts on a NASH diet improves glucose tolerance, decreases liver inflammation, and reduces fibrosis. CYR61 polarizes infiltrating monocytes promoting a pro-inflammatory/pro-fibrotic phenotype through an IRAK4/SYK/NFκB signaling cascade. In vitro, CYR61 activates a pro-fibrotic program including PDGFa/PDGFb expression in macrophages in an IRAK4/SYK/NFκB-dependent manner. Furthermore, targeted-antibody blockade reduces CYR61-driven signaling in macrophages in vitro and in vivo, reducing fibrotic development. This study demonstrates that in NASH, CYR61 is a key driver of liver inflammation and fibrosis.
One Sentence Summary:
Cyr61 drives NASH fibrosis by polarizing pro-fibrotic monocyte-derived macrophages through an IRAK4/SYK/NFκB signaling cascade.
INTRODUCTION
Obesity has been increasing worldwide over the past few decades. It is linked to poor diet, lack of exercise, social class, and some genetic factors (1). Obesity leads to many complications including cardiac hypertension, type 2 diabetes mellitus, pancreatitis, and non-alcoholic fatty liver disease (NAFLD) (2, 3). NAFLD is the most common chronic liver disease worldwide and if left untreated can progress to more severe diseases such as non-alcoholic steatohepatitis (NASH) and cancer (4). The development of cardio-metabolic disorders such hypertension and diabetes is intimately linked to the presence of NAFLD/NASH and are common causes of morbidity and mortality in these patients (5-7). Currently, the best treatment options for NAFLD and NASH target the source of metabolic symptoms: dietary changes, increased exercise, or bariatric surgery (8, 9). However, a proportion of individuals with NAFLD/NASH are lean (10-12) indicating a need to improve our understanding of the biochemical steps leading to NAFLD/NASH.
Development of NAFLD/NASH is believed to occur through multiple hits, including insulin resistance, toxic fatty acid metabolites, inflammation, and fibrosis (13). Fibrosis in NASH is the primary determinant of mortality (14, 15), leading to the expectation that limiting its development may reduce the burden of disease. Therefore, understanding fibrogenesis in NAFLD/NASH is vital to developing treatments for it.
Previously, we demonstrated that hepatocyte CYR61 expression is a driver of liver fibrosis and inflammation during injury (16). CYR61 (cysteine-rich angiogenic inducer 61), also known as cellular communication network factor 1 (CCN1), is a secreted protein that signals to both fibroblasts and macrophages (17). It supports recruitment and adhesion of monocytes and macrophages in vitro and in vivo (18, 19). When treated with CYR61 in vitro, macrophages produce pro-inflammatory cytokines such as TNFα, IL-1β, and IL-6 (20). However, the effect of CYR61 on fibroblasts is less clear. Expression of CYR61 is upregulated in human livers with cirrhosis and NASH as well as in murine fibrotic liver injury models such as bile duct ligation and carbon tetrachloride (CCl4) (16, 21-23). However, other work shows that direct stimulation of fibroblasts with CYR61 in vitro results in senescence or apoptosis (23-25), pointing to a role for CYR61 in the resolution of fibrosis. Thus, the in vivo mechanism for CYR61-driven liver fibrosis, particularly in NASH liver fibrosis, remains to be uncovered.
Here, we explore the mechanism(s) of inflammatory and fibrotic remodeling caused by CYR61 during NASH injury. We find that CYR61 modulates NASH fibrosis through the activation and polarization of infiltrating monocyte-derived macrophages, which in turn secrete factors that activate fibroblasts. Furthermore, we find that hepatocytic CYR61 exacerbates glucose homeostasis in fatty liver disease. Ultimately, we demonstrate that CYR61 blockade reduces the inflammation and fibrosis during liver injury. Together, these findings uncover a potential mechanism of CYR61 during NASH injury that could be targeted to ameliorate aspects of this disease.
RESULTS
Loss of CYR61 ameliorates injury in a murine NASH model
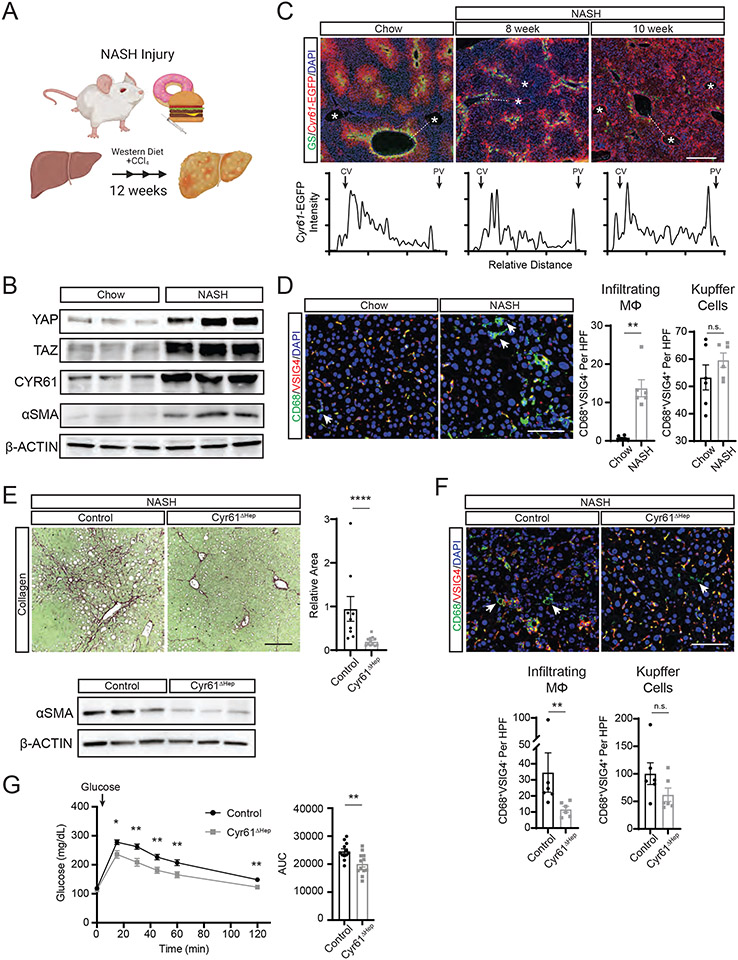
Previously, we examined a small cohort of liver biopsies from healthy individuals and patients with NASH, finding a direct relationship between the expression of YAP/TAZ/CYR61 (16). To directly determine if chronic expression of CYR61 influences NASH pathology, we employed a mouse model using high fat, high cholesterol, high sucrose diet (Western diet, WD) with a weekly injection of low-dose carbon tetrachloride (CCl4) resulting in histologic and transcriptional features that closely resembles human NASH injury (Fig 1A) (26). Livers from mice on this diet for 12 weeks show increased YAP, TAZ, CYR61, and αSMA expression compared to mice fed normal chow (Fig 1B). Cyr61-EGFP reporter mice (16) were placed on this diet to identify changes in Cyr61 expression during NASH injury. In controls, Cyr61-EGFP was expressed in a graded pattern radiating outward from the central veins whereas NASH-treated mice showed disorganization of CYR61 expression across the liver parenchyma (Fig 1C). Progression of NASH injury resulted in increased Cyr61-EGFP expression in endothelial cells of the portal vein. Liver zonation was also disrupted as indicated by the loss of glutamine synthetase (GS)-expressing hepatocytes around the central vein.
Figure 1. Cyr61 drives fibrotic injury in NASH.
(A) Cartoon of 12-week NASH injury in mice and associated biological analyses. (B) Immunoblot of indicated protein expression in whole livers of mice on chow diet or 12 weeks NASH diet. (C) Representative immunofluorescence imaging of GS (green) and Cyr61-EGFP (red) in livers from Cyr61-EGFP reporter mice on chow diet or on 8 or 10 weeks of NASH diet. Asterisks (*) indicate portal regions. Scale bar=400μm. Below: intensity of Cyr61-EGFP staining across dotted line from central vein (CV) to portal vein (PV). (D) Representative immunofluorescence imaging of specified proteins in livers from mice on chow diet (n=6) or 12 weeks of NASH diet (n=6). Scale bar=100μm. Quantification of infiltrating MΦs (CD68+VSIG4−) and Kupffer cells (CD68+VSIG4+) to the right. HPF=high power field. Arrows indicate CD68+VSIG4− cells. (E) Representative picrosirius red staining of livers from Cyr61 fl/fl mice treated with AAV8-TBG-GFP (control, n=9) or AAV8-TBG-CRE (Cyr61ΔHep, n=12) on NASH diet for 12 weeks. Scale bar=200μm. Quantification to the right. Below: Immunoblot of αSMA expression in whole livers of control or Cyr61ΔHep mice on 12 weeks of NASH diet. (F) Representative immunofluorescence imaging of specified proteins in livers from control (n=6) or Cyr61ΔHep (n=6) NASH mice. Arrows indicate CD68+VSIG4− cells. Scale bar=100μm. Quantification of infiltrating MΦs (CD68+VSIG4−) and Kupffer cells (CD68+VSIG4+) below. (G) Serum glucose measured in control (n=12) or Cyr61ΔHep (n=12) mice at indicated time points after glucose challenge (indicated by arrow). Fasting glucose was measured at 0 minutes. Area under the curve (AUC) calculation plotted to the right. Mean and SEM plotted; p value calculated with Mann-Whitney U test. *p<0.05, **p<0.01, ****p<0.0001
Next, we examined macrophage expansion and their infiltration into the liver during NASH injury. Macrophage populations greatly increase in response to liver injury (27, 28). In NASH, fibroblasts are activated by contributions from Kupffer cells (KC) (29, 30) and monocyte-derived macrophages (MoMΦs) (29, 31-37). We examined the relative macrophage populations during NASH injury using the KC marker VSIG4 (34) and the general macrophage marker CD68. Macrophages expressing CD68 but lacking VSIG4 were deemed infiltrating MoMΦs (38). In chow-fed control livers, few CD68+VSIG4− MoMΦs were present, however significantly more appeared after 12 weeks of NASH injury (0.67 vs 13.7 per high powered field (HPF), p<0.01; Fig 1D). The number of KCs (CD68+VSIG4+) between chow and NASH livers was not significantly different after 12 weeks of NASH injury (53.3 vs 59.6 per HPF, p=0.37; Fig 1D). We confirmed this using Clec4F and CD163 staining (Fig S1A).
To elucidate the effect of hepatocytic Cyr61 in NASH injury, we infected Cyr61fl/fl mice with either AAV8-TBG-EGFP (control) or AAV8-TBG-CRE (Cyr61ΔHep) and placed them on NASH injury for 12 weeks. Cyr61 deletion from hepatocytes was confirmed with RNA in-situ hybridization and Western blots (Fig S1B/C). Liver to body weight measurements were similar across all groups (Fig S1D). Pathologic analysis showed reduced histologic severity scores in Cyr61ΔHep livers compared to controls (9.75 vs 7, p=0.136, Fig S1E, Table S1). Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were modestly reduced in Cyr61ΔHep NASH mice (19%, p=0.29; 31%, p=0.12, respectively) compared to controls (Fig S1F). Cyr61ΔHep mice showed a 75% reduction in collagen deposition compared to control wildtype mice on NASH diet (p<0.0001, Fig 1E). Cyr61ΔHep NASH livers also showed reduced αSMA expression as compared to controls (Fig 1E). Furthermore, MoMΦ populations were reduced by 50% in Cyr61ΔHep compared to control NASH livers (23.3 vs 11.7 per HPF, p<0.01; Fig 1F). KC populations were not significantly changed in Cyr61ΔHep livers (100 vs 62 per HPF, p=0.24; Fig 1F, S1A). Thus, Cyr61 liver expression is directly related to the infiltration of macrophages and the development of fibrosis during NASH injury.
