Summary
A major goal of regenerative medicine is to generate tissue-specific mature and functional cells. However, current cell engineering protocols are still unable to systematically produce fully mature functional cells. While existing computational approaches aim at predicting transcription factors (TFs) for cell differentiation/reprogramming, no method currently exists that specifically considers functional cell maturation processes. To address this challenge, here, we develop SinCMat, a single-cell RNA sequencing (RNA-seq)-based computational method for predicting cell maturation TFs. Based on a model of cell maturation, SinCMat identifies pairs of identity TFs and signal-dependent TFs that co-target genes driving functional maturation. A large-scale application of SinCMat to the Mouse Cell Atlas and Tabula Sapiens accurately recapitulates known maturation TFs and predicts novel candidates. We expect SinCMat to be an important resource, complementary to preexisting computational methods, for studies aiming at producing functionally mature cells.
Keywords: scRNA-seq, functional cell maturation, transcription factors, cell engineering, cellular therapy
Graphical abstract

Highlights
-
•
SinCMat is the first computational method for identifying functional maturation TFs
-
•
SinCMat predicted known and novel maturation TFs
-
•
SinCMat is applicable to any mouse or human cell type
-
•
SinCMat is embedded in a user-friendly web interface
In this article, Del Sol and colleagues developed SinCMat, a computational platform for systematically predicting functional maturation TFs. Based on a model of cell maturation that integrates identity and environment components, SinCMat can predict known and novel maturation TFs required for cell functionalization. Furthermore, Del Sol and colleagues introduce SinCMatDB, a manually curated database of experimentally validated maturation TFs.
Introduction
One of the major goals in stem cell research is to generate tissue-specific mature and functional cell types for clinical applications such as drug screening, disease modeling, and cell transplantation. In that regard, huge progress has been made over the last years in our capacity to direct the differentiation of human induced pluripotent stem cells (hiPSCs) and transdifferentiation of somatic cells (Wang et al., 2021; Cieślar-Pobuda et al., 2017; Qian et al., 2012). However, while cell conversion experiments have been increasingly successful in generating desired cell types, current protocols are still unable to systematically produce fully mature and functional cells, especially in in vitro systems (Xu et al., 2015; Zhang et al., 2021; Boshans et al., 2021; Sun et al., 2021). Namely, such immature cells exhibit the proper cell-type-specific markers but are unable to entirely perform their specialized physiological functions. Nonetheless, the in vitro generation of functionally mature cells is crucial for modeling the development and maintenance of tissues/organs and for cell/tissue replacement therapies for disease treatment.
The advent of transcriptomics technologies and the concomitant increase in the amount of available data have made possible the generation of a number of sophisticated computational approaches for identifying the transcriptional regulators for cell-fate specification and cell conversion (D’Alessio et al., 2015; Cahan et al., 2014; Xu et al., 2021; Rackham et al., 2016; Ribeiro et al., 2021). However, while these computational approaches rigorously address the conversion of cell identity from one cell type into another, they do not explicitly take into account the cell maturation process. Indeed, a large body of evidence indicates that the acquisition of cell identity is necessary but not sufficient (Alvarez-Dominguez and Melton, 2022). Recent studies demonstrated that environmental signals play a significant role in the cell maturation process by modifying the gene expression through signal-dependent transcription factors (STFs), a class of TFs expressed in a broad manner across cell types and activated by extracellular stimuli (Heinz et al., 2015; Wortham and Sander, 2021). Therefore, engineering of functionally mature cells likely requires an additional, complementary set of factors to cell identity TFs.
Here, we present SinCMat, a single-cell RNA sequencing (scRNA-seq)-based computational method for systematically identifying functional cell maturation TFs. The SinCMat algorithm relies on the model of cell maturation (Heinz et al., 2015; Wortham and Sander, 2021) and first identifies a set of identity TFs (ITFs) controlling the target cell identity and then predicts ITF-STF pairs that co-target the genes necessary for functional maturation. Therefore, the output of SinCMat consists of a list of ITF-STF pairs, which can be experimentally overexpressed either together with more traditional cell differentiation/reprogramming factors or later in the protocol when cells exhibit desired cell identity but are still missing required functional properties.
