Summary
Mutations in the LRRK2 gene cause familial Parkinson’s disease presenting with pleomorphic neuropathology that can involve α-synuclein or tau accumulation. LRRK2 mutations are thought to converge upon a pathogenic increase in LRRK2 kinase activity. A subset of small RAB GTPases has been identified as LRRK2 substrates, with LRRK2-dependent phosphorylation resulting in RAB inactivation. We used CRISPR-Cas9 genome editing to generate a novel series of isogenic iPSC lines deficient in the two most well-validated LRRK2 substrates, RAB8a and RAB10, from deeply phenotyped healthy control lines. Thorough characterization of NGN2-induced neurons revealed opposing effects of RAB8a and RAB10 deficiency on lysosomal pH and Golgi organization, with isolated effects of RAB8a and RAB10 ablation on α-synuclein and tau, respectively. Our data demonstrate largely antagonistic effects of genetic RAB8a or RAB10 inactivation, which provide discrete insight into the pathologic features of their biochemical inactivation by pathogenic LRRK2 mutation in human disease.
Keywords: Rab8a, Rab10, LRRK2, iPSC, synuclein, Tau, Parkinson Disease
Graphical abstract

Highlights
-
•
RAB8a and RAB10 deficiency induce divergent lysosomal and Golgi defects
-
•
RAB8a and RAB10 KO neurons show isolated effects on α-synuclein and tau, respectively
-
•
Inactivation of different RAB GTPases can induce distinct disease-relevant phenotypes
Here, LaVoie and colleagues characterize isogenic iPSC-derived neurons deficient in the two most well-characterized LRRK2 substrates, RAB8a and RAB10. Data revealed independent Parkinson’s disease-relevant phenotypes, with frequent opposing effects associated with RAB8a and RAB10 KO. While proteostasis was affected in both cases, dominant effects were notably seen in for α-synuclein and tau in RAB8a and RAB10 KO neurons, respectively.
Introduction
Missense mutations in the LRRK2 gene are the most common cause of inherited Parkinson’s disease (PD), with this locus further linked to idiopathic PD risk (Barrett et al., 2008; Bonifati, 2006; Paisán-Ruíz et al., 2004). LRRK2 is a large protein with functional GTPase and kinase domains within which PD-linked mutations largely cluster (Kluss et al., 2019). LRRK2-PD can manifest with pure nigral degeneration or classic Lewy body pathology typical of idiopathic PD, but surprisingly a slight majority of cases show deposition of tau into neurofibrillary tangles (Henderson et al., 2019; Herbst et al., 2022; Ujiie et al., 2012; Zimprich et al., 2004). How LRRK2 pathobiology might provoke distinct and independent neuropathological features is one of the greatest unanswered questions in the field.
A subset of ∼14 RAB GTPases are among the most well-established LRRK2 substrates. LRRK2-dependent phosphorylation at a highly conserved threonine residue in their GTPase switch domain results in a net inhibition of RAB activity (Pfeffer, 2018; Steger et al., 2016). RAB GTPases regulate intracellular membrane and protein trafficking and confer membrane identity, organelle morphology, and organelle protein content (Pfeffer, 2018; Stenmark, 2009). RAB8a and RAB10 are the most well-validated LRRK2 substrates to date. RAB8a has been linked to neurodegeneration via its interactions with α-synuclein and PINK1 (Vieweg et al., 2020; Yin et al., 2014), in addition to phosphorylation by LRRK2 (McFarland et al., 2018; Steger et al., 2016; Vieweg et al., 2020). Both RAB8a and RAB10 have been implicated in cellular responses to lysosomal injury (Bonet-Ponce et al., 2020; Eguchi et al., 2018; Mamais et al., 2021). Interestingly, opposing roles have been proposed for RAB8a and RAB10 in normal ciliogenesis (Dhekne et al., 2018; Madero-Pérez et al., 2018), but such antagonistic properties have not been explored in other contexts. RAB mutations have been linked to diseases including cancer, Charcot-Marie-Tooth disease type 2B, and Warburg Micro syndrome (Banworth and Li, 2018). Thus, their inactivation by mutant LRRK2 is well-positioned to broadly impact cellular proteostasis and may contribute to neurodegenerative disease.
Here, we used CRISPR-Cas9 genome editing to ablate RAB8a or RAB10 expression in iPSCs derived from deeply phenotyped (clinically and genetically) wild-type healthy control human subjects (Bennett et al., 2018; Lagomarsino et al., 2021). Isogenic clones from two independent control lines were differentiated into cortical neurons by NEUROGENIN-2 (NGN2) expression to produce induced neurons (iNs). We report a significant decrease in lysosomal number across both RAB8a knockout (KO) and RAB10 KO neurons compared with their isogenic controls. However, RAB8a KO neurons exhibited mild lysosome acidification, whereas RAB10 KO lysosomes were mildly alkaline. Altered glycosylation of LAMP1 was noted in both KO models, prompting an investigation into Golgi integrity that revealed a compressed, or clustered Golgi distribution in RAB8a but a more dispersed Golgi in RAB10 KO iPSCs. Interestingly, LAMP1 retention at the Golgi was only observed in RAB8a KO cells.
