Abstract
Lipopolysaccharide (LPS), the major outer membrane component of gram-negative bacteria, is a potent endotoxin that triggers cytokine-mediated systemic inflammatory responses in the host. Plasma lipoproteins are capable of LPS sequestration, thereby attenuating the host response to infection, but ensuing dyslipidemia severely compromises this host defense mechanism. We have recently reported that Escherichia coli J5 and Re595 LPS chemotypes that contain relatively short O-antigen polysaccharide side chains are efficiently redistributed from high-density lipoproteins (HDL) to other lipoprotein subclasses in normal human whole blood (ex vivo). In this study, we examined the role of the acute-phase proteins LPS-binding protein (LBP) and phospholipid transfer protein (PLTP) in this process. By the use of isolated HDL containing fluorescent J5 LPS, the redistribution of endotoxin among the major lipoprotein subclasses in a model system was determined by gel permeation chromatography. The kinetics of LPS and lipid particle interactions were determined by using Biacore analysis. LBP and PLTP were found to transfer LPS from HDL predominantly to low-density lipoproteins (LDL), in a time- and dose-dependent manner, to induce remodeling of HDL into two subpopulations as a consequence of the LPS transfer and to enhance the steady-state association of LDL with HDL in a dose-dependent fashion. The presence of LPS on HDL further enhanced LBP-dependent interactions of LDL with HDL and increased the stability of the HDL-LDL complexes. We postulate that HDL remodeling induced by LBP- and PLTP-mediated LPS transfer may contribute to the plasma lipoprotein dyslipidemia characteristic of the acute-phase response to infection.
Lipopolysaccharide (LPS), a major outer membrane constituent of gram-negative bacteria, is a potent endotoxin that, through the activation of cellular immunity, induces a cytokine-mediated systemic inflammatory response in the host (5). Lipopolysaccharide-binding protein (LBP) is an acute-phase protein responsible for the binding and transport of LPS in circulation (15). The acute-phase (AP) response is characterized by increased plasma LBP levels, from 5 to 15 mg/liter to 50 to 100 mg/liter (14), and a 5- to 20-fold decrease in plasma lipoprotein cholesterol levels (10). Delivery of LPS by LBP to macrophage receptors initiates signal transduction pathways (20) that lead to the increased release of proinflammatory cytokines (23). However, the delivery of LPS to high-density lipoprotein (HDL) (3) by LBP, which has been found to reside exclusively on HDL (22), results in the attenuation of the immune response to infection. LBP has indeed been detected in association with low-density lipoprotein (LDL) only under conditions of dyslipidemia, such as that observed in septic patients (1). The observation that patients with high circulating LBP levels have a better prognosis than those with lower levels (15) suggests that LBP plays an essential role in LPS transfer in addition to phospholipid transport (24). Phospholipid transfer protein (PLTP) is essential in HDL metabolism for the regulation of phospholipid transfer from cell membranes to HDL (18) and is therefore capable of modulating HDL size and composition (19). These properties indicate that PLTP protein is essential for the maintenance of normal plasma HDL levels (6). In addition, PLTP has been shown to transfer LPS from artificial vesicles to reconstituted HDL in a model system (4). Both LBP and PLTP, therefore, promiscuously bind phospholipids and LPS and, on the basis of structural homology, belong to a family of lipid transfer proteins which includes bactericidal permeability-increasing protein and cholesteryl ester transfer protein (8).
Systemic inflammatory reactions are generally associated with significant changes in lipoprotein homeostasis, including a reduction in cholesterol concentrations and extensive remodeling of HDL (7). During the acute-phase response, apolipoprotein (Apo) A-I is displaced by serum amyloid A, which results in a modified HDL population termed AP-HDL (2, 7). PLTP, in turn, generates pre-beta HDL from AP-HDL which is depleted of ApoA-I more effectively than from normal HDL in vitro (17), indicating a rapid turnover of AP-HDL.