We then investigated the effect of Cyr61 on metabolic aspects of NASH. Control and Cyr61ΔHep mice were placed in metabolic cages to measure energy expenditure and food intake, as well as analyzed for body composition after 12 weeks of NASH injury. There were no significant differences in food/water intake, energy expenditure, or body composition between the groups (Fig S1G). After glucose challenge, Cyr61ΔHep mice demonstrated a 20% improvement in glucose tolerance compared to controls (24,000 vs 19,700 AUC, p<0.01, Fig 1G) without any significant difference in plasma insulin concentrations (Fig S1H). Therefore, hepatocytic Cyr61 reduction in the context of NASH injury improves glucose tolerance, likely through improved insulin sensitivity.
Transcriptional Profiling Cyr61ΔHep in the Context of NASH Diet
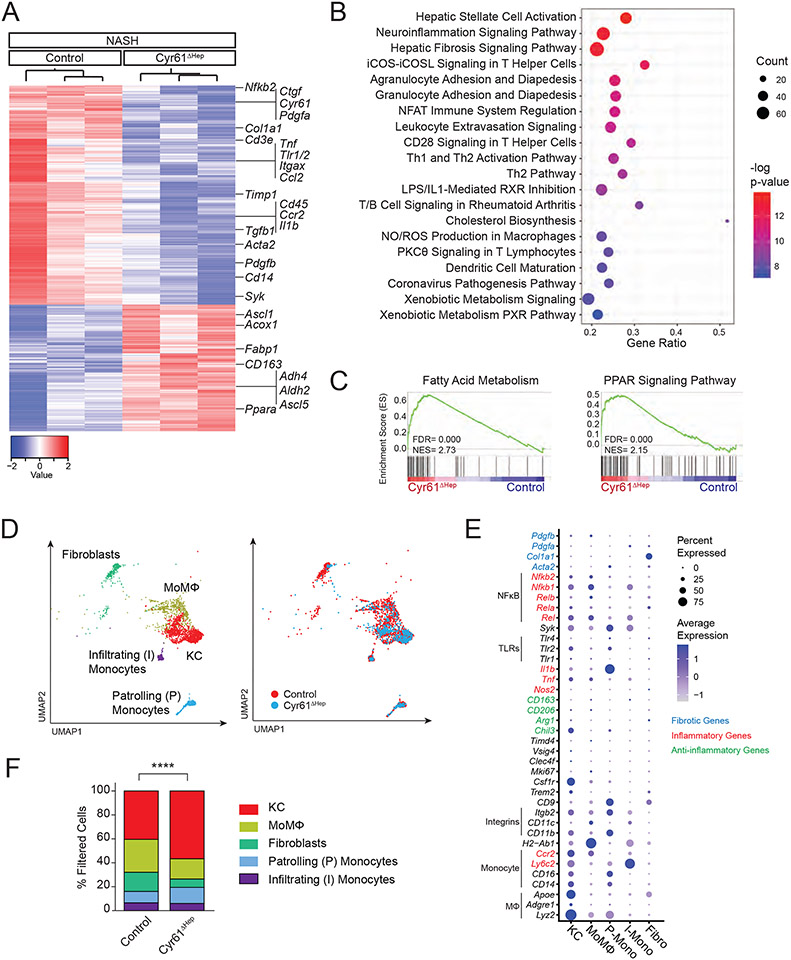
To understand the mechanisms driving differences in Control and Cyr61ΔHep mice on a NASH diet, we examined the liver transcriptional profiles by RNA sequencing (RNA-seq). Principal component analysis (PCA) demonstrated that chow-fed control and Cyr61ΔHep livers were highly similar whereas NASH-treated Control and Cyr61ΔHep livers differed substantially (Fig S1I). Of 2,198 differentially expressed genes, 805 genes were upregulated and 1,393 genes were downregulated in Cyr61ΔHep NASH livers compared to control NASH livers. Differential gene expression analysis showed increased expression of metabolic genes (Acox1, Fabp1, Ppara), and decreased expression of fibrotic genes (Ctgf, Col1a1, Tgfb1, Acta2) and inflammatory genes (Tnf, Tlr1/2, Ccl2) in Cyr61ΔHep livers (Fig 2A). Ingenuity pathway analysis showed changes in hepatic fibrosis (hepatic stellate cell activation; hepatic fibrosis signaling pathway) and metabolic pathways (cholesterol biosynthesis; xenobiotic metabolism signaling/PXR pathway). We additionally found a number of immune pathways altered in Cyr61ΔHep livers, especially those involved in inflammation (NO/ROS production in macrophages) and infiltration and adhesion of immune cells (agranulocyte/granulocyte adhesion and diapedesis; leukocyte extravasation signaling, Fig 2B). Gene set enrichment analysis (GSEA) in Cyr61ΔHep livers also showed increases in PPARα signaling and fatty acid metabolism (Fig 2C). Overall, the NASH injury in Cyr61ΔHep mice is associated with gene signatures associated with improved glucose and fat metabolism as well as reduced inflammation and fibrosis compared to controls.
Figure 2. Loss of Cyr61 improves fibrotic, inflammatory, and metabolic signaling in NASH.
(A) Heatmap of 2198 differentially expressed genes between male control and Cyr61ΔHep NASH livers (FDR<0.05, Fold change > 1.5). (B) Top significantly enriched pathways called by differentially expressed genes comparing control and Cyr61ΔHep NASH livers. (C) Gene set enrichment analysis of indicated metabolic KEGG Pathway gene sets (NES= normalized enrichment score). (D) Uniform Manifold Approximation and Projection (UMAP) plots of filtered cells in control (n=2; 1,755 cells) and Cyr61ΔHep (n=2; 697 cells) NASH livers. Left: cell type identification labeled; Right: UMAP colored by condition. (E) Dot plot of selected genes expressed in each cluster. (F) Quantification of filtered populations in control and Cyr61ΔHep NASH livers as a percent of total cells. P value calculated with Fisher’s Exact test (****p<0.0001).
Cyr61 expression alters macrophage and monocyte signaling in NASH-injured livers
To assess the cellular composition of control and Cyr61ΔHep NASH-injured livers, we performed single-cell RNA sequencing (scRNA-seq), recovering 18,695 cells. Seurat analysis of these cells with Uniform Manifold Approximation and Projection (UMAP) dimension reduction identified 9 clusters (Fig S2A). Whole liver RNA-seq analysis suggested that Cyr61 activity primarily affects fibroblasts and macrophages/monocytes, so we limited our analysis to these cell types. Upon re-clustering on the filtered macrophage (MΦ), monocyte, and fibroblast populations (2,452 cells, Fig S2B-D), we identified five distinct clusters: KCs (Adgre1, Csf1r, Clec4f, Vsig4), MoMΦ (H2-Ab1, Cd11c), infiltrating monocytes (Ly6c2), patroling monocytes (Cd11b), and fibroblasts (Col1a1) (Fig 2D, E). Fibroblast populations were diminished in Cyr61ΔHep livers compared to control. Additionally, MΦs in the Cyr61ΔHep livers skewed towards KCs and away from MoMΦ populations (Fig 2D, F). The increase in KC populations in Cyr61ΔHep livers was confirmed by flow cytometry (Fig S2E). This suggests that Cyr61 drives MΦs toward the MoMΦ phenotype. Fibrotic (Pdgfb) and inflammatory (Nfkb1/2) genes were relatively increased in MoMΦs compared to KC, whereas anti-inflammatory genes (Chil3) were enriched in KC compared to monocytes/MoMΦs (Fig 2E).
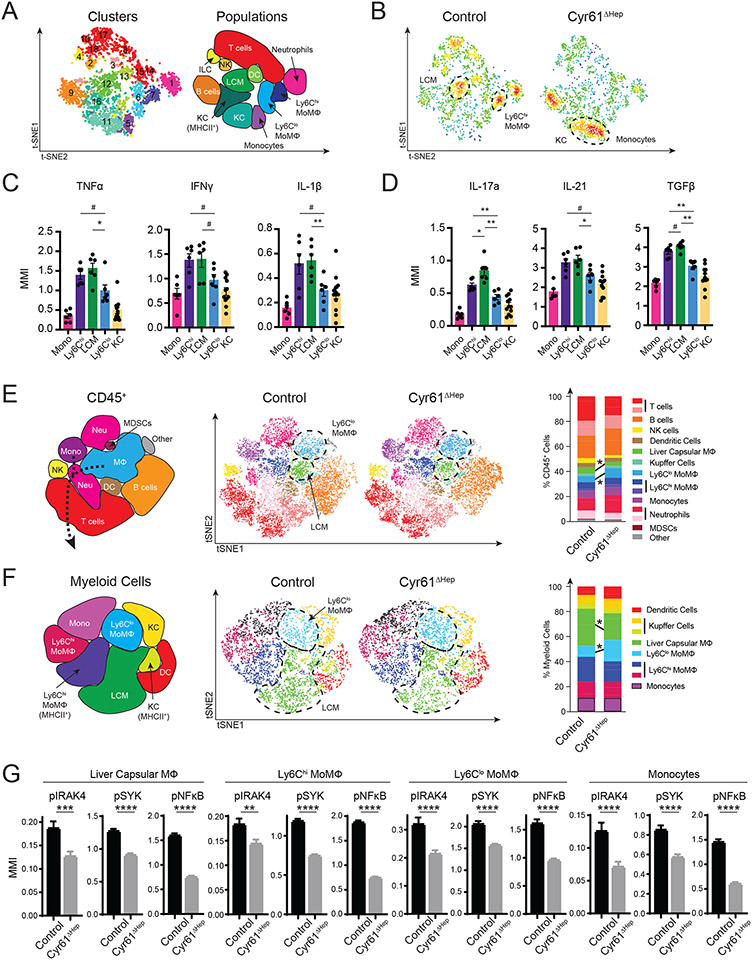
We complemented these studies by using mass cytometry by time of flight (CyTOF) to further characterize the immune cells in these livers. We performed CyTOF on control (n=3; 650 cells/sample) and Cyr61ΔHep NASH livers (n= 3; 650 cells/sample) using a cell phenotyping and cytokine expression panel (Fig S2F, Table S2). We identified populations of T cells, B cells, and natural killer (NK) cells that were also seen in scRNA-seq (Fig S2A, B, G). CyTOF improved our ability to profile dendritic cells (DCs) and MΦs/monocytes as well as to detect neutrophils, which were not effectively sampled by scRNA-seq. The additional cell surface markers facilitated MΦ populations to be more specifically identified as liver-resident KCs, Ly6Chi MoMΦs, Ly6Clo MoMΦs, and a third type of MoMΦs known as liver capsular MΦ (LCM) (Fig 3A, S2G). Control livers were enriched for LCM and Ly6Chi MoMΦs; Cyr61ΔHep livers shifted away from these cell types and toward KCs and monocytes (Fig 3B, S2H). These changes seen in Cyr61ΔHep by CyTOF are consistent with the shift to KCs seen by scRNA-seq.
Figure 3. Loss of Cyr61 reduces inflammatory signaling in macrophages.