LRRK2-PD most commonly manifests with either classic Lewy body inclusions composed of α-synuclein, or neurofibrillary tangles composed of hyper-phosphorylated tau, prompting us to look at the consequence of RAB8a or RAB10 deficiency on these two proteins. Consistent with other opposing phenotypes linked to these two LRRK2 substrates, we found features of α-synuclein homeostasis altered in RAB8a but not RAB10 KO neurons, with selective effects on tau in RAB10 KO neurons. These data identify distinct cellular pathways that can contribute to the unique pleiomorphic pathology associated with LRRK2-PD and may provide a framework to begin to understand why co-morbid α-synuclein and tau pathology is not commonly reported (Henderson et al., 2019; Herbst et al., 2022; Ujiie et al., 2012; Zimprich et al., 2004).
Results
Divergent lysosomal alterations in RAB8a KO and RAB10 KO human neurons
To establish neuronal models of RAB8a and RAB10 deficiency we used CRISPR-Cas9 gene editing to generate homozygous-null lines of the two genes in two independent wild-type (WT) healthy control iPSC lines. Successful KO of RAB8a and RAB10 were initially tested by immunoblotting on different clones from the two control lines, BR01 and BR33 (Figure S1). Sanger sequencing validated two homozygous-null RAB8a clones on the BR33 background while the BR01 RAB8a KO clones were heterozygous null and thus were not used for further experiments. In turn, homozygous-null RAB10 clones were validated on one of each BR01 and BR33 background lines and used for subsequent experiments (Figure S1). Cells were differentiated into mature cortical glutamatergic neurons by forced expression of NGN2 via lentiviral gene delivery, according to published protocols (Sanyal et al., 2020; Zhang et al., 2013). Neurite and presynaptic phenotyping were performed to characterize differentiation and synaptic density across the different lines. MAP2 staining revealed no difference in longest neurite or total neurite length across the different RAB KO and WT control lines (Figures S2A–S2C). Furthermore, while a trend of a divergence was noted between the density of the presynaptic marker Bassoon in RAB8a KO and RAB10 KO lines, there was no significant difference compared with WT controls (Figures S2D and S2E). Initially, we set out to determine whether loss of these RAB GTPases affects lysosome numbers, by LAMP1 and LysoTracker staining (Figure 1). LAMP1 staining indicated a drop in lysosome numbers in both RAB8a KO and RAB10 KO neuron models (Figure 1A). High-content imaging was used to assay lysosomal integrity readouts across the four RAB KO lines and compared with the common WT control line BR33. High-content imaging of the LysoTracker deep red cell dye showed a significant decrease in RAB8a KO iNs (∼60%) in the total number of lysosomes (Figures 1B and 1C), while the average lysosomal area of individual lysosomes remained unaffected (Figure 1D). Lysosomal homeostasis was investigated further by assaying lysosomal pH using LysoSensor, a ratiometric pH-sensitive dye. RAB8a KO neurons exhibited significantly more acidic lysosomal lumen compared with isogenic controls (Figure 1E). In a similar line of experiments, we observed a significant decrease in lysosomal numbers in RAB10 KO compared with WT neurons, while lysosomal size was largely unaffected, as in RAB8a KO neurons (Figures 1F–1H). In contrast, we observed alkalinization of lysosomal pH in RAB10 KO, suggesting a divergent effect to RAB8a deficiency (Figure 1I).
Figure 1.
RAB8a KO and RAB10 KO human neurons have altered lysosomal morphology and function
(A) Representative confocal images of RAB8a KO, RAB10 KO, and isogenic WT control neurons stained with LAMP1, MAP2, and DAPI.
(B–I) Lysosomal parameters including lysosomal count, average lysosomal area, and lysosomal pH were assessed by high-content imaging of LysoTracker red or LysoSensor staining.
(J and K) Western blot analysis of levels of glycosylated LAMP1. All lysosomal analyses were collated from three to four independent differentiations in separate 96-well plates, with more than eight wells per genotype per plate, on 21-day-old iNs. Replicates in the graphs represent wells across four independent differentiation batches (>32 per line). ∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001; one-way ANOVA Tukey’s post hoc.
As a marker of lysosomal integrity, we quantified the levels of LAMP1 by immunoblotting. RAB8a KO and RAB10 KO neurons exhibit lower levels of LAMP1 protein, compared with the isogenic control lines, with altered migration of the glyco-protein observed independent of background (Figures 1J and 1K). Our data indicate a parallel effect of RAB8a and RAB10 deficiency in lysosomal numbers and levels of glycosylated LAMP1 protein, but a divergent effect on lysosomal pH in human neurons. We have reported altered LAMP1 glycosylation in in vivo models of LRRK2 activity as well as GCase1 heterozygous-null human neurons previously (Kluss et al., 2020; Sanyal et al., 2020). RAB8a and RAB10 are involved in vesicle trafficking between the trans-Golgi network (TGN), a central organelle for protein glycosylation, and the plasma membrane. Dysregulation of LAMP1 glycosylation in our RAB GTPase KO cell models suggested impairment in TGN-related processes, prompting us to assay Golgi integrity. The morphology of the cis-Golgi network (CGN; GM130) and TGN (TGN46) network, as well as the distribution of individual Golgi stacks and colocalization with LAMP1, were assayed by confocal microscopy. After 3D rendering of z stack images (Imaris platform; Bitplane), CGN and TGN stack size, sphericity, ellipticity, relative proximity to each other, and proximity to the nucleus were measured in WT BR33 and the isogenic null RAB8a KO and RAB10 KO iPSCs (Figures 2 and S3). RAB8a KO iPSCs exhibited a clustered Golgi morphology with significantly reduced (−16%) mean distance between CGN stacks compared with WT cells, while an opposing phenotype was observed in RAB10 KO cells with increased (+8%) mean intra-stack distance reflecting a more dispersed morphology (Figures 2A and S3). Both RAB GTPase null models exhibited a decrease in total Golgi volume per cell and closer proximity to the nucleus of CGN and TGN stacks (Figures 2C–2F and S3). Given the impact of RAB8a and RAB10 KO on lysosomal pH, we considered LAMP1 localization. Analysis of LAMP1 colocalization with TGN revealed an increase in LAMP1 association with the Golgi in RAB8a KO cells but not RAB10 KO cells (Figure 2G), while both KO models showed a significant decrease in lysosomal numbers (Figure 2H). Taken together, these data support an overall model whereby inactivation of RAB8a or RAB10 drives opposing effects on Golgi and lysosome function.