We have previously demonstrated that all plasma lipoprotein subclasses sequester LPS under simulated physiological conditions, that HDL has the highest binding capacity for LPS, and that HDL-bound LPS is redistributed to LDL and very-low-density lipoprotein (VLDL) (11). The rapid uptake of LPS by HDL appears to serve as a first line of defense against the sustained activation of cellular immunity by LPS in the host, whereas the mechanism and consequences of LPS binding to LDL and VLDL remain to be determined.
In the present study, we examined the role of LBP and PLTP in the transfer of LPS from HDL in a model system containing isolated lipoprotein fractions by using high-performance gel permeation chromatography (HPGC) and determined the interaction kinetics and dependence of LPS transfer by using surface plasmon resonance. In view of the known LPS-binding properties of LPB and PLTP, we propose that lipid transport proteins may regulate LPS shuttling between plasma lipoproteins.
MATERIALS AND METHODS
Materials.
Escherichia coli serotype J5 LPS was from List Biological Laboratories (Campbell, Calif.), and LBP was from the Xoma Corporation (Berkeley, Calif.) and Hycult Biotechnology (Uden, The Netherlands). The PLTP was a generous gift from Matti Jauhiainen (Department of Biochemistry, Public Health Institute, Helsinki, Finland). Rabbit antiserum to ApoA-I was kindly provided by Jean-Charles Fruchard (University of Lille, Lille, France), and LBP was provided by Peter S. Tobias (Scripps Research Center, La Jolla, Calif.). Anti-ApoB was from Beckman (Mijdrecht, The Netherlands). The fluorescent label, 7-nitrobenz-2-oxa-1,3 diazole fluoride (NBD-F), was from Molecular Probes Inc. (Eugene, Oreg.). All buffers were reconstituted with pyrogen-free water (Braun Medical AG, Melsungen, Germany). Endotoxin contamination in the buffers was found to be <1.2 endotoxin units/ml. The Biacore 2000 biosensor system, the amine coupling kit, and the CM-5 sensor chips (research grade) were from Biacore AB (Uppsala, Sweden).
NBD labeling of LPS.
E. coli J5 LPS was labeled with NBD-F as previously described (11). The stoichiometry of labeling was found to be approximately one molecule of NBD per four molecules of LPS, and the concentration of NBD-LPS was 0.73 μg/ml as determined by a KDO (2-keto-3-deoxyoctulosonic acid) assay (9). The biological activities of labeled and unlabeled LPS were previously found to be essentially similar (11).
Lipoprotein isolation by HPGC.
Native HDL, LDL, and VLDL, which represent the major lipoprotein subclasses, were quantitatively isolated from plasma by size exclusion chromatography in a system consisting of a Superose 6 HR 10/30 column (Pharmacia Biotech, Uppsala, Sweden), a PU-980 ternary pump, an LG-980-02 linear degasser, an FP-920 fluorescence detector, and a UV-975 UV-visible light detector (Jasco, Tokyo, Japan). An extra P-50 pump (Pharmacia Biotech) was used for in-line cholesterol detection with PAP enzymatic reagent (Biomerieux, Marcy l'Etoile, France) at a flow rate of 0.1 ml/min. Lipoprotein fractions from 100-μl aliquots of citrated plasma were eluted from the matrix at a flow rate of 0.3 ml/min with Tris-buffered saline (10 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing 0.005% (vol/vol) Tween 20. Fractions were collected, and the separated lipoproteins were concentrated to a final volume of approximately 100 μl by using Centricon-100 filters (Amicon, Beverly, Mass.). The protein concentration of the combined LDL and VLDL fractions was 6 mg of protein/ml, and that of the HDL fraction was 10 mg/ml. The total cholesterol recovery of the isolated lipoprotein fractions was >90% of that in plasma. Samples were processed immediately or frozen in liquid nitrogen and stored at −80°C. A computer analysis of chromatograms for qualitative lipoprotein analysis was performed by means of Borwin Chromatographic software, version 1.23 (JMBS Developments, Le Fontanil, France).
Lipoprotein model system.