(A) T-distributed stochastic neighbor embedding (tSNE) plots of CD45+ populations in control (n=3) and Cyr61ΔHep (n=3) NASH livers using cytokine CyTOF panel. Right, identification of populations. (B) t-SNE plot showing density of cells in each condition. (C to D) Mean metal intensity (MMI) for indicated inflammatory (C) and fibrotic (D) cytokines in monocyte and macrophage populations. (E) Representative t-SNE plots of control (n=4) and Cyr61ΔHep (n=4) NASH livers using phosphorylation CyTOF panel. Significantly changed (p<0.05) populations circled. Right: plot of indicated populations as a percent of CD45+ cells. (F) Representative t-SNE plots of Ly6G− myeloid cells (CD45+CD3−CD19−NKp45−Ly6G−F4/80+ and/or CD11b+) in control (n=3) and Cyr61ΔHep (n=3) NASH livers. Significantly changed populations circled. Right: plot of indicated populations as a percent of Ly6G− myeloid cells. Mean and SEM plotted; p value calculated with Mann-Whitney U test. #p<0.1, *p<0.05, **p<0.01. (G) MMI of phosphorylated proteins in cells of indicated populations. Liver capsular MΦs: control n=1334 cells, Cyr61ΔHep n=1901; Ly6Chi MoMΦs: control n=1512, Cyr61ΔHep n=2636, Ly6Clo MoMΦs: control n=396, Cyr61ΔHep n=1522; and monocytes: control n=474, Cyr61ΔHep n=960. Mean and SEM plotted; p value calculated with Student’s T test. ***p<0.001, ****p<0.0001.
To understand the roles played by the different monocyte/MΦ populations during NASH injury, we examined the expression of inflammatory and fibrotic cytokines. As expected, Ly6Clo MoMΦs, an anti-fibrotic/pro-repair phenotype (39), consistently expressed less inflammatory (TNFα, IFNγ, IL-1β) and fibrotic (IL-17a, IL-21, TGFβ) cytokines than both Ly6Chi MoMΦs and LCMs (Fig 3C, D). Of note, LCMs had higher expression of fibrotic IL-17a and TGFβ than Ly6Chi MoMΦs (Fig 3D).
After confirming cytokine expression in the different MΦ clusters, we used a phenotyping and phosphorylation detection panel (Table S2) to examine the signaling pathways activated in control (n=4; 3000 cells/sample) and Cyr61ΔHep livers (n=7; 3000 cells/sample) by CyTOF during NASH injury. Automated clustering of all CD45+ cells based on surface marker expression in this analysis identified similar populations of cells as in the cytokine analysis (Fig 3E, S3A, B). Consistent with the cytokine panel, we saw a significantly reduced proportion of LCMs in Cyr61ΔHep livers (3.3% vs 5.4%, p<0.05, Fig 3E, S3C). Furthermore, we detected an increase in the population of Ly6Clo MoMΦs between control and Cyr61ΔHep (7.6% vs. 4.9%, p<0.05, Fig 3E, S3C). With a greater number of cells, we were able to isolate MΦ/monocyte data for more in-depth analysis.
To investigate signaling changes in MΦ/monocytes, we examined a panel of eleven well-known phosphorylated proteins differentially activated during inflammation (Table S2). We isolated 1,400 non-lymphocyte cells expressing CD11b or F4/80 per sample (control n=3, Cyr61ΔHep n=6; Fig S3D). The resulting Ly6G− myeloid cells were identified as DCs, monocytes, KCs, Ly6Chi MoMΦs, Ly6Clo MoMΦs, and LCMs (Fig 3F, S3E). Consistent with the analysis of all CD45+ cells, Ly6G− myeloid cells in Cyr61ΔHep livers showed increased Ly6Clo MoMΦs and decreased LCM populations (17% vs 8.8% and 21.3% vs 29.6% respectively, p<0.05, Fig 2F, S3F). Monocytes and MoMΦs (LCMs, Ly6Chi MoMΦs, Ly6Clo MoMΦs) in Cyr61ΔHep livers showed significantly less phosphorylated IRAK4 (p<0.01), SYK (p<0.0001), and NFκB (p<0.0001) (Fig 3G). This suggests that Cyr61 stimulates an IRAK4/SYK/NFκB signaling cascade in infiltrating monocytes and macrophages.
Cyr61 differentiates infiltrating monocytes
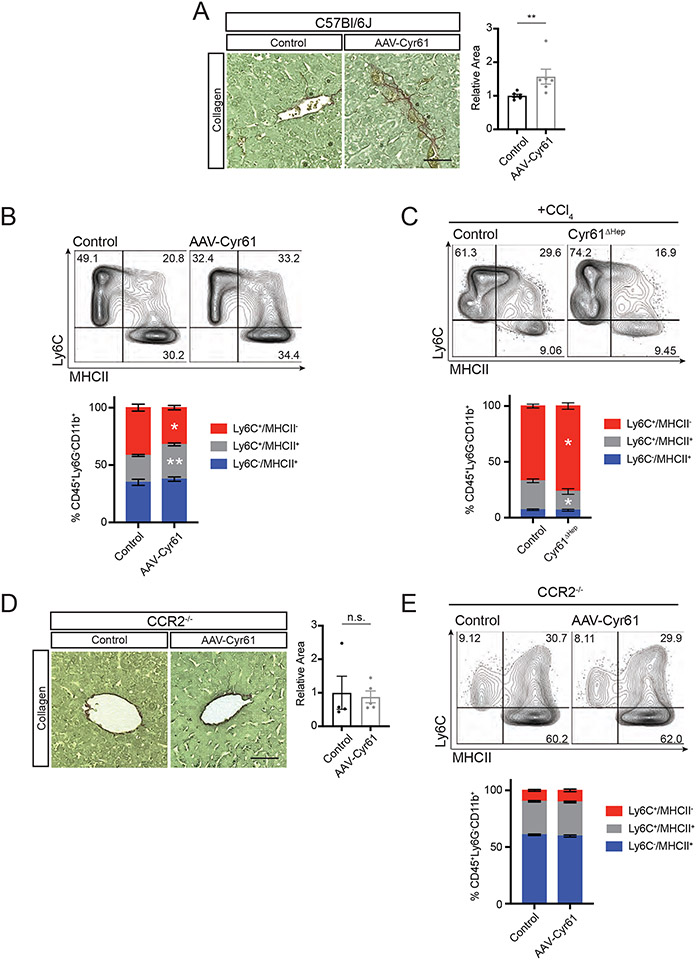
We next examined whether Cyr61 directly attracts monocytes, polarizes infiltrating monocytes into inflammatory MΦs, or plays a combination of both roles. To assess a Cyr61 gain-of-function phenotype, C57Bl/6J mice were transduced with either AAV8-TBG-Null (control) or AAV8-TBG-Cyr61 (AAV-Cyr61) for 2 weeks. AAV-Cyr61 mice showed no change in liver weight from control mice (Fig S4A). Collagen deposition in AAV-Cyr61 livers was increased by 56.8% as compared to controls (p<0.01, Fig 4A). We then examined monocyte differentiation to MΦ in these livers through graded expression of Ly6C and MHCII (38). AAV-Cyr61 livers showed fewer monocytes (Ly6C+/MHCII−, 31.5% vs 42.65%, p<0.05), but a greater proportion of monocyte/MΦ intermediate cells (Ly6C+/MHCII+, 31.1% vs 22.9%, p<0.01) compared to controls (Fig 4B, S4B). Next, we examined if our loss-of-function Cyr61ΔHep phenotype would confirm our gain-of-function phenotype and impair monocyte recruitment or their transition into intermediate cells after acute CCl4 injury. Cyr61ΔHep mice showed no change in liver weight from control mice after CCl4 injury (Fig S4C). Injured Cyr61ΔHep livers show a greater proportion of monocytes (76.54% vs 67%, p<0.05), but decreased monocyte/MΦ intermediate cells (25.9% vs 16.8%, p<0.05, Fig 4C) suggesting that Cyr61 is not directly involved in monocyte recruitment, but it is involved in their differentiation once they arrive.
Figure 4. Cyr61 drives monocyte-to-macrophage differentiation.
(A) Representative picrosirius red staining of livers from C57Bl/6J mice treated with AAV8-TBG-Null (control, n=6) or AAV8-TBG-CYR61 (CYR61, n=6). Quantification to the right. (B) Representative flow cytometry plots of CD11b+ cells, excluding Ly6C−/MHCII−, of control (n=6) and CYR61 (n=6) treated livers with percentage of parent population labeled. Quantification below. (C) Representative flow cytometry plots of CD11b+ cells, excluding Ly6C−/MHCII−, of control (n=5) and Cyr61ΔHep (n=5) acute CCl4-treated livers with percentage of parent population labeled. Quantification below. (D) Representative picrosirius red staining of livers from CCR2−/− mice treated with AAV8-TBG-Null (control, n=4) or AAV8-TBG-CYR61 (CYR61, n=5). Quantification to the right. (E) Representative flow cytometry plots of CD11b+ cells, excluding Ly6C−/MHCII−, of control (n=4) and CYR61 (n=5) treated CCR2−/− livers with percentage of parent population labeled. Quantification below. Scale bars=50μm. Mean and SEM plotted; p value calculated with Mann-Whitney U test. *p<0.05, **p<0.01.
Monocyte infiltration is necessary for Cyr61-mediated inflammation and fibrosis
Our models thus far (NASH, CCl4, AAV-Cyr61) demonstrate a role for Cyr61 in monocyte and MoMΦ activation. Next, we investigated the requirement for peripheral monocyte recruitment to the liver for the development of fibrosis. The CCR2 receptor traffics monocytes to injured tissues expressing CCL2 (40). Using CCR2-deficient mice, we expressed Cyr61 in hepatocytes to examine its effects on liver fibrosis. CCR2−/− mice treated with AAV-Cyr61 for 2 weeks showed no change in liver weight compared to control CCR2−/− mice (Fig S4D). There was no significant difference in fibrosis in CCR2−/− mice given control or AAV-Cyr61 virus (Fig 4D). Furthermore, AAV-Cyr61 CCR2−/− livers showed no change in the proportion of monocytes, MΦs, or intermediates in the liver (Fig 4E). This points to the necessity of monocyte infiltration as a key step in Cyr61-mediated inflammation and fibrosis.