Figure 2.
RAB8a KO and RAB10 KO cells exhibit alterations in Golgi distribution
RAB8a KO, RAB10 KO, and isogenic WT control iPSCs were fixed and stained for cis-Golgi (GM130), trans-Golgi (TGN46), and LAMP1 (A and D). z stack confocal images were 3D reconstructed in Imaris (Bitplane) and the mean distance between CGN stacks (B), distance of CGN to nucleus (C), TGN volume (E), TGN distance to nucleus (F), colocalization between TGN46 and LAMP1 (G), and number of lysosomes per cell were analyzed (H). (B, C, E, and F represent individual Golgi stacks data points across N > 50 cells over three independent experiments; G and H represent per-cell data). ∗∗p < 0.001; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001; one-way ANOVA Tukey’s post hoc.
RAB8a KO human neurons accumulate insoluble α-synuclein
Accumulation of highly insoluble α-synuclein is a pathological hallmark of postmortem PD tissue and indicative of Lewy body pathology. We, and others, have shown accumulation of α-synuclein in different neuronal models of genetic PD, including LRRK2 mutant primary mouse neurons and GBA1 heterozygous-null human neurons (Mazzulli et al., 2016; Sanyal et al., 2020; Schapansky et al., 2018). To assess α-synuclein metabolism in RAB8a and RAB10 null cells, we sequentially extracted total cellular proteins from iNs in isotonic buffer containing 1% (w/v) NP40 and determined the levels of detergent-soluble and insoluble α-synuclein (dissolved in 2% [w/v] SDS). We observed an accumulation of insoluble α-synuclein in RAB8a KO neurons compared with isogenic controls while no difference was observed in RAB10 KO neurons (Figures 3A–3C), suggesting RAB8a-specific effects on the capacity of these neurons to degrade α-synuclein. Here, we used β-actin as a loading control and validated our data by normalizing on β3-tubulin as well, as a neuronal-specific loading control (Figure S3).
Figure 3.
RAB8a KO human neurons accumulate insoluble α-synuclein and secrete α-synuclein in media
RAB8a KO, RAB10 KO, and WT neurons were collected at D21 and extracted in NP-40 containing lysis buffer (soluble fraction), followed by SDS resuspension of the pellet (insoluble). Densitometry analysis represents α-synuclein levels normalized to actin (A–C: ∗p < 0.05; ∗∗p < 0.001; one-way ANOVA Tukey’s post hoc, n = 3 differentiations).
(D and E) Medium was collected at D21 and analyzed by dot-blot for total and oligomeric α-synuclein. Protein yields of neuronal lysates were used to normalize for variability in cell density and analyze equivalent amounts of conditioned media (∗∗∗p < 0.0001; ∗∗∗∗p < 0.0001; one-way ANOVA Tukey’s post hoc; n = 4 differentiations).
One of the prominent models of how pathology may be propagated in PD supports the release of oligomeric α-synuclein from cells and direct cell-to-cell transfer (Volpicelli-Daley et al., 2014). Previously, we have shown that GBA1 heterozygous-null neurons secrete higher levels of oligomeric α-synuclein in the media compared with isogenic controls (Sanyal et al., 2020), suggesting that lysosomal dyshomeostasis has a direct effect on α-synuclein secretion in human neurons. Here, we asked whether the increase in insoluble α-synuclein observed in RAB8a KO neurons is accompanied by changes in α-synuclein release, by immunoblotting of conditioned media. RAB8a KO and RAB10 KO neurons secreted higher levels of α-synuclein in the media compared with isogenic controls, while a trend toward an increase was observed in oligomeric α-synuclein in RAB8a KO cells (Figures 3D and 3E). These data suggest that RAB8a and RAB10 deficiency may induce insufficient degradation and augmented secretion of α-synuclein species in neuronal models.