Isolated native HDL (30 μl containing 10 mg of total protein/ml) was loaded with NBD-LPS (6 μg/ml) in Tris-buffered saline (10 mM Tris-HCl, pH 7.4, 150 mM NaCl) by incubation for 1 h at 37°C. Isolated VLDL (1 mg of total protein/ml) and LDL (5 mg of total protein/ml) were subsequently added in aliquots of 30 μl followed by the addition of LBP to final concentrations of 35, 70, or 135 mg/liter. The final reconstituted mixture contained lipoprotein populations in the same proportions as in plasma but at 10-fold-lower concentrations. Samples were drawn after 1, 2, 12, and 24 h of incubation at 37°C. Lipoprotein-associated LPS fluorescence and cholesterol profiles were determined by HPGC analysis as previously described (11). PLTP was added to the mixture to a final concentration of 475, 780, or 1,460 nmol/ml/h, and samples were drawn for HPGC analysis after 1, 12, and 24 h of incubation at 37°C.
Biacore analysis.
Surface plasmon resonance was used to determine the optimal binding of HDL to rabbit anti-human ApoA-I. ApoA-I antibodies were coupled at different concentrations to an activated CM-5 sensor chip according to the manufacturer's instructions. A control channel on each sensor chip was coated with a nonspecific rabbit IgG antibody that yielded a baseline binding signal of 4,500 to 5,000 reading units (RU). Specific binding to the anti-ApoA-I channels was always corrected for nonspecific binding to the control channels. Isolated native HDL at a protein concentration of 1 mg/ml in Hopes buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 3.4 mM EDTA) was injected for 600 s at a flow rate of 5 μl/min, and HDL capture was monitored in real time. E. coli J5 LPS (average monomer size, 2,300 kDa), at 2.5, 5.0, and 20 μM in HEPES buffer, was subsequently injected at a flow rate of 20 μl/min for 120 s, and the binding of LPS to HDL was monitored in real time. Regeneration of the sensor chip was achieved by a 5-min wash with a solution of 3 M potassium isothiocyanate followed by a 2-min wash with 10 mM glycine, pH 2.0, and it was finally equilibrated with HEPES buffer. All analyses were done at a constant temperature of 25°C. Similarly, LDL at a protein concentration of 0.5 mg/ml in HEPES buffer in the absence or presence of LBP (0 to 17 nM) or PLTP (0 to 1,460 nmol/ml/h) were injected for 2 min at a flow rate of 20 μl/min for the determination of binding to immobilized HDL.
RESULTS
LBP mediates transfer of LPS from HDL and induces particle remodeling.
The transfer of fluorescent LPS from HDL in our model system after incubation for 24 h is depicted in Fig. 1A. In the absence of added LBP, a minimal amount of LPS fluorescence appears to localize with LDL or VLDL. In the presence of LBP at the highest concentration examined (135 mg/liter), <95% of the decrease in the HDL-associated LPS fluorescence signal was recovered in the LDL and VLDL fractions. That the transfer of LPS from HDL apparently results in the remodeling of the HDL particles is evident by changes in the average size distribution of the HDL population, indicated by shoulders in the HDL fluorescence profile, with average molecular weights of 450 and 150 kDa (in addition to the reduced normal HDL peak of 300 kDa). The cholesterol profile, however, only reveals HDL with an average molecular weight of 400 kDa. This result indicates that a proportion of the HDL population which still contained residual fluorescent LPS has been reduced in size from 300 to 150 kDa and contained a significantly reduced amount of cholesterol.
FIG. 1.
A representative example of chromatographic profiles showing the LBP-induced (A) and PLTP-induced (B) changes in cholesterol and LPS distribution in HDL, LDL, and VLDL. After the loading of HDL with LPS, the LPS fluorescence and cholesterol distribution among the lipoprotein subclasses were measured in the absence or presence of LBP or PLTP after 24 h of incubation.