In addition to the prediction algorithm, we also introduce SinCMatDB, a manually curated database compiling experimentally validated cell maturation TFs. The application of SinCMat identifies known maturation TFs and predicts novel candidates for experimental validation. Furthermore, we show through two case studies on dopaminergic (DA) neurons and limbal SCs (LSCs) SinCMat’s applicability to both post-mitotic and progenitors/SCs. Finally, we show that SinCMat more accurately predicts cell maturation TFs than preexisting computational methods that rely only on gene expression profiles and gene networks (D’Alessio et al., 2015; Rackham et al., 2016; Cahan et al., 2014).
In summary, SinCMat is, to our knowledge, the first computational tool specifically designed for predicting functional cell maturation TFs. We expect SinCMat to be an important resource for studies aiming at producing functionally mature cells, especially for clinical applications. The SinCMat web interface is user friendly and facilitates easy browsing of SinCMatDB. Furthermore, the SinCMat web interface allows users to upload their own scRNA-seq data and run the tool for any mouse or human cell type. SinCMat is freely available for academic, non-profit use at https://sincmat.lcsb.uni.lu/.
Results
Method overview
SinCMat’s model is designed based on a recently proposed concept by Wortham and Sander (2021) in which ITFs, along with epigenetic modifiers, drive cell differentiation and set a specific epigenetic landscape. Later, during the functional maturation, STFs are triggered by environmental signals to fine-tune the expression of functional genes (Figure 1A).
Figure 1.
Schematic outline of SinCMat
(A) Model of functional cell maturation used to design SinCMat.
(B) The workflow of SinCMat. From scRNA-seq data of target cell, SinCMat identifies TFs required for functional maturation.
(C) 4 major stages of SinCMat algorithm.
(D and E) Histogram of genes extracted from functional GO categories binned by normalized expression ranks calculated in (D) the MCA and (E) the TS.
(F) Enrichment analysis of functional GO genes for the 10%, 20%, 30%, 40%, and 50% most highly expressed genes in 18 MCA cell types and 16 TS cell types. Odds ratio (OR) is reported for each bin in each cell type. Categories with the highest OR were counted (frequency indicated by the y axis).
In line with this model, SinCMat uses scRNA-seq data of the target cell type as input to identify pairs of ITFs and STFs that can bind to the regulatory regions of the same functional genes (see below for the identification of candidate functional gene set) (Figure 1B) (Heinz et al., 2010; Mullen et al., 2011; Wortham and Sander, 2021). Note, genome-wide binding analyses unveiled that, here, the “co-binding/targeting” of ITFs and STFs does not necessitate direct physical binding, but rather these two types of TFs have to bind to the regulatory regions of common functional genes independently at some point during development and cell maturation (Heinz et al., 2010; 2015; Zhang and Glass, 2013). The SinCMat algorithm is composed of 4 major stages (Figure 1C; experimental procedures), which aim at modeling the stepwise acquisition of cell functionality (Heinz et al., 2015; Wortham and Sander. 2021) where (1) cell identity is set by ITFs that initiate the expression of cell-type-specific genes during differentiation and, along with epigenetic modifiers, open up the chromatin of the regulatory regions of functional genes and (2) STFs are activated by environment signals and fine-tune the expression of those functional genes. In stage 1, SinCMat identifies cell-type-specific ITFs by single-cell Jensen-Shannon divergence (scJSD) by identifying the most specifically expressed TFs. In this context, JSD is a method used to measure the deviance between an ideal and the actual gene expression for a particular TF. The idealized TF expression is characterized by high expression in a target cell and no expression in any of the background cells. The resulting divergence score is used to rank TFs, with the top predicted TFs being those with the lowest divergence (indicating the highest specificity) (D’Alessio et al., 2015; Ribeiro et al., 2021). In stage 2, starting with the 20 ITFs generated in stage 1, SinCMat generates all possible ITF-STF pairs (6,380 pairs in total). The pairs that are the most often co-expressed (top 10 percentile) in the target cell type are kept for downstream steps. In stage 3, the functional score is computed for each ITF-STF pair. This score consists of two components to leverage the co-targeting property of ITF and STF: (1) the Jaccard similarity index score computed on the common functional target genes and (2) the sum of Pearson’s correlation scores between ITF/STF and each co-target functional gene. For this purpose, we compiled a TF-Target database from multiple sources (experimental procedures) for mouse and human. In stage 4, SinCMat returns as output a list of ITF-STF pairs ranked by the functional score. The normalization step of the functional score components performed in stage 3 allows the users to compare the quality of TF pairs.