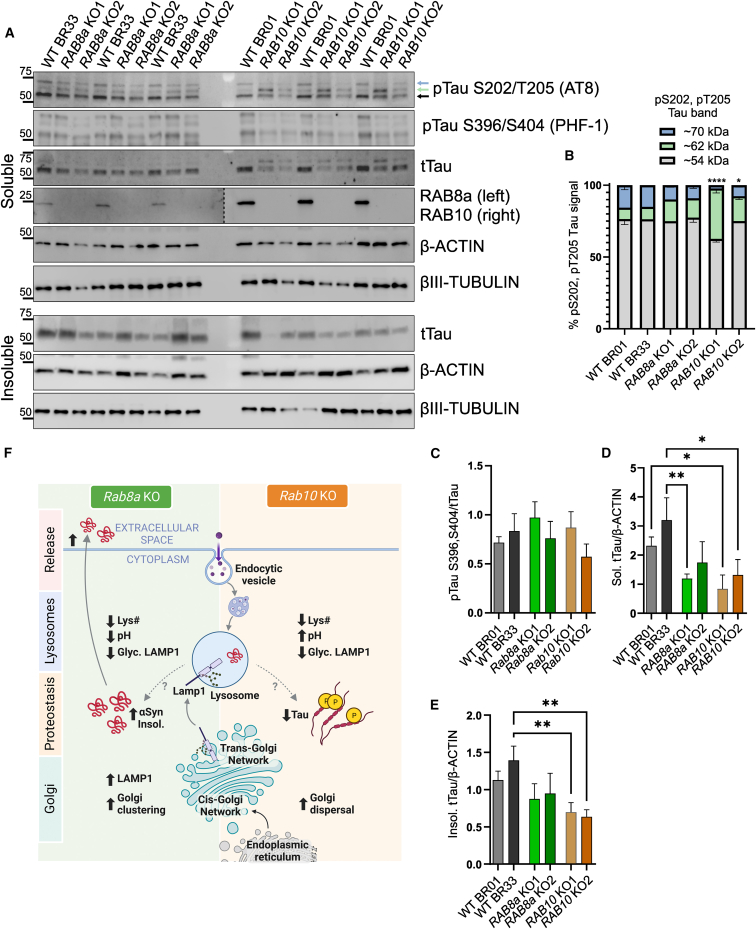
RAB10 KO human neurons show altered tau levels and phosphorylation
Tau pathology in the form of intracellular inclusions of hyperphosphorylated and aggregated tau protein are a common occurrence in LRRK2 PD brain, and studies in cell models have highlighted a mechanistic interplay between tau and RAB GTPase pathways downstream of LRRK2 activity (Guerreiro et al., 2016; Sobu et al., 2021). To characterize the effects of RAB8a and RAB10 deficiency on the biochemical properties of tau, we extracted proteins from mature cortical neurons and compared the levels of Ser202/Thr205-phosphorylated tau between the KO lines and isogenic controls. Previous studies have reported detection of distinct phospho-Tau bands, between ∼54 kDa and 68 kDa (Falcon et al., 2015; Jackson et al., 2016), by immunoblot analysis of human tissue. In fact, a strong correlation with tau aggregation propensity has been shown for pTau species running close to 62 kDa but not the lower molecular weight pTau band (∼54 kDa) (Jackson et al., 2016). Here, we probed with a commercial pS202, pT205 tau (AT8) antibody and detected three bands approximately at 54 kDa, 62 kDa, and 70 kDa (Figure 4A). A significant increase in phosphorylated tau (∼62 kDa) was observed in RAB10 KO neurons compared with WT or RAB8a KO neurons (Figures 4A and 4B). We also assessed tau phosphorylation at residues S396/S404 (PHF-1) that have been linked to Alzheimer’s disease (AD) and other Tauopathies and observed no difference in phosphorylation across lines (Figure 4C). We observed an overall decrease in total tau levels in RAB10 KO neurons compared with WT controls and tested whether this reflects a shift toward insolubility. Here, we found a similar trend of lower tau levels in the detergent-insoluble fraction (Figures 4D and 4E). These data demonstrate a divergence between pathways regulating disease-relevant post-translational modifications of tau downstream of the two RAB GTPases.
Figure 4.
RAB10 KO increases phospho-Tau species in human neurons
Human cortical neurons were collected on D21 and phosphorylated (AT8: pS202/T205 Tau and PHF-1: pS396/S404) and total soluble and insoluble Tau levels were assessed in RAB8a KO and RAB10 KO cells compared with two isogenic control lines (A and B). The ratio of each pTau band (54 kDa, 62 kDa, and 70Da) out of the total pTau signal was plotted (62 kDa groups: one-way ANOVA Tukey post hoc, BR01RAB10KO vs. BR01 or BR33: ∗∗∗∗p < 0.0001, BR33 RAB10 KO vs. BR01 or BR33: ∗p < 0.05, F (5, 12) = 29.83). Densitometry analysis represents PHF-1/tTau (C), soluble tTau normalized to actin (D) and detergent-insoluble tTau over actin (E) (n = 3 differentiations, one-way ANOVA Tukey’s post hoc). Schematic of RAB8a KO and RAB10 KO cellular phenotypes and proposed model (F) (created with BioRender.com).
Discussion
LRRK2 is a multi-domain kinase genetically linked to multiple neurologic diseases, which most commonly present with the pathologic accumulation of either α-synuclein or tau protein. A subset of the RAB GTPase protein family are bona fide LRRK2 substrates with PD-linked mutations increasing their phosphorylation (Liu et al., 2018; Steger et al., 2016), but how this contributes to disease is unknown. Given the converging evidence of endolysosomal dysfunction in PD (Kluss et al., 2019; Mamais et al., 2023), we focused attention on the two most well-validated substrates of LRRK2, RAB8a and RAB10, and their roles in endolysosomal biology and the proteostasis of α-synuclein and tau.