LPS transfer from HDL mediated by LBP is depicted in Fig. 2. The decrease in HDL-bound LPS is paralleled by an almost identical dose-dependent increase in LDL- and VLDL-associated LPS fluorescence at all time points after 1 h. The bulk of the LPS transferred from HDL (55%) appears to have been transferred within 2 h, while the remaining LPS (13%) appears to have been transferred in the subsequent 8 to 24 h.
FIG. 2.
Percent change (compared to the total signal) of LBP-dependent LPS transfer from HDL (A) to LDL and VLDL (B) at different LBP concentrations over time. Corrections were made for baseline differences (<10% of the total signal) in LPS distribution without added exogenous LBP. All data points were normalized against the fluorescence signal measured after 1 h of incubation and represent the means of duplicates ± standard errors of the means (error bars).
PLTP mediates shuttling of LPS from HDL and induces particle remodeling.
The transfer of LPS from HDL in our model system after incubation for 24 h is depicted in Fig. 1B. In the absence of additional PLTP, LPS is recovered almost exclusively in the HDL fraction with only a minimal amount detectable in the LDL and VLDL fractions. The presence of PLTP at the highest concentration examined (1,460 nmol/ml/h) resulted in a 25% decrease in HDL-associated LPS fluorescence, of which approximately 50% is recovered in the LDL fraction after 24 h. Apart from LPS transfer, the presence of additional PLTP also resulted in a decreased HDL retention time, indicative of an increase in the average size of the HDL population from 300 to 400 kDa, which is apparent in both the fluorescence and cholesterol chromatograms. LPS transfer from HDL mediated by PLTP also appears to be dose- and time-dependent (Fig. 3). The 13 and 25% decreases in the LPS signal associated with the HDL fraction (Fig. 3A) at time points 12 and 24 h (Fig. 3A) are accompanied by 4 and 7% increases in LDL- and VLDL-bound LPS, respectively (Fig. 3B).
FIG. 3.
Percent change (compared to the total signal) of PLTP-dependent LPS transfer from HDL (A) to LDL and VLDL (B) at different PLTP concentrations over time. Corrections were made for baseline differences in LPS distribution (<5% of the total signal) without added exogenous PLTP. All data points were normalized to the fluorescence signal measured after a 1-h incubation time and represent the means of duplicates ± standard deviations (error bars).
Kinetic analysis of HDL interactions.
To study the interactions that occur during LBP- and PLTP-mediated LPS transfer from HDL in more detail, HDL was captured on the surface of a sensor chip (CM-5) via immobilized polyclonal anti-ApoA-I antibodies (Fig. 4A). Quantitative binding of HDL was found to be proportional to the concentration of antibody present on the sensor chip and dissociation of HDL was found to be negligible (Kd = 10−5/s). Capture of HDL could therefore be efficiently achieved by binding to immobilized ApoA-I antibodies. The observation that anti-ApoB gave no positive signal demonstrated the specificity of HDL capture.
FIG. 4.
Sensorgrams representing immobilization of HDL by anti-ApoA-I coated on a CM-5 sensor chip (A). Immobilization of HDL was with 5,150 (line 1), 2,000 (line 2), and 580 (line 3) RU bound anti-ApoA-I antibody at a flow rate of 5 μl/min. (B) Capture of LPS by HDL at three different concentrations. HDL was immobilized with a 5,150-RU anti-ApoA-I. All sensorgrams were corrected for nonspecific background.
LPS binding to HDL.
Immobilized HDL was exposed to E. coli J5 LPS at concentrations of 2.5, 5.0, and 20 μM, and binding was determined in real time (Fig. 4B). The calculated association and dissociation constants are given in Table 1. HDL appeared to bind LPS in a dose-dependent manner, with the exception of the highest concentration of 20 μM LPS, which resulted in non-Langmuirian kinetics. LPS was therefore used at a concentration of 5 μM in all further experiments.
TABLE 1.