Identification of candidate functional gene set
In order to unbiasedly obtain candidate functional genes that need to be targeted by ITF-STF pairs, genes associated with functional GO terms of 18 and 16 cell types available in the Mouse Cell Atlas (MCA) and Tabula Sapiens (TS) datasets, respectively, were collected from the Cell Ontology database and complemented with prior literature knowledge (Table S1; experimental procedures) (Diehl et al., 2016; Han et al., 2018; Jones et al., 2022). Then, all expressed genes (mean expression >0 in the target cell type) were ranked by mean expression, and the ranks were normalized between 0 and 100. We observed that the most highly expressed genes showed the highest enrichment of genes extracted from the functional GO terms (Figures 1D and 1E). Fisher’s exact test also showed that the top 10% most highly expressed genes had the highest enrichment of functional GO terms among the first five cumulative decile bins (i.e., top 10%, 20%, 30%, 40%, and 50%) (Figure 1F). In addition, the top 10% most highly expressed genes of well-known cell types also accurately recapitulated their main cellular functions (Figures S1A–S1L). Thus, we defined our cell-type-specific functional gene set as the top 10% most highly expressed genes in each cell type.
Large-scale application of SinCMat to the MCA and TS datasets
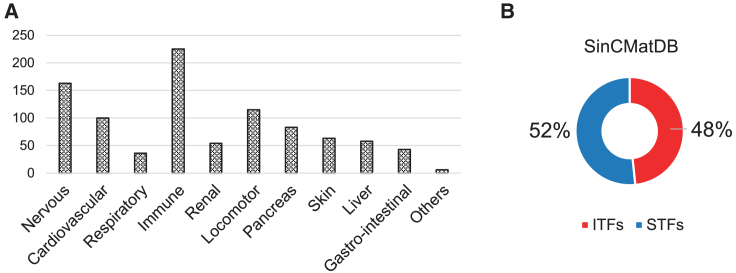
To assess the performance and demonstrate the applicability of SinCMat, we first created SinCMatDB, a manually curated database with experimentally validated cell maturation cues from literature. SinCMatDB is composed of a total of 950 TFs among 70 cell types (Figures 2A and 2B; Table S2). We then applied SinCMat to 82 and 64 cell (sub)types from the MCA and TS. Among the 25 cell types in both MCA and TS, for which at least 5 TFs had literature evidence in SinCMatDB, 53% and 56% TFs among the top 5 ITF-STF pairs (experimental procedures) have been previously validated (Figures 3A and 3C; Table S3). Fisher’s exact test revealed that in 49 cell types out of 50, SinCMat’s predictions were significantly enriched for validated TFs (Figures 3B and 3D; Table S4). Of note, a non-significant p value was obtained with osteoclasts; however, mature osteoclasts are large, multinucleated cells with a size that exceeds the usual compatible size for standard sequencers. Therefore, it is likely the annotated osteoclasts in the MCA are rather smaller immature osteoclasts, which led to the prediction of TFs related to immature osteoclasts functions (Tsukasaki and Takayanagi, 2022). For example, NR3C1 and ATF3 are both TFs well known for their function in pre-osteoclasts and osteoclast differentiation (Fukasawa et al., 2016; Jiang et al., 2020). Additionally, the in silico double knockout (KO) of some predicted TFs using scTenifold (Osorio et al., 2022) demonstrated that their virtual inactivation leads to perturbation of genes involved in essential cell-type-specific functions (Figure S2).
Figure 2.
SinCMatDB characteristics
(A) Number of experimentally validated maturation TF evidences in SinCMatDB across body systems.
(B) Proportion of ITFs/STFs in SinCMatDB.
Figure 3.
Performance evaluation of SinCMat on MCA and TS
(A and C) Average number of validated maturation TFs in SinCMat predictions applied to (A) MCA and (C) TS.
(B and D) Distribution of Fisher’s exact test p value for enrichment in validated TFs with respect to the total numbers of predicted TFs and validated TFs for (B) MCA and (D) TS.