We and others have shown that PD-linked mutations in LRRK2 impair lysosomal acidification and reduce basal autophagic flux (Schapansky et al., 2018; Mamais et al., 2021; Eguchi et al., 2018; Henry et al., 2015; Obergasteiger et al., 2020). However, the contribution of individual LRRK2 substrates to these effects remained unexplored. We used CRISPR-Cas9 gene editing to ablate RAB8a and RAB10 expression in iPSCs and differentiated these into NGN2-induced neurons. While we observed a decrease in lysosomal number in both RAB KO neurons, luminal lysosome pH was reduced in RAB8a KO iNs and increased in RAB10 KO iNs, compared with control.
RAB8a and RAB10 can both localize to endosomal compartments and the TGN and regulate the dynamic interface between late Golgi membranes and endolysosomes (Pfeffer, 2018), with new evidence of a presynaptic role for RAB10 in neurons (Singh et al., 2023). Glycosylation of LAMP1 is necessary for its transport from the endoplasmic reticulum via the Golgi to the lysosome and its role in lysosomal integrity (Schwake et al., 2013). Irregular LAMP1 glycosylation has been linked to PD and the tauopathy, Niemann-Pick disease (Cawley et al., 2020; Fernandes et al., 2016). Here, we observed irregularities in LAMP1 glycosylation in RAB8a and RAB10 KO neurons supporting impairment in LAMP1 post-Golgi trafficking. We observed a decrease in total Golgi volume in both RAB GTPase KO cell models while only RAB8a KO cells showed LAMP1 retention in the Golgi. Opposing effects were noted, with RAB8a KO Golgi being more compacted and RAB10 KO Golgi more dispersed than WT. We surmise that both deviations from WT morphology mutually impair glycosylation at the Golgi and post-Golgi trafficking. There is a precedent for RAB8a and RAB10 to hold opposing actions, as Dhekne and colleagues (2018) reported that RAB8a stimulates cilia formation while RAB10 inhibits this process. Thus, this antagonistic relationship between RAB8a and RAB10 with respect to lysosomal pH and Golgi morphology may further represent a canonical feature of their biology.
We found that the levels of detergent-insoluble and secreted α-synuclein were increased in RAB8a KO, but not in RAB10 KO iNs. These new findings are consistent with previous reports from our group and others linking LRRK2 mutations to α-synuclein solubility and release, and suggest that LRRK2-RAB8a interactions are sufficient for these effects (Schapansky et al., 2018; Volpicelli-Daley et al., 2016). The distinction between RAB8a and RAB10 here is interesting, as RAB8a but not RAB10 has been linked to another PD gene (PINK1) (Vieweg et al., 2020), which presents with classic α-synuclein-rich Lewy body pathology (Gandhi et al., 2006). Furthermore, RAB8a has been found to physically associate with α-synuclein (Yin et al., 2014). Thus, these new data and the larger available literature would endorse a special relationship between the LRRK2 substrate RAB8a and α-synuclein.
While Lewy bodies are a main pathological hallmark of idiopathic PD, there is an overrepresentation of tau pathology in LRRK2 PD (Henderson et al., 2019; Herbst et al., 2022; Ujiie et al., 2012; Zimprich et al., 2004). Furthermore, a recent genome-wide association study linked genetic variation in the LRRK2 locus to progressive supranuclear palsy (PSP), a formal tauopathy (Jabbari et al., 2021). How LRRK2 might drive independent and distinct neuropathological features remains unexplored. It is noteworthy that RAB10 phosphorylation has been reported in PSP and AD brains with colocalization between pRAB10 and phosphorylated tau in neurons bearing tau-positive neurofibrillary tangles (Yan et al., 2018) and that RAB10 has been genetically linked to altered risk for AD (Andrews et al., 2019; Le Guen et al., 2021; Ridge et al., 2017; Tavana et al., 2018). Here, we report complex changes in tau in RAB10 KO but not RAB8a KO iNs. We observed a decrease in total tau and relative increase in Ser202/Thr205 phosphorylation in RAB10 KO neurons compared with isogenic controls, with neither occurring in RAB8a KO iNs. Given the multiple links between RAB10 and tau, and our new data, allelic variation affecting RAB10 function may influence the manifestation of α-synuclein or tau pathology in patients. Although altered tau phosphorylation has been reported in LRRK2 models (Bailey et al., 2013; Melrose et al., 2010; Nguyen et al., 2018; Schapansky et al., 2018), the mechanisms remain unclear, and these data indicate a likely role for RAB10.
Our study presents critical further evidence of opposing functions for RAB8a and RAB10 (Figure 4F). While many of the same organelles are implicated, RAB8a and RAB10 ablation consistently manifested distinct phenotypes within the secretory pathway. In addition, we identify novel and independent roles for RAB8a and RAB10 in modulating the homeostasis of α-synuclein and tau, respectively, the two proteins implicated in the neurological disorders linked to the RAB kinase, LRRK2. This work aligns well with independent studies involving RAB8a and RAB10 and brings novel resources and a new conceptual framework to consider the downstream consequences of LRRK2-dependent phosphorylation of individual RAB proteins.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for reagents should be directed to the corresponding author Dr. Matthew J. LaVoie (mlavoie@ufl.edu).
Materials availability
Materials and additional details can be made available by the corresponding author upon request.
Data and code availability
All data generated and analyzed during this study are included in the article and supporting files.
Generation of RAB8a KO and RAB10 KO human iPSCs
CRISPR-Cas9 genome editing was performed as before (Sanyal et al., 2020). For information on single guide RNA sequences and validation of clones see supplemental experimental procedures.