Kinetic parameters of LPS binding to HDLa
| Parameterb | Value at LPS concn (μM) of:
|
||
|---|---|---|---|
| 2.5 | 5.0 | 20.0 | |
| ka (M−1 s−1) | 9.38e3 | 5.36e3 | 1.51e3 |
| kd (s−1) | 2.9e−3 | 2.3e−3 | 3.0e−3 |
| Rmax (RU) | 54.5 | 94.0 | 128.0 |
| KD (M) | 3.1e−7 | 4.3e−7 | 2.0e−6 |
Calculations were performed with BIA evaluation software (version 3.2).
ka, association constant; kd, dissociation constant; KD = kd/ka.
Effect of LBP and LPS on HDL and LDL interactions.
The interaction between HDL and LDL and the influence of LBP and LPS on a complex formation were examined in detail (Fig. 5). LDL clearly bound to HDL (Fig. 5, sensorgram 1, region A). The addition of a mixture of LDL and LBP (8.4 nM) enhanced complex formation (Fig. 5, sensorgram 2, region A). In both instances, the subsequent elution with buffer resulted in a nearly complete dissociation of the HDL-LDL complex during the 300-s washing period, as illustrated by a return of the binding signal to the baseline level. The presence of LPS on HDL further enhanced LDL binding (Fig. 5, sensorgram 3, region A). A mixture of LDL and LBP (8.4 nM) gave maximum complex formation with HDL preloaded with LPS (Fig. 5, sensorgram 4, region A). LPS therefore appeared to increase the stability of the HDL-LDL complex, because the washing step resulted in only an approximately 40% dissociation of the complex, as illustrated by a sustained binding signal after 350 s relative to the maximum signals at 100 s (Fig. 5, sensorgrams 3 and 4). The presence of LDL in the complex was unequivocally demonstrated with the use of anti-ApoB antibodies. Both sensorgrams 3 and 4 displayed a 25- to 50-RU signal of antibody binding, illustrated by the increase in the slope of the signal after the injection of the antibody solution (Fig. 5, region B), while no interaction was apparent in sensorgrams 1 and 2.
FIG. 5.
A representative example of the sensorgrams of LDL complexation with HDL in the absence (broken lines) or presence (solid lines) of LPS and the association of LDL with HDL in the absence (curves 1 and 3) or presence of LBP (curves 2 and 4). A second injection with anti-ApoB for LDL identification in the LDL-HDL complex is indicated by region B. All sensorgrams were corrected for nonspecific background.
Effect of LBP and PLTP on HDL and LDL interactions.
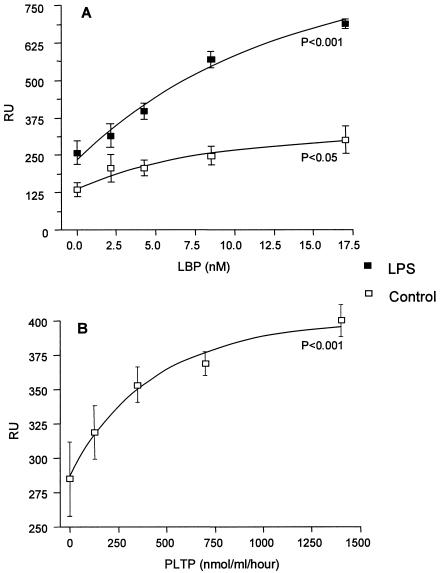
Steady-state binding of LDL to HDL in the presence of increasing concentrations of LBP or PLTP were determined. The binding of LDL by HDL was clearly dependent on LBP and PLTP concentrations (Fig. 6). HDL-LDL complexation in the presence of LBP appeared to be further enhanced by the presence of LPS on HDL (Fig. 6A). An enhancement of HDL-LDL complexation by LPS in the presence of PLTP was, however, not apparent under our experimental conditions.
FIG. 6.
Effects of LBP (A) and PLTP (B) on LDL complexation with HDL, measured by Biacore analysis. The average binding signals (RU) of the association of LDL with HDL or with HDL containing bound LPS in the presence of increasing concentrations of LBP or PLTP are shown. Data are presented as means ± standard errors of the means (error bars). The P values were calculated by using the analysis of variance test for repeated measures and are indicated in the graphs.