(E) Comparison of mean coverage of function GO genes (%) between 5 ITFs predicted by SinCMat and random samples (n = 1,000) of 5 ITFs from the initial 20 ITFs obtained in stage 1. Significance was calculated by one-sample Wilcoxon test. ∗∗∗p < 0.001.
Finally, the ability to target GO functional genes was compared between the top 5 predicted ITFs and 1,000 random samples of 5 ITFs taken from the 20 ITFs identified in stage 1. One-sample Wilcoxon test unveiled that in all the 7 cell types for which >3 cell-type-specific functional GO terms were available, predicted ITFs significantly (p < 0.001) target more GO functional genes than the random samples (Figure 3E), highlighting the ability of SinCMat to prioritize ITFs that possess important functional roles.
Case study 1: SinCMat recapitulates maturation TFs for DA neurons and predicts novel candidates
Parkinson’s disease (PD) is a common neurological disorder resulting from the loss of midbrain DA neurons, leading to motor symptoms such as bradykinesia, rigidity, and resting tremor (Alexander, 2004). Recent advancements in SC research have made it promising to generate DA neurons from iPSCs for potential cell therapy (Kikuchi et al., 2017; Gilmozzi et al., 2021; Lebedeva et al., 2023). However, a key challenge is the maturation time required for in-vitro-differentiated DA neurons (Mahajani et al., 2019). We therefore applied SinCMat to scRNA-seq data of human healthy DA neurons to predict TFs that could improve current protocols (Kamath et al., 2022).
We first integrated scRNA-seq data of healthy DA neurons (Kamath et al., 2022) into our human background (Figure 4A). SinCMat predicted the well-known pre-B cell leukemia homeobox 1 (PBX1), which, in addition to being involved in the specification and development of DA neurons, is also required for the survival of DA neurons (Villaescusa et al., 2016) (Figure 4B). Inactivation experiments of HIF1A, the hypoxia-responsive STF associated with PBX1, previously demonstrated the importance of oxygen tension for the proper survival and complete differentiation of midbrain-derived neural precursors cells (Milosevic et al., 2007). Baek and colleagues showed that overexpression of EBF3, predicted in the second pair, led to a significant increase in the maturation level of developing DA neurons (Baek et al., 2014). Finally, Kang et al. (2017) used metformin to activate the ATF2/CREB pathway to confer protection to DA neurons and improved dopamine-related motor performance in a PD animal model. Although the other predicted TFs do not have direct evidence for DA neuron maturation, studies demonstrated their roles in global neuronal maturation and functions and their importance for more specific DA functions such as dopamine synthesis (Hashizume et al., 2018; Puymirat et al., 1983; Yamada et al., 2013; Zhu et al., 2018).
Figure 4.
Application of SinCMat to DA neurons
(A) Schematic analysis outline (left) and predicted ITF-STF pairs (right).
(B) Predicted TFs with literature support for the maturation of DA neurons by direct TF expression modulation (overexpression/KO experiments) or indirectly via exogenous treatment.
(C) GO biological process enrichment (p value cutoff = 0.01 and q value cutoff = 0.05) for functional co-target genes of predicted ITF-STF pairs.
(D) DA-specific functional genes targeted by predicted ITF-STF pairs.
(E) Venn diagram showing overlapping co-target genes among 3 different ITFs paired with NFE2L1.
GO enrichment analysis of co-targeted functional genes by each ITF-STF pair resulted in an enrichment for global neuronal functions like “regulation of trans-synaptic signaling” and “modulation of chemical synaptic transmission” (Figure 4C). Furthermore, in addition to global neuronal functions, predicted ITF-STF pairs also co-target genes involved in dopamine metabolism, secretion, and release, such as DOC, PINK1, and COMT (Figure 4D) (Bus et al., 2020; Lepack et al., 2020; Lohr et al., 2014). Finally, we observed that a given predicted STF can contribute to the activation of complementary sets of functional genes by associating with different ITFs. For example, among the total predictions (Table S5), SinCMat associated NFE2L1, a stress-dependent TF, with CUX2, MYT1L, and HIVEP3, and these 3 pairs co-target distinct sets of functional genes (Figure 4E).
Overall, the application of SinCMat to the scRNA-seq data of human DA neurons predicted known maturation TFs along with novel promising candidates for further experimental validation.