Differentiation of human iPSCs
iPSCs were differentiated into cortical neurons using established protocols (Zhang et al., 2013). See supplemental experimental procedures for further information on iPSC culture, transduction, and differentiation methods.
Biochemical assays and cell imaging
Several biochemical assays, including analysis of soluble/insoluble cell lysate fractions, dot-blot, immunocytochemistry, and lysosomal characterization were performed. For further information on these assays and their analysis refer to supplemental experimental procedures.
Statistical analyses
All experiments were conducted at least three independent times for three differentiations. Error bars indicate mean ± SD. Statistical analysis was performed using GraphPad Prism software, using a one-way ANOVA with Tukey’s post hoc test.
Acknowledgments
The authors thank Dr. Matthew Farrer (UF/CTRND) for access to the Olympus Fluoview FV-1000 confocal microscope and Dr. Mark S. Moehle for thoughtful discussion.
This work was supported in part by the Michael J. Fox Foundation for Parkinson’s Research (MJFF) grants MJFF-004932 (M.J.L.) and MJFF-023425 (A.M., M.J.L.) and National Institutes of Health grants NS110188 (M.J.L.) and AG077269 (A.M.).
Author contributions
M.J.L., A.S., A.M.: Conceptualization, Methodology, Data Analysis, Writing - Original draft preparation. A.M., A.S., A.F., C.G.Z., M.G., L.R.D., N.S., W.G., S.L.: Visualization, Investigation. A.M., M.J.L.: Supervision. A.F., A.M., M.J.L.: Writing - Reviewing and Editing.
Declaration of interests
M.J.L. serves on the scientific advisory board for SPARC, Ltd.
Published: February 1, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.01.001.
Supplemental information
References
- Andrews S.J., Fulton-Howard B., Goate A. Protective Variants in Alzheimer’s Disease. Curr. Genet. Med. Rep. 2019;7:1–12. doi: 10.1007/s40142-019-0156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey R.M., Covy J.P., Melrose H.L., Rousseau L., Watkinson R., Knight J., Miles S., Farrer M.J., Dickson D.W., Giasson B.I., Lewis J. LRRK2 phosphorylates novel tau epitopes and promotes tauopathy. Acta Neuropathol. 2013;126:809–827. doi: 10.1007/s00401-013-1188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banworth M.J., Li G. Consequences of RAB GTPase dysfunction in genetic or acquired human diseases. Small GTPases. 2018;9:158–181. doi: 10.1080/21541248.2017.1397833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.A., Buchman A.S., Boyle P.A., Barnes L.L., Wilson R.S., Schneider J.A. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 2018;64:S161–S189. doi: 10.3233/JAD-179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet-Ponce L., Beilina A., Williamson C.D., Lindberg E., Kluss J.H., Saez-Atienzar S., Landeck N., Kumaran R., Mamais A., Bleck C.K.E., et al. LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci. Adv. 2020;6:eabb2454. doi: 10.1126/sciadv.abb2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V. The pleomorphic pathology of inherited parkinson’s disease: Lessons from LRRK2. Curr. Neurol. Neurosci. Rep. 2006;6:355–357. doi: 10.1007/s11910-996-0013-z. [DOI] [PubMed] [Google Scholar]
- Cawley N.X., Sojka C., Cougnoux A., Lyons A.T., Nicoli E.-R., Wassif C.A., Porter F.D. Abnormal LAMP1 glycosylation may play a role in Niemann-Pick disease, type C pathology. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhekne H.S., Yanatori I., Gomez R.C., Tonelli F., Diez F., Schüle B., Steger M., Alessi D.R., Pfeffer S.R. A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. Elife. 2018;7 doi: 10.7554/eLife.40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi T., Kuwahara T., Sakurai M., Komori T., Fujimoto T., Ito G., Yoshimura S.I., Harada A., Fukuda M., Koike M., Iwatsubo T. LRRK2 and its substrate RAB GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc. Natl. Acad. Sci. USA. 2018;115:E9115–E9124. doi: 10.1073/pnas.1812196115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B., Cavallini A., Angers R., Glover S., Murray T.K., Barnham L., Jackson S., O’Neill M., Isaacs A.M., Hutton M.L., et al. Conformation Determines the Seeding Potencies of Native and Recombinant Tau Aggregates. J. Biol. Chem. 2015;290:1049–1065. doi: 10.1074/jbc.M114.589309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes H.J.R., Hartfield E.M., Christian H.C., Emmanoulidou E., Zheng Y., Booth H., Bogetofte H., Lang C., Ryan B.J., Sardi S.P., et al. ER Stress and Autophagic Perturbations Lead to Elevated Extracellular α-Synuclein in GBA-N370S Parkinson’s iPSC-Derived Dopamine Neurons. Stem Cell Rep. 2016;6:342–356. doi: 10.1016/j.stemcr.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Muqit M.M.K., Stanyer L., Healy D.G., Abou-Sleiman P.M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A.J., et al. PINK1 protein in normal human brain and Parkinson’s disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- Guerreiro P.S., Gerhardt E., Lopes da Fonseca T., Bähr M., Outeiro T.F., Eckermann K. LRRK2 Promotes Tau Accumulation, Aggregation and Release. Mol. Neurobiol. 2016;53:3124–3135. doi: 10.1007/s12035-015-9209-z. [DOI] [PubMed] [Google Scholar]