DISCUSSION
The plasma lipid transport proteins LBP and PLTP transfer LPS and phospholipids to plasma lipoproteins (21). During gram-negative bacterial infection, the immunostimulatory activity of LPS is attenuated by delivery and binding to lipoproteins which have been proposed to form an integral part of the innate host defense system. We have previously shown that HDL is the primary endotoxin-scavenging lipoprotein in whole blood or plasma (ex vivo) and that HDL-associated LPS is subsequently transferred to LDL and VLDL in a time-dependent manner (11). We hypothesized that the transfer of LPS between different lipoproteins would require specific lipid transfer proteins and designed the present study to investigate the involvement of the LBP and PLTP in a well-defined model system. Using purified native lipoproteins that contain a full complement of associated proteins and fluorescently labeled biologically active J5 LPS, we found that the transfer of LPS from HDL to LDL is enhanced under simulated acute-phase conditions by the addition of exogenous LBP or PLTP. Antibiotic treatment of patients with gram-negative bacteremia results in the release of heterogeneous LPS molecules containing O-antigenic polysaccharide side chains of various lengths, including the J5 LPS employed in this study (13).
Our results indicate that LBP is capable of transferring LPS from HDL to LDL and, to a lesser extent, to VLDL. Almost all of the LBP-mediated decrease in LPS fluorescence associated with HDL was recovered in the LDL and VLDL fractions at each time point from 2 to 24 h, as opposed to approximately 50% with PLTP-mediated LPS transfer, indicating that LBP is a more efficient LPS transporter. At the highest concentration of LBP employed (135 mg/liter), a level often detected in the sera of septic patients (14), the generation of both large- and small-HDL subpopulations is evident. The cholesterol-poor 150-kDa HDL subpopulation is reminiscent of lipid-poor pre-beta-like particles described as being able to bind LPS (19). This fact appears to indicate that HDL is remodeled into larger and smaller subpopulations by the LPS transfer activity of LBP, which is in agreement with the finding that LBP is colocalized on particles containing phospholipids and ApoA-I (16). Our data suggest that HDL remodeling may be a consequence of LPS transfer by LBP and PLTP.
Surface plasmon resonance was used to investigate the interaction between LDL and HDL in the presence or absence of lipid transport proteins. Both LBP and PLTP were found to enhance the association of LDL with HDL in a dose-dependent manner. Only minor residual binding was observed in the absence of added transport protein, indicating that no permanent association between the lipoproteins had occurred. We assume that under these conditions, LPS and phospholipid exchange are prerequisites for lipid particle remodeling. The presence of LPS on HDL promoted complex formation with LDL that was further enhanced by increasing concentrations of LBP. In the absence of added LBP, complex formation between LDL and HDL occurred, but it occurred at a significantly lower level. This outcome suggests that endogenous LBP, present on HDL particles isolated by HPGC, is still active in LPS transfer, and it confirms the previously described intimate association of LBP with HDL (16). The residual association of LDL with HDL together with detectable ApoB in the absence of added LBP (Fig. 6A) indicates a prolonged LDL-HDL interaction or perhaps a transient fusion of LDL and HDL particles. It seems reasonable to assume that LPS exchange and particle remodeling are more efficient during prolonged contact between the lipoprotein populations.
Surprisingly LBP was not immunologically detectable in the LDL-HDL complexes (data not shown). However, this fact does not preclude the presence of LBP in the complex. A plausible explanation for this phenomenon may be that LBP adopts a specific conformation, when bound to HDL, which masks epitopes normally exposed on the surface of the protein in free solution. Alternatively, the lipoproteins themselves may shield LBP from the antibodies.