Case study 2: SinCMat applied to LSCs
SCs and progenitors play a multifaced role within the body. In addition to their primary cell replacement function, they contribute to tissue regeneration and repair by releasing growth factors, cytokines, extracellular matrix molecules, and exosomes. Furthermore, SCs possess a crucial migratory ability that enables them to sustain tissue balance and facilitate the processes of repair and regeneration (de Lucas et al., 2018). Therefore, to perform their entire set of functions, similarly as differentiated cells, SCs and progenitors must undergo a functional maturation phase (Figure 5A).
Figure 5.
Application of SinCMat to LSCs
(A) Application of the maturation concept to SCs/progenitors and acquisition of cell functionality.
(B) Differentiation path of LSCs in CECs.
(C) Top 5 ITF-STF pair predictions for LSCs from TS.
(D) Top 10 GO biological processes enriched for functional co-target genes of predicted ITF-STF pairs.
(E) GO cellular component enrichment (p value cutoff = 0.01 and q value cutoff = 0.05) for functional target genes of TP63-RELA pair.
(F) Functional roles of the ITF PAX6 in association with NCOA1 or NFKB1 identified in LSCs and CECs by GO biological process enrichment on distinct sets of co-target genes (see also Figure S3).
Here, we broaden the scope of the maturation process and demonstrate SinCMat’s applicability to the case of LSCs. LSCs reside in the crypts of the limbus and replace the senescent or damaged cells of the cornea epithelium (Figure 5B) (Sacchetti et al., 2018; Collin et al., 2021). Dysfunction or loss of LSCs and/or their niche leads to LSC deficiency (LSCD), and currently there is no cure available for bilateral LSCDs. The in vitro generation of functional LSCs enabling successful cell therapy remains to be a challenge in the field (Elhusseiny et al., 2022). In this regard, we applied SinCMat to human LSCs from the TS to identify the TFs required for the proper function of LSCs.
Among SinCMat predictions (Figure 5C; Table S5), the well-known LSC marker TP63 was associated with RELA (Pellegrini et al., 2001). TP63 is involved in epithelial SC maintenance, and its expression was positively correlated with successful engraftment therapies (Bhattacharya et al., 2019; De Luca et al., 2006). PAX6, another master regulator responsible for LSC characteristics, was also predicted (Li et al., 2015). Although little evidence exists for the other predicted TFs, they find some support for their functional roles in closely related cells. For example, ASCL2 and FOXP2 are known to have a role in the maintenance of the quiescent state of SCs (Leishman et al., 2013; Schuijers et al., 2015).
Beyond experimental literature support, GO enrichment analyses revealed that functional genes targeted by the top 5 ITF-STF pairs were associated with LSC properties such as “stem cell population maintenance,” “keratinocyte differentiation,” and “regulation of epithelial cell proliferation” (Figure 5D). In addition to those functions, LSCs possess the ability to migrate from their crypts as they start to differentiate toward the cornea (Puri et al., 2020). Consistent with this, the enrichment of GO cellular component terms included “cell leading edge,” “ruffle,” and “cell-substrate junction” for TP63-RELA target genes (Figure 5E). Overall, these results identify candidate TFs required for LSCs to be fully functional and demonstrate the applicability of SinCMat to progenitors/SCs.
Finally, we interrogated the capacity of SinCMat to accurately associate relevant STFs to PAX6, a lineage-specific TF that targets different functions at different time points of the differentiation path. More specifically, PAX6 initially determines the LSC lineage but is also essential for the corneal cell-fate specification and is also known to be important for the proper function of corneal epithelial cells (CECs) (Sunny et al., 2022). Application of SinCMat to CECs indeed predicted PAX6. While PAX6 was paired with NCOA1 in the LSC prediction, it is paired with NFKB1 in CECs (Table S5). Functional GO analysis of the unique sets of co-targeted functional genes of each of these pairs resulted in an enrichment for distinct processes (Figure S3). In LSCs, PAX6-NCOA1 regulates genes involved in SC differentiation, cell-cell junction organization, and proliferation through the leukemia inhibitory factor response (Ramaesh et al., 2004) (Figure 5F). On the other hand, PAX6-NFKB1 controls macroautophagy, an essential guard of cornea homeostasis (Dias-Teixeira et al., 2022). These observations demonstrate that in the case of lineage-specific rather than cell-type-specific TFs, SinCMat can accurately associate STFs that, together with the ITFs, will co-target functional genes necessary for the function of each cell type.