- Henderson M.X., Sengupta M., Trojanowski J.Q., Lee V.M.Y. Alzheimer’s disease tau is a prominent pathology in LRRK2 Parkinson’s disease. Acta Neuropathol. Commun. 2019;7:183. doi: 10.1186/s40478-019-0836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A.G., Aghamohammadzadeh S., Samaroo H., Chen Y., Mou K., Needle E., Hirst W.D. Pathogenic LRRK2 mutations, through increased kinase activity, produce enlarged lysosomes with reduced degradative capacity and increase ATP13A2 expression. Hum. Mol. Genet. 2015;24:6013–6028. doi: 10.1093/hmg/ddv314. [DOI] [PubMed] [Google Scholar]
- Herbst S., Lewis P.A., Morris H.R. The emerging role of LRRK2 in tauopathies. Clin. Sci. 2022;136:1071–1079. doi: 10.1042/CS20220067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari E., Koga S., Valentino R.R., Reynolds R.H., Ferrari R., Tan M.M.X., Rowe J.B., Dalgard C.L., Scholz S.W., Dickson D.W., et al. Genetic determinants of survival in progressive supranuclear palsy: a genome-wide association study. Lancet Neurol. 2021;20:107–116. doi: 10.1016/S1474-4422(20)30394-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.J., Kerridge C., Cooper J., Cavallini A., Falcon B., Cella C.V., Landi A., Szekeres P.G., Murray T.K., Ahmed Z., et al. Short Fibrils Constitute the Major Species of Seed-Competent Tau in the Brains of Mice Transgenic for Human P301S Tau. J. Neurosci. 2016;36:762–772. doi: 10.1523/JNEUROSCI.3542-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluss J.H., Mamais A., Cookson M.R. LRRK2 links genetic and sporadic Parkinson’s disease. Biochem. Soc. Trans. 2019;47:651–661. doi: 10.1042/BST20180462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluss J.H., Mazza M.C., Li Y., Manzoni C., Lewis P.A., Cookson M.R., Mamais A. 2020. Preclinical Modeling of Chronic Inhibition of the Parkinson’s Disease Associated Kinase LRRK2 Reveals Altered Function of the Endolysosomal System in Vivo. (In Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino V.N., Pearse R.V., Liu L., Hsieh Y.-C., Fernandez M.A., Vinton E.A., Paull D., Felsky D., Tasaki S., Gaiteri C., et al. Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron. 2021;109:3402–3420.e9. doi: 10.1016/j.neuron.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guen Y., Belloy M.E., Napolioni V., Eger S.J., Kennedy G., Tao R., He Z., Greicius M.D., Alzheimer’s Disease Neuroimaging Initiative A novel age-informed approach for genetic association analysis in Alzheimer’s disease. Alzheimer's Res. Ther. 2021;13:72. doi: 10.1186/s13195-021-00808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Bryant N., Kumaran R., Beilina A., Abeliovich A., Cookson M.R., West A.B. LRRK2 phosphorylates membrane-bound RABs and is activated by GTP-bound RAB7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 2018;27:385–395. doi: 10.1093/hmg/ddx410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madero-Pérez J., Fdez E., Fernández B., Lara Ordóñez A.J., Blanca Ramírez M., Gómez-Suaga P., Waschbüsch D., Lobbestael E., Baekelandt V., Nairn A.C., et al. Parkinson disease-associated mutations in LRRK2 cause centrosomal defects via RAB8a phosphorylation. Mol. Neurodegener. 2018;13:3. doi: 10.1186/s13024-018-0235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamais A., Kluss J.H., Bonet-Ponce L., Landeck N., Langston R.G., Smith N., Beilina A., Kaganovich A., Ghosh M.C., Pellegrini L., et al. Mutations in LRRK2 linked to Parkinson disease sequester RAB8a to damaged lysosomes and regulate transferrin-mediated iron uptake in microglia. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamais A., Wallings R., Rocha E.M. Disease mechanisms as subtypes: Lysosomal dysfunction in the endolysosomal Parkinson’s disease subtype. Handb. Clin. Neurol. 2023;193:33–51. doi: 10.1016/B978-0-323-85555-6.00009-6. [DOI] [PubMed] [Google Scholar]
- Mazzulli J.R., Zunke F., Tsunemi T., Toker N.J., Jeon S., Burbulla L.F., Patnaik S., Sidransky E., Marugan J.J., Sue C.M., Krainc D. Activation of β-Glucocerebrosidase Reduces Pathological α-Synuclein and Restores Lysosomal Function in Parkinson’s Patient Midbrain Neurons. J. Neurosci. 2016;36:7693–7706. doi: 10.1523/JNEUROSCI.0628-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland N., Parmar M., Park H.-J., Ryu D., Powell L., Foels R., Anagnostis S. RAB8a protects against alpha-synuclein toxicity in a rat model of Parkinsonism (P3.049) Neurology. 2018;90 [Google Scholar]
- Melrose H.L., Dächsel J.C., Behrouz B., Lincoln S.J., Yue M., Hinkle K.M., Kent C.B., Korvatska E., Taylor J.P., Witten L., et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol. Dis. 2010;40:503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.P.T., Daniel G., Valdés P., Islam M.S., Schneider B.L., Moore D.J. G2019S LRRK2 enhances the neuronal transmission of tau in the mouse brain. Hum. Mol. Genet. 2018;27:120–134. doi: 10.1093/hmg/ddx389. [DOI] [PubMed] [Google Scholar]