Cyr61 drives monocyte-derived macrophages toward inflammatory/fibrotic phenotypes in vitro
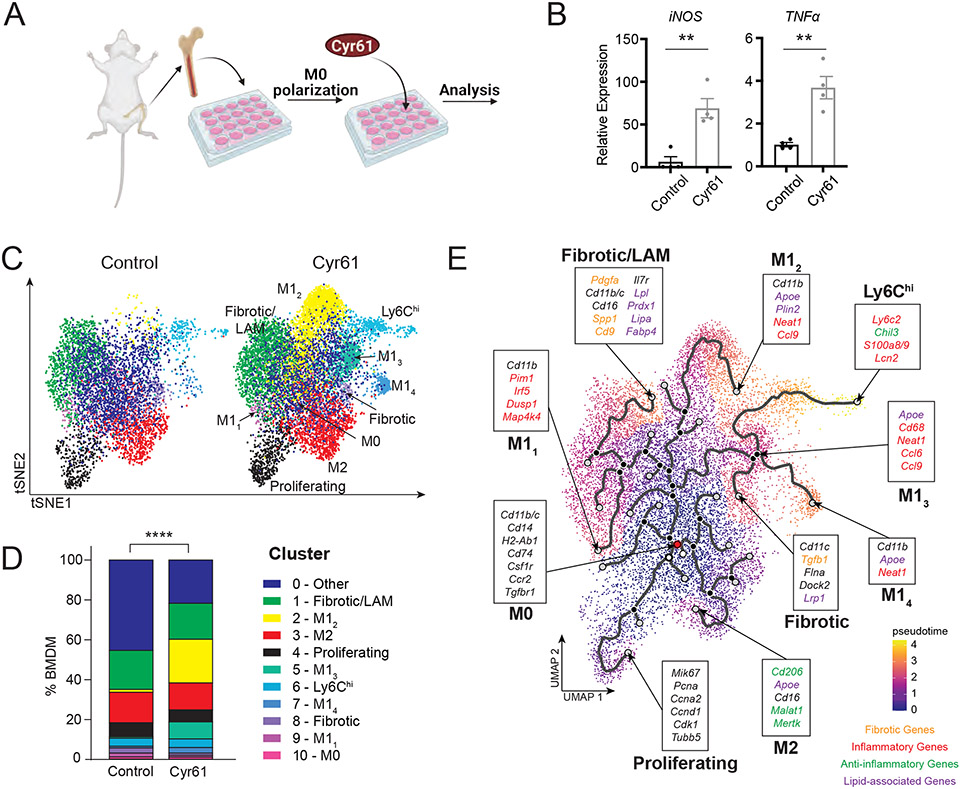
Having observed the contribution of Cyr61 to inflammation and fibrosis in several injury models in vivo, next we investigated the signaling activated by Cyr61 in MoMΦ. Bone marrow-derived MΦs (BMDMs) were isolated from leg bones of mice and treated with Cyr61 (Fig 5A). After 24 hours of Cyr61 treatment, BMDMs showed increased expression of the inflammatory genes iNOS and TNFα (Fig 5B). To examine gene expression more closely, we used scRNA-seq on Cyr61-treated BMDMs for 24 hours. Seurat UMAP analysis of control (n=4; 4,702 cells) and Cyr61-treated BMDMs (n=4; 8,733 cells) resulted in 11 distinct macrophage clusters (Fig S5A, B). A cluster of lipid-associated MΦs (LAM; Apoe, Plin2) and 2 clusters of M1 MΦs (M12, M13; Apoe, Cd68, Neat1) appeared after Cyr61 treatment (Fig 5C, D, S5B). Using Monocle3 trajectory analysis, we sought to understand the process of BMDM polarization by Cyr61 (Fig 5E). The most undifferentiated macrophages (M0) in the presence of Cyr61 differentiated towards macrophages expressing inflammatory (M11, M12, M13), fibrotic, and lipid-responsive phenotypes (LAM). Cyr61 activation culminated in Ly6Chi MΦs (Fig 5E). Consistent with our in vivo results, in vitro polarization of BMDMs with Cyr61 drove undifferentiated monocytes toward MΦs with a Ly6Chi, inflammatory, and fibrotic phenotypes. Macrophages stimulated by Cyr61 express inflammatory genes (20), but whether fibrotic signaling pathways are activated in a similar fashion is unclear.
Figure 5. Stimulation with Cyr61 induces inflammatory and fibrotic phenotypes in bone marrow-derived macrophages.
(A) Cartoon of bone marrow-derived macrophage (BMDM) workflow. (B) Quantitative PCR analysis of indicated gene expression in control BMDMs (n=4) or BMDMs treated with Cyr61 protein (n=4). Mean and SEM plotted; p value calculated with Student’s T-test. *p<0.05, **p<0.01. (C) UMAP plots of macrophages in control (n=4) and CYR61 treated (n=4) BMDM. Cell type identification labeled. (D) Quantification of populations in control (n=4) and Cyr61-treated (n=4) BMDMs as a percent of total cells. P value calculated with Fisher’s Exact test (****p<0.0001). (E) Trajectory plot using Monocle3. Populations and key genes labeled. Starting node labeled in red.
Cyr61 signals through IRAK4, SYK, and NFκB to activate fibrotic cytokine expression in macrophages
In the context of NASH injury, liver MoMΦs had significant reductions in phospho-IRAK4/SYK/NFKβ upon Cyr61 loss (Fig 3G). Notably, the promoter regions of PDGFa and PDGFb contain putative NFκB (Rela/p65) binding sites that are conserved across multiple species (Fig S6A), suggesting that Cyr61 polarizes MoMΦs through an IRAK4/SYK/NFκB signaling cascade. This may result in production of profibrotic cytokines (for example PDGFa/b) that stimulate HSCs. To directly test this hypothesis, we examined the signaling pathways that are activated in Cyr61-treated BMDMs, their downstream products, and how liver MoMΦs affect fibroblast activation in the context of Cyr61.
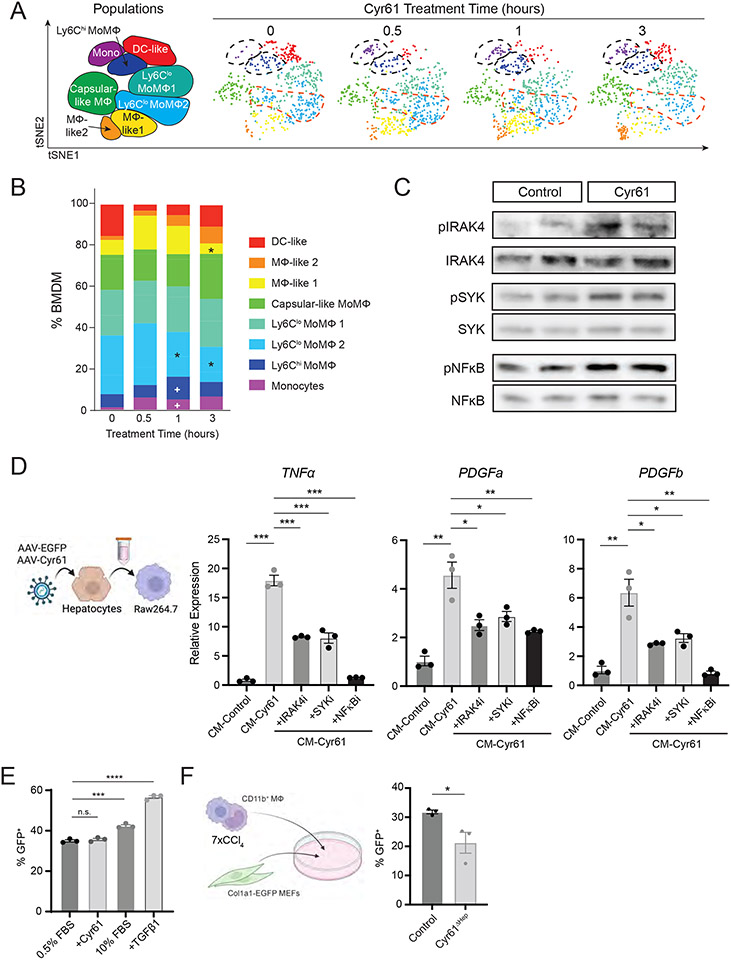
Using CyTOF, Cyr61-treated BMDMs were assessed by surface marker expression (n=3 per timepoint, 2,400 cells each) and characterized through automated clustering (Fig S5C-E). Proportions of Ly6Chi populations, monocytes, and MoMΦs increased 1 hour after Cyr61 treatment (5% vs 1.2% and 11.2% vs 6.5% respectively, p<0.05, Fig 6A, fig. S5F). Proportions of Ly6Clo MoMΦ (CD11c−) populations were decreased at 1 hour and 3 hours (22.3% and 17.3% vs 30.33%, p<0.05), and proportions of MΦ-like 1 were decreased at 3 hours after Cyr61 treatment (16.7% vs 5.2% respectively, p<0.05, Fig 6A, fig. S5F). We then examined intracellular signaling proteins; phosphorylation of IRAK4 was increased at 0.5 hours, and phosphorylation of SYK and NFκB were increased at 3 hours after Cyr61 treatment (Fig S5G). We confirmed phosphorylated IRAK4, SYK, and NFκB increased after Ad-Cyr61 treatment in J774A.1 cells (Fig 6C).
Figure 6. Cyr61 activates transcription of fibrotic cytokines through NFκB.
(A) t-SNE plot of CD45+ Ly6G− F4/80+ and/or CD11b+ BMDMs over the course of Cyr61 treatment. Populations labeled to the left. (B) Plot of indicated populations as a percent of CD45+/Ly6G− BMDMs. p value calculated with Mann-Whitney U test: +p<0.05, change from 0 hours; *p<0.05, change from 0.5 hours. (C) Immunoblot of indicated phosphorylated signaling proteins in control (Ad-GFP) and Ad-Cyr61-treated J774A.1 cells. (D) Cartoon indicating process of treating Raw264.7 cells. Right - Quantitative PCR analysis of indicated gene expression in conditioned media control (CM-control), conditioned media Cyr61 (CM-Cyr61) or CM-Cyr61 plus inhibitors to IRAK4 (zimlovisertib), SYK (lanraplenib) or NFkB (JSH-23). (E) Percent GFP+ as assessed by flow cytometry of Col1a1-EGFP MEFs with indicated treatments. (F) Cartoon indicating co-culture of CD11b+ FACS-sorted liver monocytes with Col1a1-EGFP MEFS. Mean and SEM plotted; p value calculated with Students T-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
BMDMs polarized towards either an M1 (Fig S6B) or M2 (Fig S6C) phenotype lacked responsiveness to Cyr61 treatment. Last, we purified liver macrophages from quiescent livers to examine their responsiveness to Cyr61. In vitro cultured liver macrophages showed robust response to Cyr61 with respect to TNFα expression, but neither PDGFa/b were detectable with or without Cyr61 treatment after 24 hours (Fig S6D).
Like the BMDM response to Cyr61, RAW264.7 cells treated with Cyr61 induced expression of TNFα (p<0.001), PDGFa, and PDGFb (p<0.01, Fig S6E). We then generated conditioned media from cultured hepatocytes infected with either EGFP (CM-Control) or Cyr61 (CM-Cyr61). CM-Cyr61 stimulated RAW264.7 cells to upregulate their expression of TNFα (p<0.001), PDGFa, and PDGFb (p<0.01, Fig 6D). Inhibitors to IRAK4, SYK, and NFkB reduced the ability of CM-Cyr61 to activate these genes in RAW264.7 cells (Fig 6D; TNFα, p<0.001; PDGFa/b, p<0.05).
We then tested if liver macrophages in the context of injury could activate fibroblasts in vitro. We generated mouse embryonic fibroblasts (MEFs) from the Col1A1-EGFP mouse, a mouse reporter commonly used to demonstrate fibroblast activation in the context of fibrotic injury (41). These MEFs demonstrated robust activation to 10% FBS and TGFβ1, but minimal response to Cyr61 (Fig 6E, S6F). Using FACS-purified liver macrophages (CD45+Ly6G−CD11b+) from CCl4 injured livers, these were co-cultured with Col1A1-EGFP MEFs. There was a 75% increase in EGFP fluorescence as compared to control (Fig S6G, p<0.001). We then co-cultured Col1A1-EGFP MEFs with monocytes/macrophages from either control or Cyr61ΔHep CCl4 livers. Liver macrophages from Cyr61ΔHep mice had (33%) less activity in activating Col1A1-EGFP MEFs than controls (p<0.05, Fig 6F).