Comparison with existing computational methods for prediction of reprogramming TFs
SinCMat is, to our knowledge, the first computational method specifically designed for predicting cell maturation TFs rather than more classical cell reprogramming TFs and is therefore more complementary to than competition for other existing computational methods. Nevertheless, the performance of SinCMat on maturation TF prediction was compared to the approaches commonly used for reprogramming TF prediction including CellNet, Mogrify, and the entropy-based method from D’Alessio et al. (Rackham et al., 2016; Cahan et al., 2014; D’Alessio et al., 2015). We first compared SinCMat with the D’Alessio et al. method, which relies on TFs that are uniquely expressed in the target cell type with respect to the heterogeneous background using JSD. Overall, the result showed that in almost all cases, SinCMat outperformed with a significantly higher total number of recovered known maturation TFs (Figures 6A, 6B, and S4A; Table S6).
Figure 6.
Comparison with preexisting methods
(A and B) Number of recapitulated known maturation TFs by SinCMat (n = 10) and D’Alessio et al. (n = 10), with predictions (A) cell type by cell type and (B) with all cell types combined. Significance was calculated by Wilcoxon rank-sum test. ∗∗p < 0.01.
(C and D) Number of recapitulated known differentially expressed (p < 0.05 and log2FC > 1) maturation TFs by SinCMat (n = 10), CellNet (n = 10), and Mogrify (n = 8), with predictions (C) cell type by cell type and (D) with all cell types pooled (Mogrify predictions are multiplied by 1.25 to elude any bias linked to the difference in total number of predictions). Significance was calculated by Wilcoxon rank-sum test. ∗∗p < 0.01, ∗p < 0.05.
(E) Proportion of the 10% most highly expressed DE and non-DE genes (p < 0.05 and log2FC > 1) between the target cell type and fibroblasts.
(F) Total number of TFs targeting cardiac maturation genes based on TF-Target database.
Next, we compared SinCMat with Mogrify and CellNet, which, unlike SinCMat, both consider only differentially expressed TFs (DETFs) between the initial and target cell types as potential candidate reprogramming TFs. Therefore, to establish a comparison as fair as possible, we applied SinCMat as described above, with the constraint in stage 4 to select DETFs with fibroblasts as starting cell type. The results showed that in this comparison too, SinCMat overall significantly outperformed Mogrify and CellNet (Figures 6C, 6D, S4B, and S4C).
The performance difference between SinCMat and the preexisting methods may come from the fact that our algorithm incorporates a biological model of cell maturation. Indeed, to predict TFs for cell conversion, the D’Alessio et al. method collects gene expression data from a wide range of cell types and identifies uniquely expressed TFs in target cell types. While the first stage of SinCMat is similar to this, SinCMat complements cell-type-specific ITFs with broadly expressed STFs and bases its predictions on co-target functional genes. Mogrify and CellNet rank candidate reprogramming TFs based on the gene expression differences of both TFs and their target genes between the initial and target cell types. However, we observed that functional genes are often not DE even between distantly related cell types. For example, DE genes between various target cell types and fibroblasts showed that a low percentage of target genes used by SinCMat are DE (Figure 6E).
Finally, we selected a widely studied case—cardiomyocytes (CMs)—and investigated the extent to which the different methods can target CM functional genes. During CM maturation, CMs acquire specific morphological, electrophysiological, and contractile characteristics that make mature CMs distinguishable from their immature counterparts (Karbassi et al., 2020). The results demonstrated that overall, TFs predicted by SinCMat can target to a greater extent functional genes related to these CM maturation processes than the other methods (Figure 6F; Table S6).
One of the strengths of SinCMat lies in its ability to predict functional maturation TFs from only the information on the target cell type without requiring any additional input. Nonetheless, we compared its performance with FateCompass, a tool that computes TF activity over time (Jiménez et al., 2023), by using time-series datasets of maturing cells/organs. Interestingly, our results unveiled that TF activity over the cell maturation time course does not correlate with the concept of functional cell maturation considered in this study, supporting the need for a model that takes into account the co-binding property of ITFs/STFs. Indeed, our results demonstrated that SinCMat is not only able to recover a higher number of known maturation TFs, but it also predicts TFs that target a significantly higher proportion of functional genes (Tables S1 and S6; Figures S4D and S4E).