PLTP has previously been reported to transfer LPS to HDL and reconstituted HDL (4). Our findings clearly demonstrate that PLTP is also capable of shuttling LPS from HDL to LDL. Approximately half of the 25% decrease in HDL-associated LPS fluorescence was recovered in the LDL fraction. We assume that the remaining amount of LPS transported by PLTP may be present in the form of PLTP-LPS complexes or micelles that are known to quench fluorescence. Alternatively, a portion of the LPS transported by PLTP from HDL may be internalized in the LDL particles in such a manner that it would quench the fluorescence signal. The transport of LPS by PLTP, therefore appears to predispose HDL to remodeling. The overall increase in HDL size indicates that HDL fusion had probably taken place as described by Settasatian et al. (19). The addition of PLTP to HDL without bound LPS also resulted in an enlargement of the HDL particles (data not shown). It is now clear that PLTP is not only capable of transferring phospholipids to HDL but is also able to transfer LPS from HDL, both processes resulting in the remodeling of the HDL population. The generation of a smaller HDL subpopulation by PLTP has also been reported (12) but was not evident in our model system.
Taken together, our data indicate that both LBP and PLTP shuttle LPS from HDL predominantly to LDL in a dose-dependent manner, which concurrently induces HDL remodeling. The presence of LPS on HDL promotes LDL-HDL complex formation or lipid particle fusion, which is further increased by additional LBP and presumably PLTP under appropriate experimental conditions. We propose that the accelerated transfer of LPS from HDL to LDL by increased levels of LBP and PLTP that occur during systemic inflammation predisposes HDL to remodeling, which may be one of the contributory factors in the dyslipidemia characteristic of acute infection.
Editor: J. T. Barbieri
REFERENCES
- 1.Barlage, S., D. Fröhlich, A. Böttcher, M. Jauhiainen, H. P. Müller, F. Noetzel, G. Rothe, C. Schütt, R. P. Linke, K. J. Lackner, C. Ehnholm, and G. Schmitz. 2001. Apo E-containing high density lipoproteins and phospholipid transfer protein activity increased in patients with a systemic inflammatory response. J. Lipid Res. 42:281-290. [PubMed] [Google Scholar]
- 2.Cabana, V. G., J. N. Siegel, and S. M. Sabesin. 1989. Effects of the acute phase response on the concentration and density distribution of plasma lipids and apolipoproteins. J. Lipid Res. 30:39-49. [PubMed] [Google Scholar]
- 3.Eggesbo, J. B., T. Lyberg, T. Aspelin, I. Hjermann, and P. Kierulf. 1996. Different binding of 125I-LPS to plasma proteins from persons with high or low HDL. Scand. J. Clin. Lab. Investig. 56:533-543. [DOI] [PubMed] [Google Scholar]
- 4.Hailman, E., J. J. Albers, G. Wolfbauer, A. Tu, and S. D. Wright. 1996. Neutralisation and transfer of lipopolysaccharide by phospholipid transfer protein. J. Biol. Chem. 271:12172-12178. [DOI] [PubMed] [Google Scholar]
- 5.Harris, R. L., D. M. Musher, K. Bloom, J. Gathe, L. Rice, B. Sugarman, T. W. Williams, and E. J. Young. 1987. Manifestation of sepsis. Arch. Intern. Med. 1477:1895-1906. [PubMed] [Google Scholar]
- 6.Huuskonen, J., V. M. Olkkonen, M. Jauhiainen, and C. Ehnholm. 2001. The impact of phospholipid transfer protein (PLTP) on HDL metabolism. Atherosclerosis 155:269-281. [DOI] [PubMed] [Google Scholar]
- 7.Khovidhunkit, W., K. Kim, A. Memon, J. Sigenaga, A. Moser, K. R. Feingold, and C. Grunfeld. 2004. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 45:1169-1196. [Online.] [DOI] [PubMed] [Google Scholar]
- 8.Kirschning, C. J., J. Au-Young, N. Lamping, D. Reuter, D. Pfeil, J. J. Seilhamer, and R. R. Schumann. 1997. Similar organization of the lipopolysaccharide-binding protein (LBP) and phospholipid transfer protein (PLTP) genes suggests a common gene family of lipid-binding proteins. Genomics 46:416-425. [DOI] [PubMed] [Google Scholar]