Taken together, these data support the hypothesis that Cyr61 preferentially acts upon BMDMs of the undifferentiated MoMΦ (M0) phenotype early in the injury process. Cyr61 signals through a pathway involving IRAK4/SYK/NFκB resulting in the production of fibrotic cytokines such as PDGFa/b. These Cyr61-stimulated BMDMs then activate fibroblasts to begin the process of collagen deposition.
Blocking Cyr61 during fibrotic liver injury reduces fibrosis
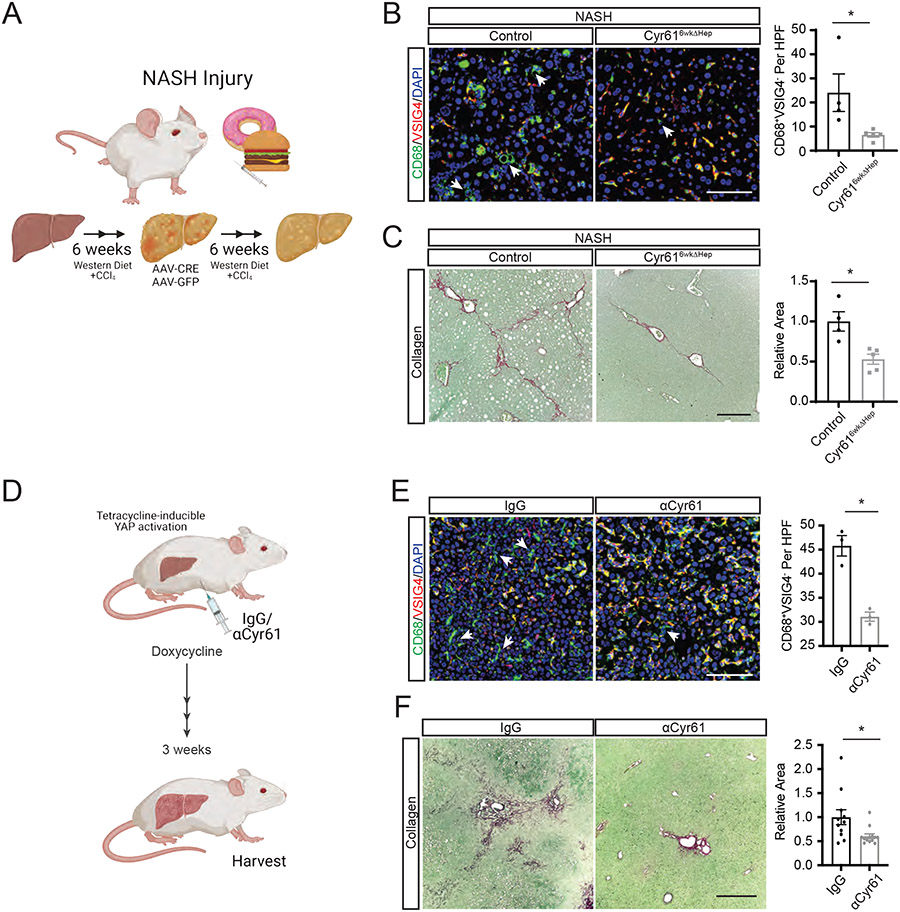
We next examined if Cyr61 intervention might be effective in the context of preexisting NASH injury. Cyr61fl/fl mice were placed on a NASH diet for 6 weeks. At week 6, mice were given either AAV8-TBG-EGFP (control) or AAV8-TBG-CRE (Cyr616wkΔHep). Mice remained on this diet for 6 more weeks completing a 12-week treatment (Fig 7A). Liver to body mass ratios were similar between all the NASH treatment groups (Fig S7A). Compared to controls, mice receiving the Cyr616wkΔHep intervention showed a significant reduction in infiltrating MoMΦs (6.55 vs 22.1 per HPF; p<0.01, Fig 7B) and a 47.2% reduction in fibrosis (p<0.01, Fig 7C). Mice on the NASH injury protocol for 6 weeks prior to their randomization into control and treatment groups showed a similar degree of fibrosis as 12 week-treated mice, but the collagen displayed less organization and a lack of bridging compared to 12 week-treated mice (Fig S7B).
Figure 7. Blocking Cyr61 in YAP-induced Fibrosis Blunts Macrophage Recruitment and Fibrosis.
(A) Cartoon of Cyr616wkΔHep intervention after 6 weeks of NASH diet. (B) Representative immunofluorescence imaging of specified proteins in control (n=6) or Cyr616wkΔHep-intervened (n=6) NASH livers. Scale bar=100μm. Quantification of CD68+VSIG4− macrophages to the right. HPF=high power field. Arrows indicate CD68+VSIG4− cells. (D) Cartoon of tetracycline-inducible YAP liver fibrosis (YAP-Tg) with αCyr61 treatment. (D) Representative immunofluorescence imaging of specified proteins in YAP-Tg livers treated with IgG (n=3) or αCyr61 (n=3). Scale bar=100μm. Quantification of CD68+VSIG4− macrophages to the right. HPF=high power field. Arrows indicate CD68+VSIG4− cells. (F) Representative picrosirius red staining of livers from YAP-Tg mice treated with IgG (n=11) or αCyr61 (n=13) antibody. Scale bar=500μm. Quantification to the right. Mean and SEM plotted; p values calculated with Mann-Whitney U test. *p<0.05, **p<0.01, ***p<0.001.
We then developed a Cyr61 blocking antibody targeting the N-terminal region of Cyr61 (αCyr61), a region known to bind to integrin αvβ5 (17). Integrin αvβ5 binding to Cyr61 was disrupted by αCyr61 in a dose-dependent manner. A CTGF/CCN2 antibody targeting the same region showed minimal interaction (42) (Fig S7C). αCyr61 recognizes human, mouse, and xenopus Cyr61 protein with high specificity (Fig S7D). To test αCyr61’s efficacy in blocking MΦ activation, we examined TNFα expression in human MΦs after Cyr61 treatment with or without αCyr61. Addition of αCyr61 in a dose-dependent manner reduced TNFα secretion compared to treatment with control IgG (Fig S7E). A similar reduction in TNFα production was observed in mouse BMDMs (p<0.01, Fig S7F).
Using the YAP-Tg genetic model of liver fibrosis (16), we examined if αCyr61 could blunt fibrotic development in vivo. The liver to body mass ratio of IgG and αCyr61 YAP-Tg were similar (Fig S7G). YAP-Tg mice were pre-treated with either non-immune human IgG or αCyr61. YAP was then induced in these mice for 21 days (Fig 7D). αCyr61 treatment reduced CD68+VSIG4− MoMΦ infiltration in YAP-Tg mice by 33% (45 vs 30 per HPF; p<0.05, Fig 7D). Expression of PDGFa and PDGFb were reduced after αCyr61 treatment by 75% and 50% respectively (p<0.05, Fig S7H). Lastly, αCyr61 treatment reduced collagen deposition by 44% compared to control IgG (p<0.05, Fig 7F). In summary, reducing Cyr61 after the initiation of NASH injury can limit the development of NASH liver fibrosis. Cyr61 reduction or interference are potentially effective means of limiting pathologic liver remodeling mediated by Cyr61 through its interference of MoMΦ infiltration/activation.
DISCUSSION
As NAFLD progresses to the more severe disease NASH, the key features that emerge are inflammation and fibrosis. Currently the best standard of care is lifestyle change (i.e. dietary change, increased exercise and/or bariatric surgery). However, even these are not fully effective in treating the sequelae of NASH. In non-responders to these treatments, the dominant signaling pathways include increased expression of inflammatory genes (43). As such, inflammation and fibrosis need to be addressed when investigating therapies for this disease. In this study we identified CYR61 as a regulator of fibrosis through activation of monocyte-derived macrophages (MoMΦ). We found dysregulation and increased expression of CYR61 in NASH injured livers. During NASH injury, loss of CYR61 limits the inflammatory and fibrotic signature of MoMΦs, reduces fibrosis, and improves glucose tolerance.
Monocytes and macrophages (MΦs) have become a center of focus for fibrotic signaling. Indeed, we previously demonstrated that clodronate ablation of liver resident and circulating MΦ/monocytes significantly reduces fibrotic development in a fibrotic liver injury model (16). However, as more MΦ subtypes are identified in the liver, it is important to understand their separate contributions to fibrosis. Ly6Chi MoMΦs are the initial phenotype of infiltrating monocytes differentiating into macrophages and are pro-inflammatory, expressing TNFα and IL-1β (44). These are recruited to the liver after injury and contribute to activation of stellate cells (27). Ly6Clo MoMΦs are thought to be anti-fibrotic and pro-repair, expressing matrix metalloproteinases and aiding in recovery from injury (39, 42). LCMs, the newly classified but still poorly understood MoMΦs, respond to pathogens and elicit a neutrophil response (45).
In this study, we find that CYR61 polarizes infiltrating monocytes towards inflammatory Ly6Chi MoMΦs and LCM phenotypes and away from pro-repair Ly6Clo MoMΦs. The specific role LCMs play in liver injury is largely unknown (44). Our analysis shows that CD11c+ LCMs have higher fibrotic IL-17a and TGFβ expression than Ly6Chi MoMΦs, pointing to a role for LCMs in fibrosis. Of note, CD11c-expressing macrophages have been implicated as major contributors to NASH fibrosis (46). Thus, LCMs could be an even greater contributor to fibrosis than Ly6Chi MoMΦs during NASH. CD11c+ bone marrow-derived macrophages along with myofibroblasts are central players in the NASH histological hallmark “crown-like structures” (CLS) that surround steatotic hepatocytes (47). In fact, IL-17a drives steatosis (48). However, the factor that activates CD11c+ macrophages and IL-17a signaling in CLS is unknown. We found that CYR61 activity in NASH promotes increased CD11c+ LCM populations thus providing a potential answer to this question.
There is some debate over the MΦ cell types (namely KCs or MoMΦs) which drive NASH liver fibrosis (29-35, 49). Here, we show that NASH injury increases populations of infiltrating MoMΦs over chow-fed mice whereas KC proportions remain similar after 12 weeks of injury. This suggests a predominant role for MoMΦs in driving early NASH fibrosis. Others have demonstrated that embryonically-derived KCs are gradually replaced by MoMΦs in the context of NASH injury (36, 37), further supporting the idea that bone-marrow derived MoMΦs are a main contributor to fibrosis. Indeed, loss of CYR61 during NASH injury reduces fibrosis while also reducing populations of infiltrating MoMΦs, with minimal effect on KC populations. CYR61 over-expression in chow-fed mice independently increases fibrosis, only in the presence of CCR2+ infiltrating monocytes. Although KCs may still play a role in NASH, CYR61 predominantly influences infiltrating monocytes to coordinate fibrosis.
Thus far, CYR61 is mainly known as an activator of inflammatory signaling in MΦs through NFκB/p65 (20). Here, we identified that IRAK4/SYK are upstream of NFκB/p65 activity. IRAK4/NFκB signaling is a well-known transducer of inflammatory signals, activating genes such as TNFα (50). SYK, a tyrosine kinase that transduces signals from intracellular immune receptor tyrosine-based activation motifs in MΦs, can also contribute to NFκB activation through the IRAK4 pathway (51). This pathway targets PDGFa and PDGFb, potent activators of hepatic fibroblasts and stellate cells which are key drivers of liver fibrosis (49, 52). Conserved NFκB binding sites in the promoters of PDGFa and PDGFb through multiple species indicate this to be an important mechanism, not only in mice, but potentially in humans and other species as well (Fig S8).