Taken together, these results demonstrate that SinCMat, by considering the model of cell maturation, more accurately predicts cell maturation TFs than the preexisting methods.
Discussion
Generating tissue-specific functionally mature cells in in vitro systems is of importance for clinical applications. However, numerous studies have shown that current differentiation or reprogramming protocols are still unable to systematically produce fully mature cells. While computational approaches previously developed for cell conversion carefully address the conversion of cell identity, they are not specifically designed to take into account the functional cell maturation process. To address this important challenge, we have introduced the first computational method, SinCMat, for systematically predicting functional maturation TFs for any cell type identified in scRNA-seq data. Following the stepwise maturation model (Heinz et al., 2010; Wortham and Sander, 2021), SinCMat is an innovative predictive framework that integrates cell identity and maturation factors and leverages the co-targeting properties of ITFs and STFs on functional genes. In addition, we have also constructed SinCMatDB, a manually curated database for known maturation TFs that was further used to assess the performance of SinCMat.
The large-scale application of SinCMat to a set of adult mouse and human cell types in the MCA and TS accurately recapitulated known maturation TFs and predicted novel candidates. Moreover, our specific case studies demonstrated SinCMat’s applicability to both post-mitotic cells and progenitors/SCs. As SinCMat is designed specifically for maturation TF prediction rather than more traditional reprogramming TFs, SinCMat is intended to be used in complement with the preexisting computational methods. Such use cases could be to prioritize functional maturation TFs for one-step reprogramming experiments or to augment missing functionalities in in-vitro-engineered cells. It is important to note that functional cell maturation experiments may also require the signals linked to the predicted STFs.
Embedded in a user-friendly web application, SinCMat does not require any computer programming knowledge. As SinCMat only requires scRNA-seq data of the target cell, users can easily apply the tool to any mouse or human cell type of their interest. We foresee that SinCMat will constitute a valuable platform that can potentially address current cell engineering challenges in many different tissues/organs.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Antonio del Sol (antonio.delsol@uni.lu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
SinCMat was implemented in R, and the code repository is available from GitLab (https://git-r3lab.uni.lu/CBG/sincmat). The web application was developed with the PAWS framework and is available at https://sincmat.lcsb.uni.lu/.
Human and mouse background scRNA-seq data
The following publicly available processed scRNA-seq data and metadata were used in this study: GEO: GSE176063 (MCA) and GSE201333 (TS). Data from adrenal gland, brain, calvaria, heart, intestine, islet, kidney, liver, lung, pancreas, peripheral blood, rib, skin, and stomach were used from the MCA to compile the mouse background. Cell annotation was manually homogenized. Bladder, heart, eye, kidney, large intestine, liver, lung, muscle, pancreas, skin, small intestine, and tongue were obtained from the TS to compile the human background. The initial annotation from TS was kept.
Compilation of TF-Target database
Mouse and human TF-Target databases were constructed by aggregating multiple data sources such as chromatin immunoprecipitation (ChIP)-seq peak and TF binding motif information, literature-curated resources, and inference from gene expression (see also the supplemental experimental procedures). The resulting TF-Target databases contain 6,946,884 and 11,637,929 unique interactions in mouse and human, respectively.
ITF and STF classification
TFs were classified into ITFs/STFs, as described in Brivanlou and Darnell (2002), regarding their responsiveness to environment stimuli or their role in identity establishment (Table S7).
scRNA-seq data analysis
SinCMat pipeline was implemented in R (R v.4.0.5). scRNA-seq data analysis and differential gene expression tests with default settings (adjusted p value [p_val_adj] < 0.05 and log2 fold change [log2FC] > 1) were performed with R package Seurat v.4.1.1. Fisher’s exact and Wilcoxon tests were done with the R packages stats v.4.0.5 and rstatix v.0.7.0, respectively. GO overrepresentation analyses for biological processes, molecular functions, and cellular components were conducted with the R package clusterProfiler v.4.2.2 (p value cutoff = 0.01 and q value cutoff = 0.05).