- Obergasteiger J., Frapporti G., Lamonaca G., Pizzi S., Picard A., Lavdas A.A., Pischedda F., Piccoli G., Hilfiker S., Lobbestael E., et al. Kinase inhibition of G2019S-LRRK2 enhances autolysosome formation and function to reduce endogenous alpha-synuclein intracellular inclusions. Cell Death Discov. 2020;6:45. doi: 10.1038/s41420-020-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisán-Ruíz C., Jain S., Evans E.W., Gilks W.P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. LRRK2 and RAB GTPases. Biochem. Soc. Trans. 2018;46:1707–1712. doi: 10.1042/BST20180470. [DOI] [PubMed] [Google Scholar]
- Ridge P.G., Karch C.M., Hsu S., Arano I., Teerlink C.C., Ebbert M.T.W., Gonzalez Murcia J.D., Farnham J.M., Damato A.R., Allen M., et al. Linkage, whole genome sequence, and biological data implicate variants in RAB10 in Alzheimer’s disease resilience. Genome Med. 2017;9:100. doi: 10.1186/s13073-017-0486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A., Novis H.S., Gasser E., Lin S., LaVoie M.J. LRRK2 Kinase Inhibition Rescues Deficits in Lysosome Function Due to Heterozygous GBA1 Expression in Human iPSC-Derived Neurons. Front. Neurosci. 2020;14:442. doi: 10.3389/fnins.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapansky J., Khasnavis S., DeAndrade M.P., Nardozzi J.D., Falkson S.R., Boyd J.D., Sanderson J.B., Bartels T., Melrose H.L., LaVoie M.J. Familial knockin mutation of LRRK2 causes lysosomal dysfunction and accumulation of endogenous insoluble α-synuclein in neurons. Neurobiol. Dis. 2018;111:26–35. doi: 10.1016/j.nbd.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwake M., Schröder B., Saftig P. Lysosomal Membrane Proteins and Their Central Role in Physiology. Traffic. 2013;14:739–748. doi: 10.1111/tra.12056. [DOI] [PubMed] [Google Scholar]
- Singh V., Menard M.A., Serrano G.E., Beach T.G., Zhao H.T., Riley-DiPaolo A., Subrahmanian N., LaVoie M.J., Volpicelli-Daley L.A. Cellular and subcellular localization of RAB10 and phospho-T73 RAB10 in the mouse and human brain. Acta Neuropathol. Commun. 2023;11:201. doi: 10.1186/s40478-023-01704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobu Y., Wawro P.S., Dhekne H.S., Yeshaw W.M., Pfeffer S.R. Pathogenic LRRK2 regulates ciliation probability upstream of tau tubulin kinase 2 via RAB10 and RILPL1 proteins. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2005894118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M., Tonelli F., Ito G., Davies P., Trost M., Vetter M., Wachter S., Lorentzen E., Duddy G., Wilson S., et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of RAB GTPases. Elife. 2016;5 doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. RAB GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Tavana J.P., Rosene M., Jensen N.O., Ridge P.G., Kauwe J.S., Karch C.M. RAB10: an Alzheimer’s disease resilience locus and potential drug target. Clin. Interv. Aging. 2018;14:73–79. doi: 10.2147/CIA.S159148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujiie S., Hatano T., Kubo S.-I., Imai S., Sato S., Uchihara T., Yagishita S., Hasegawa K., Kowa H., Sakai F., Hattori N. LRRK2 I2020T mutation is associated with tau pathology. Parkinsonism Relat. Disord. 2012;18:819–823. doi: 10.1016/j.parkreldis.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Vieweg S., Mulholland K., Bräuning B., Kachariya N., Lai Y.-C., Toth R., Singh P.K., Volpi I., Sattler M., Groll M., et al. PINK1-dependent phosphorylation of Serine111 within the SF3 motif of RAB GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72. Biochem. J. 2020;477:1651–1668. doi: 10.1042/BCJ20190664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley L.A., Gamble K.L., Schultheiss C.E., Riddle D.M., West A.B., Lee V.M.-Y. Formation of α-synuclein Lewy neurite–like aggregates in axons impedes the transport of distinct endosomes. Mol. Biol. Cell. 2014;25:4010–4023. doi: 10.1091/mbc.E14-02-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley L.A., Abdelmotilib H., Liu Z., Stoyka L., Daher J.P.L., Milnerwood A.J., Unni V.K., Hirst W.D., Yue Z., Zhao H.T., et al. G2019S-LRRK2 Expression Augments α-Synuclein Sequestration into Inclusions in Neurons. J. Neurosci. 2016;36:7415–7427. doi: 10.1523/JNEUROSCI.3642-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Wang L., Gao J., Siedlak S.L., Huntley M.L., Termsarasab P., Perry G., Chen S.G., Wang X. RAB10 Phosphorylation is a Prominent Pathological Feature in Alzheimer’s Disease. J. Alzheimers Dis. 2018;63:157–165. doi: 10.3233/JAD-180023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Lopes da Fonseca T., Eisbach S.E., Anduaga A.M., Breda C., Orcellet M.L., Szegő É.M., Guerreiro P., Lázaro D.F., Braus G.H., et al. α-Synuclein interacts with the switch region of RAB8a in a Ser129 phosphorylation-dependent manner. Neurobiol. Dis. 2014;70:149–161. doi: 10.1016/j.nbd.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 Cause Autosomal-Dominant Parkinsonism with Pleomorphic Pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in the article and supporting files.