CYR61 has previously been shown to inhibit fibrosis when expressed in portal fibroblasts (21) or used to stimulate fibroblasts directly (23, 24). Our data demonstrate that MΦs act as an intermediary between CYR61-expressing hepatocytes and stellate cells. Therefore, we propose hepatocyte stress or injury stimulates CYR61 expression which in turn activates fibroblasts. Whereas direct activation of fibroblasts by CYR61 may be anti-fibrotic, CYR61-MΦ-fibroblast signaling is fibrotic. It is worth noting that in our Cyr61ΔHep livers where CYR61 is selectively lost from hepatocytes, it is present in other cell types such as cholangiocytes and endothelial cells. Thus, presentation of CYR61 from hepatocytes may be a key step in determining the course of NASH liver fibrosis.
Reduction of hepatic CYR61 after NASH injury has started limits the degree of liver fibrosis, suggesting this could be a viable target for NASH fibrosis. Whether this also improves metabolic NASH pathology after substantial injury has occurred should be investigated. CYR61 reduction prior to the introduction of a NASH diet improved glucose tolerance in this study through transcriptional rewiring. Several reports suggest that liver inflammation alters hepatic glucose and lipid homeostasis due to direct effects of inflammatory macrophages, like those activated by CYR61 (53-55). Alternatively, there may be direct effects on hepatocyte metabolism that CYR61 exerts in the context of injury, as has been shown for other cell types (24, 56, 57). Although deletion of CYR61 has beneficial effects on fibrosis, inflammation, and metabolic signaling, its known roles in ductular reaction (22), angiogenesis (58, 59) and cellular senescence (60, 61) suggest there may be deleterious long-term aspects of repair and regeneration that should be considered.
Effective pharmacologic therapy has yet to be approved for NAFLD/NASH treatment, most likely due to the complex nature of this disease. Current therapeutics in development target specific aspects of the metabolic syndrome such as fibrosis, inflammation, glucose metabolism, or other features. Thus far, none have proceeded past Phase III (62). CYR61 antagonism may represent a strong candidate for a multi-dimensional therapeutic. Notably, PPARα signaling was improved in CYR61 knockout livers. Several drugs that increase PPARα activity have been tested in NAFLD/NASH, like saroglitazar (63, 64) and elafibranor (65). However, these have shown little to no effect on fibrosis in clinical trials (66). As such, more complete therapeutics are needed to combat a variety of NAFLD/NASH symptoms. CYR61 antagonism in patients with fatty liver disease potentially could improve glucose tolerance and fatty acid metabolism through reduction of liver inflammation and fibrosis.
There are several limitations to this study. Here, we focused on liver fibrosis and its improvement after CYR61 reduction because fibrosis is consistently linked to improvement in long-term outcomes. Whether fibrotic reduction in this manner leads to reduced morbidity and extended lifespan should be directly explored. Furthermore, reduction of hepatocytic CYR61 prior to the introduction of a NASH diet leads mice with improved glycemic tolerance. If reduction of CYR61 can improve glucose tolerance in the context of preexisting liver disease needs to be explored. Other aspects of metabolic syndrome associated with NASH such as hypertension, hyperlipidemia and hepatocellular carcinoma were not followed in this studied, but should be considered as CYR61 is investigated as a potential target in this area.
MATERIALS AND METHODS
Study design
In this study, we used scRNA-seq and CyTOF to identify critical cell types and cellular mechanisms that emerge in NASH that may serve as therapeutic targets for advanced fibrosis. We as well as others had previously identified Cyr61 as a potential critical target in the pathogenesis of liver fibrosis, although its mechanism was unclear. In the context of Cyr61 and NASH, scRNA-seq and CyTOF identified important cellular and biochemical mechanisms such as the development of pro-fibrotic monocytes and the SYK/IRAK4/NFκB signaling cascade. All results in this study were reproduced in independent cohorts of mice and in biological replicates for cell culture. Activity of the Cyr61 blocking antibody was validated in multiple samples of discarded patient peripheral monocytes.
Mouse lines
C57BL/6J, Cyr61-enhanced green fluorescent protein (EGFP) reporter mice (http://gensat.org/), conditional Cyr61 floxed (Cyr61fl/fl) (23), Ccr2gfp KI/KO (CCR2−/−) (Jackson Laboratories), and tetracycline-inducible Yap-S127A (TetOYAP/YAP-Tg) (67) were used in this study. Experiments were started at 8-12 weeks of age. Male and female mice were used for collagen analysis and AAV-TBG-CYR61 studies. Sequencing and CyTOF analysis were performed only with male mice. A minimum of 3 mice were used for all displayed results. All mouse procedures and protocols were approved by an Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility.
To simulate fibrotic injury, CCl4 was diluted to a 25% solution in corn oil and delivered intraperitoneally at a dosage of 2μL per gram. Mice were injected every third day for 21 days and sacrificed on the 21st day for a total of 7 injections (chronic injury) or once and sacrificed 24 hours later (acute injury).
NASH injury modeling
To simulate the steatosis, inflammation, and fibrosis of non-alcoholic steatohepatitis, mice were fed a high-fat, high-cholesterol diet (Envigo TD.120528) and drinking water with 23.1g/L D-fructose (Millipore-Sigma) and 18.9g/L D-glucose (Millipore-Sigma) ad libitum. Once per week, mice were injected with 2μL of 10% carbon tetrachloride diluted in corn oil per gram intraperitoneally for 12 weeks (26).
AAV gene delivery, YAP overexpression, and Cyr61 expression
Adeno-associated virus (AAV8)- thyroid hormone-binding globulin (TBG)- cyclization recombinase (CRE) (AAV-CRE; Addgene) was delivered to the indicated genotypes retro-orbitally at 1011 pfu/mouse; AAV8-TBG-GFP (Addgene) or AAV8-TBG-NULL (Addgene) were used as controls. To induce expression of TetOYAP, 3 days after AAV-CRE injections, mice were given 1mg/mL doxycycline ad libitum in drinking water. These mice are referred to as “YAP-Tg” in the text. To induce CYR61 expression, AAV8-TBG-CYR61 (16) was administered retro-orbitally at 3.5x1011pfu/mouse.
Tissue preparation, immunohistochemistry, and RNA in situ hybridization
Tissue was fixed overnight in 10% buffered formalin (Fisher-Scientific) and embedded in paraffin for sectioning. For picrosirius red staining, tissue sections were rehydrated and incubated with 0.1% Direct Red (Millipore-Sigma) and 0.08% Fast Green FCF (Millipore-Sigma) in saturated picric acid (Millipore-Sigma) for 1 hour. For immunohistochemistry, tissue sections were rehydrated and Citrate-based Unmasking Solution (Vector Labs) was used prior to staining. RNA in situ hybridization of Cyr61 was carried out using RNAscope kits (ACD Bio). Images taken with Revolve microscope (Echo).
CyTOF staining
Single cell suspensions were made from frozen livers (preserved in FBS with 10% DMSO) and digested using Liver Digest Medium (Gibco) on the gentleMACS Octo Dissociator (Miltenyi-Biotec). Inter-hepatic leukocytes (IHL) were isolated using a 37.5% Percoll (Millipore-Sigma) solution. Cell viability was labeled using Intercalator-Rh103 (Fluidigm). TruStain FcX (Biolegend) was used as a blocking agent before surface antibody stain (Table S2). Phosphorylation panel: cells were permeabilized with methanol. Cytokine panel: cells were permeabilized with FoxP3 fixation and permeabilization solution (ThermoFisher) before incubation with antibodies. Samples were labeled with Intercalator-Ir (Fluidigm) and normalized using EQ Four Element Calibration Beads (Fluidigm). Samples were run on the Helios, a CyTOF system (Fluidigm). All washes, dilutions, and stains were performed in cell staining buffer (CSB: DPBS, 0.5% bovine serum albumin (BSA, Millipore-Sigma) and 0.02% sodium azide (Millipore-Sigma)).
Using Premium Cytobank, normalized FCS files were gated on DNA+, live cells (Rh103−), bead− prior to analysis. Automatic clustering was performed on gated populations using the Cytofkit R package (doi: 10.18129/B9.bioc.cytofkit) (68-70). Data were transformed with cytofAsinh and merged with ceil. t-distributed stochastic neighbor embedding (tSNE) was used for dimensionality reduction. Phenograph was selected from clustering for all data using the preset k=30. Data were visualized using the ShinyApp feature in Cytofkit. Data displayed were extracted from Phenograph clustering output. Cell type identification of clusters was performed using marker intensity heatmaps. Mean metal intensity (MMI) was calculated using Cytofkit Phenograph clustering output.
BMDM isolation and cell culture
Leg bones of C57Bl/6J mice were harvested and manually crushed and filtered to isolate bone marrow cells. Red blood cells were lysed (RBC Lysis buffer, eBioscience) and cells were resuspended in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco) supplemented with 10% FBS and 10 μg/mL recombinant murine M-CSF (Peprotech). Media was changed after 24 hours. After 72 hours, media was supplemented with 4 μg/mL recombinant human CYR61 protein (Peprotech). BMDM were analyzed at 30 minutes, 1 hour, and 24 hours after CYR61 treatment. To polarize BMDMs to a M1 and M2 phenotype, M0 BMDMs were treated for 24 hours with IMDM containing FBS/M-CSF and either, M1: LPS (Millipore-Sigma, 100 ng/mL) and IFNγ (Peprotech, 50 ng/mL) or M2: IL4 (Peprotech, 20 ng/mL), IL10 (Peprotech, 20 ng/mL), TGFβ1 (Peprotech, 20 ng/mL) with or without CYR61 (4 μg/mL) for 24 hours.
RAW264.7 or J774A.1 mouse macrophage cell lines (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% FBS (Gibco) and passaged using 0.25% Trypsin-EDTA (Gibco). For CYR61 protein treatment, media was supplemented with 4 μg/mL recombinant human CYR61 protein for 72 hours. JSH-23 was used over the course of this treatment at 30 mM (MedChemExpress). For Ad-Cyr61 treatment, cells were treated with either Ad-GFP-m-Cyr61 or Ad-GFP (Vector Biolabs) at 200 PFU/cell (RAW) or 125 PFU/cell (J7).
HIVE single-cell RNA sequencing data analysis
BMDMs from C57BL6/J mice as described above in “BMDM isolation and cell culture” were isolated. Four paired control and Cyr61 treated BMDM wells were measured at 24 hours after treatment by the HIVE scRNAseq platform (Honeycomb Biotechnologies, Inc). Libraries were sequenced on a Nova-Seq 6000 by MedGenome; 160 paired-end reads per library. BeeNet (Honeycomb Biotechnologies, Inc) was applied on the raw sequencing data to quantify the transcript count matrix per library. Downstream statistical analyses were performed by R package Seurat (71). Cells with more than 300 genes expressed and less than 12000 count were regarded as valid cells for the downstream analysis. Like the 10X scRNAseq analysis pipeline, SCT integration, UMAP dimension reduction and gene clustering/annotation analyses were performed on the valid cells. In addition, R package Monocle3 was applied to construct single-cell trajectories. Based on the Seurat processed data, a trajectory graph was learned, and the single cells were ordered by pseudotime.