- 9.Lee, C. H., and C. M. Tsai. 1999. Quantification of bacterial lipopolysaccharides by the purpald assay: measuring formaldehyde generated from 2-keto-3-deoxyoctonate and heptose at the inner core by periodate oxidation. Anal. Biochem. 267:161-168. [DOI] [PubMed] [Google Scholar]
- 10.Levels, J. H., L. C. Lemaire, A. E. van den Ende, S. J. van Deventer, and J. J. Van Lanschot. 2003. Lipid composition and lipopolysaccharide binding capacity of lipoproteins in plasma and lymph of patients with systemic inflammatory response syndrome and multiple organ failure. Crit. Care Med. 31:1647-1653. [DOI] [PubMed] [Google Scholar]
- 11.Levels, J. H. M., P. R. Abraham, A. van den Ende, and S. J. H. Van Deventer. 2001. Distribution and kinetics of lipoprotein-bound endotoxin. Infect. Immun. 68:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques-Vidal, P., M. Jauhiainen, J. Metso, and C. Ehnholm. 1997. Transformation of high density lipoprotein 2 particles by hepatic lipase and phospholipid transfer protein. Atherosclerosis 133:87-95. [DOI] [PubMed] [Google Scholar]
- 13.Morrison, D. C., and S. E. Bucklin. 1996. Evidence for antibiotic-mediated endotoxin release as a contributing factor to lethality in experimental gram-negative sepsis. Scand. J. Infect. Dis. Suppl. 101:3-8. [PubMed] [Google Scholar]
- 14.Myc, A., J. Buck, J. Gonin, B. Reynolds, U. Hammerling, and D. Emanuel. 1997. The level of lipopolysaccharide-binding protein is significantly increased in plasma patients with the systemic inflammation response syndrome. Clin. Diagn. Lab. Immunol. 4:113-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opal, S. M., P. J. Scannon, J. L. Vincent, M. White, S. F. Carroll, J. E. Palardy, N. A. Parejo, J. P. Pribble, and J. H. Lemke. 1999. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J. Infect. Dis. 180:1584-1589. [DOI] [PubMed] [Google Scholar]
- 16.Park, C. T., and S. D. Wright. 1996. Plasma lipopolysaccharide-binding protein is found associated with a particle containing apolipoprotein A-I, phospholipid, and factor H-related protein. J. Biol. Chem. 271:18054-18060. [DOI] [PubMed] [Google Scholar]
- 17.Pussinen, P. J., E. Malle, J. Metso, W. Sattler, J. G. Raynes, and M. Jauhiainen. 2001. Acute-phase HDL in phospholipid transfer protein (PLTP)-mediated HDL conversion. Atherosclerosis 155:297-305. [DOI] [PubMed] [Google Scholar]
- 18.Rao, R., J. J. Albers, G. Wolfbauer, and H. J. Pownall. 1997. Molecular and macromolecular specificity of human plasma phospholipid transfer protein. Biochemistry 36:3645-3653. [DOI] [PubMed] [Google Scholar]
- 19.Settasatian, N., M. Duong, L. Curtiss, C. Ehnholm, M. Jauhiainen, J. Huuskonen, and K. A. Rye. 2001. The mechanism of the remodeling of high density lipoproteins by phospholipid transfer protein. J. Biol. Chem. 276:26898-26905. [DOI] [PubMed] [Google Scholar]
- 20.Underhill, D. M., and A. Ozinsky. 2002. Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol. 14:103-110. [DOI] [PubMed] [Google Scholar]
- 21.Van Lenten, B. J., A. M. Fogelman, M. E. Haberland, and P. A. Edwards. 1986. The role of lipoproteins and receptor-mediated endocytosis in the transport of bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. USA 83:2704-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurfel, M. M., S. T. Kunitake, H. Lichenstein, J. P. Kane, and S. D. Wright. 1994. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J. Exp. Med. 180:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurfel, M. M., and S. D. Wright. 1997. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayers. J. Immunol. 158:3925-3934. [PubMed] [Google Scholar]
- 24.Yu, B., E. Hailman, and S. D. Wright. 1997. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. J. Clin. Investig. 99:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]