αCyr61 in vivo blockade
Two days after AAV-CRE injection, YAP-Tg mice were injected intraperitoneally with 50 mg/kg αCyr61 or control IgG. The next day mice were provided 1 mg/mL doxycycline ad libitum in drinking water. 10 days after the initial injection, mice were again injected intraperitoneally with 50mg/kg αCyr61 or control IgG. Livers from these mice were harvested on day 21 after doxycycline administration.
Mouse embryonic fibroblast isolation
Primary cultures of mouse embryonic fibroblasts (MEFs) were prepared from Col1a1-EGFP reporter mice (41) based on the protocol outlined by Durkin (72). E13.5 embryos were isolated and identified for GFP+ using a UV flashlight. Embryos were minced in a dish containing 0.25% trypsin-EDTA (Gibco) then incubated at 37°C to disassociate the fibroblasts. MEFs were resuspended in DMEM with 10% FBS and plated in T75 flasks. Media was replaced after 24 hours and the cells were frozen down after further passaging. MEF experiments were performed before passage 10.
MEF stimulation
MEFs were plated and serum starved overnight in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 0.5% FBS. Media was treated with 4 μg/mL recombinant human Cyr61 or 20 ng/mL human TGFβ1. Cells were incubated for 48 hours and prepared for flow cytometry.
Statistical analysis
Results are shown as means ± standard error of the mean unless indicated otherwise. Statistical analysis was performed using Prism unless otherwise specified. Each experiment was performed a minimum of three independent trials in triplicate with the exception of mouse models where all animals are shown in the dot plots. In vitro experimental data shown are technical replicates, in vivo experimental data shown are biological replicates. Statistical tests are identified in each figure legend. To detect the markers per cluster/ cell type in the scRNA-seq experiments, differential expression analysis was performed comparing the cells from a given cell type and all the other cells using likelihood-ratio test for single cell gene expression (73).
Supplementary Material
Acknowledgments:
We appreciate the Konnikova lab for assistance with CyTOF panel design, analysis, and immunology expertise. The authors would also like to thank the Yale CyTOF facility. This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided. Graphic illustrations created using BioRender.com through the University of Pittsburgh Institutional license.
Funding:
Funding was provided by the National Institutes of Health through the following grants: R03 DK124743 (DY), R01 DK129552 (DY), R01 DK119968 (GIS), Pittsburgh Liver Research Center (P30 DK120531), The Yale Liver Center (P30 DK034989), The Yale Diabetes Research Center (P30 DK045735), and The Yale Flow Cytometry/Cancer Center (P30 CA016359).
Footnotes
Competing interests: XY, RM and MVP are full-time employees of Kadmon Corporation, LLC. The work was supported in part by funding from Kadmon Corporation, LLC. GIS serves on the following Scientific Advisory Boards: Merck, NovoNordisk, AstraZeneca, Aegerion, iMBP, 89bio, Janssen Research and Development, Ionis, Maze Therapeutics, Levels, Equator Therapeutics, Generian, Bayer, Kriya, Ionis, OrosoBio; GIS receives Investigator-Initiated Support from: Gilead Sciences, Astra Zeneca, Merck, Maze Therapeutics, Ionis, Regeneron. GIS is an inventor on Yale patents for liver-targeted mitochondrial uncoupling agents for the treatment of NAFLD, NASH, T2D and related metabolic disorders which are licensed by OrsoBio. All other authors declare no relevant conflicts of interest.
Data and materials availability: Data for bulk RNA-seq, 10x scRNA-seq and HIVE scRNA-seq have been submitted to NCBI GEO database under accession GSE199660. Both raw and processed data can be downloaded from NCBI GEO under accession GSE199640. Figure data are available in data file S1. Yap-Tg (67), Cyr61 flox (23) and Cola1a-EGFP (74) mice are available from Drs. Fernando Camargo, Lester Lau, and David Brenner under a material transfer agreement with the Boston Children’s Hospital, University of Illinois and University of California, respectively.
References and Notes
- 1.Arroyo-Johnson C, Mincey KD, Obesity Epidemiology Worldwide. Gastroenterol Clin North Am 45, 571–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerenziani S, Guarino MPL, Trillo Asensio LM, Altomare A, Ribolsi M, Balestrieri P, Cicala M, Role of Overweight and Obesity in Gastrointestinal Disease. Nutrients 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarwar R, Pierce N, Koppe S, Obesity and nonalcoholic fatty liver disease: current perspectives. Diabetes Metab Syndr Obes 11, 533–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavrogiannaki AN, Migdalis IN, Nonalcoholic fatty liver disease, diabetes mellitus and cardiovascular disease: Newer data. International Journal of Endocrinology 2013, 8 pages (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Hadi H, Di Vincenzo A, Vettor R, Rossato M, Cardio-Metabolic Disorders in Non-Alcoholic Fatty Liver Disease. International Journal of Molecular Sciences 20, 2215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godoy-Matos AF, Silva Júnior WS, Valerio CM, NAFLD as a continuum: From obesity to metabolic syndrome and diabetes. Diabetology and Metabolic Syndrome 12, 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomba R, Friedman SL, Shulman GI, Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ, The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55, 2005–2023 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, Torres-Gonzalez A, Gra-Oramas B, Gonzalez-Fabian L, Friedman SL, Diago M, Romero-Gomez M, Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology 149, 367–378.e365 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, Bugianesi E, Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nature Reviews Gastroenterology and Hepatology 15, 11–20 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Vilarinho S, Ajmera V, Zheng M, Loomba R, Emerging Role of Genomic Analysis in Clinical Evaluation of Lean Individuals With NAFLD. Hepatology 74, 2241–2250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen KF, Dufour S, Li F, Rothman DL, Shulman GI, Ethnic and sex differences in hepatic lipid content and related cardiometabolic parameters in lean individuals. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buzzetti E, Pinzani M, Tsochatzis EA, The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 65, 1038–1048 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Taylor RS, Taylor RJ, Bayliss S, Hagström H, Nasr P, Schattenberg JM, Ishigami M, Toyoda H, Wai-Sun Wong V, Peleg N, Shlomai A, Sebastiani G, Seko Y, Bhala N, Younossi ZM, Anstee QM, McPherson S, Newsome PN, Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology 158, 1611–1625.e1612 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Vilar-Gomez E, Calzadilla-Bertot L, Wai-Sun Wong V, Castellanos M, Aller-de la Fuente R, Metwally M, Eslam M, Gonzalez-Fabian L, Alvarez-Quiñones Sanz M, Conde-Martin AF, De Boer B, McLeod D, Hung Chan AW, Chalasani N, George J, Adams LA, Romero-Gomez M, Fibrosis Severity as a Determinant of Cause-Specific Mortality in Patients With Advanced Nonalcoholic Fatty Liver Disease: A Multi-National Cohort Study. Gastroenterology 155, 443–457.e417 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Mooring M, Fowl BH, Lum SZC, Liu Y, Yao K, Softic S, Kirchner R, Bernstein A, Singhi AD, Jay DG, Kahn CR, Camargo FD, Yimlamai D, Hepatocyte Stress Increases Expression of Yes-Associated Protein and Transcriptional Coactivator With PDZ-Binding Motif in Hepatocytes to Promote Parenchymal Inflammation and Fibrosis. Hepatology 71, 1813–1830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen CC, Lau LF, Functions and mechanisms of action of CCN matricellular proteins. International Journal of Biochemistry and Cell Biology 41, 771–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schober JM, Chen N, Grzeszkiewicz TM, Jovanovic I, Emeson EE, Ugarova TP, Ye RD, Lau LF, Lam SC, Identification of integrin alpha(M)beta(2) as an adhesion receptor on peripheral blood monocytes for Cyr61 (CCN1) and connective tissue growth factor (CCN2): immediate-early gene products expressed in atherosclerotic lesions. Blood 99, 4457–4465 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Imhof BA, Jemelin S, Ballet R, Vesin C, Schapira M, Karaca M, Emre Y, CCN1/CYR61-mediated meticulous patrolling by Ly6Clow monocytes fuels vascular inflammation. Proc Natl Acad Sci U S A 113, E4847–4856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai T, Chen C-C, Lau LF, Matricellular Protein CCN1 Activates a Proinflammatory Genetic Program in Murine Macrophages. The Journal of Immunology 184, 3223–3232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borkham-Kamphorst E, Steffen BT, Van de Leur E, Haas U, Tihaa L, Friedman SL, Weiskirchen R, CCN1/CYR61 overexpression in hepatic stellate cells induces ER stress-related apoptosis. Cell Signal 28, 34–42 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kim KH, Chen CC, Alpini G, Lau LF, CCN1 induces hepatic ductular reaction through integrin alphavbeta(5)-mediated activation of NF-kappaB. The Journal of clinical investigation 125, 1886–1900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K-H, Chen C-C, Monzon RI, Lau LF, Matricellular Protein CCN1 Promotes Regression of Liver Fibrosis through Induction of Cellular Senescence in Hepatic Myofibroblasts. Molecular and cellular biology 33, 2078–2090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun JI, Lau LF, The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nature cell biology 12, 676–685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Todorovic V, Chen CC, Hay N, Lau LF, The matrix protein CCN1 (CYR61) induces apoptosis in fibroblasts. The Journal of cell biology 171, 559–568 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchida T, Lee YA, Fujiwara N, Ybanez M, Allen B, Martins S, Fiel MI, Goossens N, Chou HI, Hoshida Y, Friedman SL, A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol 69, 385–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlmark KR, Weiskirchen R, Zimmermann HW, Gassler N, Ginhoux F, Weber C, Merad M, Luedde T, Trautwein C, Tacke F, Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50, 261–274 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Hasuzawa N, Tatsushima K, Wang L, Kabashima M, Tokubuchi R, Nagayama A, Ashida K, Ogawa Y, Moriyama Y, Nomura M, Clodronate, an inhibitor of the vesicular nucleotide transporter, ameliorates steatohepatitis and acute liver injury. Scientific reports 11, 5192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Zhang X, Lau JKC, Fu K, Lau HCH, Xu W, Chu ESH, Lan H, Yu J, Bone marrow-derived macrophage contributes to fibrosing steatohepatitis through activating hepatic stellate cells. Journal of Pathology 248, 488–500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid DT, Reyes JL, McDonald BA, Vo T, Reimer RA, Eksteen B, Kupffer Cells Undergo Fundamental Changes during the Development of Experimental NASH and Are Critical in Initiating Liver Damage and Inflammation. PloS one 11, e0159524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP, Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. The Journal of clinical investigation 115, 56–65 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai B, Dongiovanni P, Corey KE, Wang X, Shmarakov IO, Zheng Z, Kasikara C, Davra V, Meroni M, Chung RT, Rothlin CV, Schwabe RF, Blaner WS, Birge RB, Valenti L, Tabas I, Macrophage MerTK Promotes Liver Fibrosis in Nonalcoholic Steatohepatitis. Cell Metabolism 31, 406–421.e407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Zhou Y, Wang H, Zhang M, Qiu P, Zhang M, Zhang R, Zhao Q, Liu J, Crosstalk Between Liver Macrophages and Surrounding Cells in Nonalcoholic Steatohepatitis. Frontiers in Immunology 11, 1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, Gatto D, Sebbel P, Beerli RR, Sonderegger I, Kopf M, Saudan P, Bachmann MF, VSIG4, a B7 family–related protein, is a negative regulator of T cell activation. The Journal of clinical investigation 116, 2817–2826 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, Bujko A, Martens L, Thoné T, Browaeys R, De Ponti FF, Vanneste B, Zwicker C, Svedberg FR, Vanhalewyn T, Gonçalves A, Lippens S, Devriendt B, Cox E, Ferrero G, Wittamer V, Willaert A, Kaptein SJF, Neyts J, Dallmeier K, Geldhof P, Casaert S, Deplancke B, ten Dijke P, Hoorens A, Vanlander A, Berrevoet F, Van Nieuwenhove Y, Saeys Y, Saelens W, Van Vlierberghe H, Devisscher L, Scott CL, Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e338 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, Bruni CM, Ouyang Z, Li RZ, Sun X, Vu BCT, Pasillas MP, Ego KM, Gosselin D, Link VM, Chong LW, Evans RM, Thompson BM, McDonald JG, Hosseini M, Witztum JL, Germain RN, Glass CK, Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 52, 1057–1074.e1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remmerie A, Martens L, Thoné T, Castoldi A, Seurinck R, Pavie B, Roels J, Vanneste B, De Prijck S, Vanhockerhout M, Binte Abdul Latib M, Devisscher L, Hoorens A, Bonnardel J, Vandamme N, Kremer A, Borghgraef P, Van Vlierberghe H, Lippens S, Pearce E, Saeys Y, Scott CL, Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity 53, 641–657.e614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamoutounour S, Henri S, Lelouard H, d. Bovis B, d. Haar C, v. d. Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D, Bain CC, Mowat AM, R. e. Sousa C, Poulin LF, Malissen B, Guilliams M, CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. European Journal of Immunology 42, 3150–3166 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, Williams MJ, Dunbar DR, Manning JR, van Rooijen N, Fallowfield JA, Forbes SJ, Iredale JP, Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A 109, E3186–3195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlmark KR, Wasmuth HE, Trautwein C, Tacke F, Chemokine-directed immune cell infiltration in acute and chronic liver disease. Expert Rev Gastroenterol Hepatol 2, 233–242 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Krempen K, Grotkopp D, Hall K, Bache A, Gillan A, Rippe RA, Brenner DA, Breindl M, Far upstream regulatory elements enhance position-independent and uterus-specific expression of the murine alpha1(I) collagen promoter in transgenic mice. Gene Expr 8, 151–163 (1999). [PMC free article] [PubMed] [Google Scholar]
- 42.Tacke F, Zimmermann HW, Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 60, 1090–1096 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Sierro F, Evrard M, Rizzetto S, Melino M, Mitchell AJ, Florido M, Beattie L, Walters SB, Tay SS, Lu B, Holz LE, Roediger B, Wong YC, Warren A, Ritchie W, McGuffog C, Weninger W, Le Couteur DG, Ginhoux F, Britton WJ, Heath WR, Saunders BM, McCaughan GW, Luciani F, MacDonald KPA, Ng LG, Bowen DG, Bertolino P, A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity 47, 374–388.e376 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Papachristoforou E, Ramachandran P, Macrophages as key regulators of liver health and disease. Int Rev Cell Mol Biol 368, 143–212 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Sierro F, Evrard M, Rizzetto S, Melino M, Mitchell AJ, Florido M, Beattie L, Walters SB, Tay SS, Lu B, Holz LE, Roediger B, Wong YC, Warren A, Ritchie W, McGuffog C, Weninger W, Le Couteur DG, Ginhoux F, Britton WJ, Heath WR, Saunders BM, McCaughan GW, Luciani F, MacDonald KPA, Ng LG, Bowen DG, Bertolino P, A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity 47, 374–388 e376 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Itoh M, Suganami T, Kato H, Kanai S, Shirakawa I, Sakai T, Goto T, Asakawa M, Hidaka I, Sakugawa H, Ohnishi K, Komohara Y, Asano K, Sakaida I, Tanaka M, Ogawa Y, CD11c+ resident macrophages drive hepatocyte death-triggered liver fibrosis in a murine model of nonalcoholic steatohepatitis. JCI Insight 2, e92902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hachiya R, Tanaka M, Itoh M, Suganami T, Molecular mechanism of crosstalk between immune and metabolic systems in metabolic syndrome. Inflammation and Regeneration 42, 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomes AL, Teijeiro A, Buren S, Tummala KS, Yilmaz M, Waisman A, Theurillat JP, Perna C, Djouder N, Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer cell 30, 161–175 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Ying HZ, Chen Q, Zhang WY, Zhang HH, Ma Y, Zhang SZ, Fang J, Yu CH, PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics. Molecular medicine reports 16, 7879 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim TW, Febbraio M, Robinet P, DuGar B, Greene D, Cerny A, Latz E, Gilmour R, Staschke K, Chisolm G, Fox PL, DiCorleto PE, Smith JD, Li X, The critical role of IRAK4-mediated NFκB activation in modified-LDL-induced atherosclerosis. Journal of Immunology (Baltimore, Md. : 1950) 186, 2871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi YS, Son YJ, Ryou C, Sung GH, Kim JH, Cho JY, Functional roles of Syk in macrophage-mediated inflammatory responses. Mediators Inflamm 2014, 270302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borkham-Kamphorst E, Weiskirchen R, The PDGF system and its antagonists in liver fibrosis. Cytokine & Growth Factor Reviews 28, 53–61 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Okin D, Medzhitov R, The Effect of Sustained Inflammation on Hepatic Mevalonate Pathway Results in Hyperglycemia. Cell 165, 343–356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Xie Z, Yu H, Du H, Wang X, Cai J, Qiu Y, Chen R, Jiang X, Liu Z, Li Y, Chen T, TLR2 inhibition ameliorates the amplification effect of LPS on lipid accumulation and lipotoxicity in hepatic cells. Annals of Translational Medicine 9, 1429–1429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Chen Y, Zhao L, Chen Y, Mei M, Li Q, Huang A, Varghese Z, Moorhead JF, Ruan XZ, Inflammatory stress exacerbates hepatic cholesterol accumulation via disrupting cellular cholesterol export. Journal of gastroenterology and hepatology 27, 974–984 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Won JH, Choi JS, Jun JI, CCN1 interacts with integrins to regulate intestinal stem cell proliferation and differentiation. Nat Commun 13, 3117 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, Elaskandrany M, Lau LF, Lazzaro D, Grant MB, Chaqour B, Interplay between CCN1 and Wnt5a in endothelial cells and pericytes determines the angiogenic outcome in a model of ischemic retinopathy. Scientific reports 7, 1405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leu SJ, Lam SCT, Lau LF, Pro-angiogenic activities of CYR61 (CCN1) mediated through integrins αvβ3 and α6β1 in human umbilical vein endothelial cells. Journal of Biological Chemistry 277, 46248–46255 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Chen N, Leu SJ, Todorović V, Lam SCT, Lau LF, Identification of a novel integrin αvβ3 binding site in CCN1 (CYR61) critical for pro-angiogenic activities in vascular endothelial cells. Journal of Biological Chemistry 279, 44166–44176 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Jun JI, Lau LF, CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat Commun 11, 1242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng M, Peng H, Yao R, Zhang Z, Mao G, Yu H, Qiu Y, Inhibition of cellular communication network factor 1 (CCN1)- driven senescence slows down cartilage inflammaging and osteoarthritis. Bone, (2020). [DOI] [PubMed] [Google Scholar]
- 62.Fraile JM, Palliyil S, Barelle C, Porter AJ, Kovaleva M, Non-Alcoholic Steatohepatitis (NASH) - A Review of a Crowded Clinical Landscape, Driven by a Complex Disease. Drug Des Devel Ther 15, 3997–4009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gawrieh S, Noureddin M, Loo N, Mohseni R, Awasty V, Cusi K, Kowdley KV, Lai M, Schiff E, Parmar D, Patel P, Chalasani N, Saroglitazar, a PPAR-alpha/gamma Agonist, for Treatment of NAFLD: A Randomized Controlled Double-Blind Phase 2 Trial. Hepatology 74, 1809–1824 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Goyal O, Nohria S, Goyal P, Kaur J, Sharma S, Sood A, Chhina RS, Saroglitazar in patients with non-alcoholic fatty liver disease and diabetic dyslipidemia: a prospective, observational, real world study. Scientific reports 10, 21117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, Serfaty L, Romero-Gomez M, Boursier J, Abdelmalek M, Caldwell S, Drenth J, Anstee QM, Hum D, Hanf R, Roudot A, Megnien S, Staels B, Sanyal A, Mathurin P, Gournay J, Nguyen-Khac E, De Ledinghen V, Larrey D, Tran A, Bourliere M, Maynard-Muet M, Asselah T, Henrion J, Nevens F, Cassiman D, Geerts A, Moreno C, Beuers UH, Galle PR, Spengler U, Bugianesi E, Craxi A, Angelico M, Fargion S, Voiculescu M, Gheorghe L, Preotescu L, Caballeria J, Andrade RJ, Crespo J, Callera JL, Ala A, Aithal G, Abouda G, Luketic V, Huang MA, Gordon S, Pockros P, Poordad F, Shores N, Moehlen MW, Bambha K, Clark V, Satapathy S, Parekh S, Reddy RK, Sheikh MY, Szabo G, Vierling J, Foster T, Umpierrez G, Chang C, Box T, Gallegos-Orozco J, Elafibranor, an Agonist of the Peroxisome Proliferator–Activated Receptor–α and –δ, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology 150, 1147–1159.e1145 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Fraile JM, Palliyil S, Barelle C, Porter AJ, Kovaleva M, in Drug Design, Development and Therapy. (Dove Press, 2021), vol. 15, pp. 3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR, YAP1 increases organ size and expands undifferentiated progenitor cells. Current Biology 17, 2054–2060 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HTK, De Lafaille MAC, Poidinger M, Newell EW, Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. Cell reports 11, 1822–1833 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KWW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, Greter M, Ginhoux F, Newell EW, High-dimensional analysis of the murine myeloid cell system. Nature Immunology 2014 15:December 15, 1181–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J, Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Comput Biol 12, e1005112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durkin ME, Qian X, Popescu NC, Lowy DR, Isolation of Mouse Embryo Fibroblasts. Bio Protoc 3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R, Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krempen K, Grotkopp D, Hall K, Bache A, Gillan A, Rippe RA, Brenner DA, Breindl M, Far upstream regulatory elements enhance position-independent and uterus-specific expression of the murine α1(I) collagen promoter in transgenic mice. Gene Expression 8, 151–163 (1999). [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells. Nature Communications 2017 8:1 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mcinnes L, Healy J, Melville J, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv e-prints 1802.03426, (2020). [Google Scholar]
- 82.Lee BT, Barber GP, Benet-Pagès A, Casper J, Clawson H, DIekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee CM, Muthuraman P, Nassar LR, Nguy B, Pereira T, Perez G, Raney BJ, Rosenbloom KR, Schmelter D, Speir ML, Wick BD, Zweig AS, Haussler D, Kuhn RM, Haeussler M, Kent WJ, The UCSC Genome Browser database: 2022 update. Nucleic Acids Research 50, D1115–D1122 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X, Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Research 31, 3651 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM, PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Krempen K, Grotkopp D, Hall K, Bache A, Gillan A, Rippe RA, Brenner DA, Breindl M, Far upstream regulatory elements enhance position-independent and uterus-specific expression of the murine α1(I) collagen promoter in transgenic mice. Gene Expression 8, 151–163 (1999). [PMC free article] [PubMed] [Google Scholar]
- 86.Charni-Natan M, Goldstein I, Protocol for Primary Mouse Hepatocyte Isolation. STAR Protoc 1, 100086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.