Construction of SinCMatDB
We constructed a database referencing TFs that have experimentally been shown to be involved in cell maturation. We obtained TF evidences from PubMed by searching a list of keywords such as “mature,” “maturation,” “function,” and “functional,” associated with “transcription factors” and names of cell types. In total, SinCMatDB contains 950 TF evidences among 70 cell types. Literature evidences are classified by cell type (Table S2). The online version of SinCMatDB also contains 457 evidences for exogeneous treatments (with natural or synthetic compounds). Treatment evidences were collected using keywords such as “promotes,” “enhances,” “function,” and “maturation.” Similarly, treatment articles are classed by cell type.
Identification of candidate functional gene set
Functional GO terms were collected from the Cell Ontology database with the keyword “capable of” (https://www.ebi.ac.uk/ols/ontologies/cl). In each cell type, all expressed genes (mean expression > 0) were ranked by expression, and the ranks were normalized between 0 and 100 following the equation
where is the rank of gene A and is the maximum rank in the cell type. See also the results.
Algorithm of SinCMat
Stage 1: Identification of cell-type-specific ITFs
SinCMat first creates a Seurat object from an input raw count matrix and normalizes the data by the total expression with the NormalizeData() function. The Seurat object is then integrated into our mouse or human background as described in Stuart et al. (2019) with the FindIntegrationAnchors() function to remove any batch effect. Then, ITFs for the target population are identified by scJSD, in which JSD is computed for each cell and each TF, and then for each TF, all JSD values over all cells are summed. The top 20 lowest summed JSD value TFs are selected as ITFs. For efficient computation in the scJSD step, human background is downsampled to set the maximum number of cells in each population to 1,000. From the normalized count matrix, non-expressed genes in the target cell type (mean expression = 0) and housekeeping genes (obtained from Hounkpe et al., 2021) are removed, genes are ranked by mean expression, and the top 10% genes are taken as the functional gene set (see above).
Stage 2: Creation of ITF-STF pairs
With the 20 ITFs identified in stage 1, SinCMat creates all possible ITF-STF pairs with the entire set of STFs. As both ITFs and STFs need to be expressed in the target cell type to confer cell identity and functions, the top 10% ITF-STF pairs with the highest number of cells expressing both TFs are kept for subsequent steps.
Stage 3: Computation of the functional score
Next, for each ITF-STF pair, SinCMat computes the functional score (FS), defined as
where represents the Jaccard similarity coefficient that gauges the similarity of functional target genes between the ITF and STF and is the Pearson correlation coefficient between each TF of the ITF-STF pair and each of the co-target functional genes, which is summed over all TF-co-target functional gene pairs. The Pearson correlation coefficient is computed in single cells where both ITFs and STFs are expressed. Finally, to normalize the sum of Pearson correlation coefficients between 0 and 1, the summed value is divided by , where is the number of co-target functional genes. The rationale behind the FS is that by computing the Jaccard similarity coefficient, we prioritize ITF-STF pairs that share high numbers of functional target genes. Then, to also give more importance to pairs that more strongly regulate their co-target functional genes in the target cell type, the normalized sum of Pearson correlation coefficient is added. The normalization is performed to attribute the same weight as the Jaccard similarity coefficient, which also ranges from 0 to 1.
Stage 4: Ranking of ITF-STF pairs
Finally, SincMat returns as output a list of ITF-STF pairs that are ranked by the FS and provides the number of functional co-target genes for each pair. In addition, the web interface informs the user of which predicted TFs have previously been experimentally validated.
Evaluation of SinCMat
The MCA and TS datasets were used to evaluate the performance of SinCMat. SinCMat’s performance was assessed with cell types for which at least 5 TFs had literature evidence (Table S2). Given the decreasing number of co-target functional genes of predicted ITF-STF pairs, which on average falls below 50% at the 6th pair, we benchmarked SinCMat with the 5 top pairs (Figure S4). Fisher’s exact test was used to assess the enrichment of SinCMat’s predictions in validated TFs (Table S4). In silico double KO was performed with the R package scTenifoldKnk v.1.0.2 on raw TS data with default parameters and nc_q = 0.999. One-sample Wilcoxon test was used to compare the mean coverage of functional GO genes (Table S1) between the top 5 ITFs predicted by SinCMat and random sets of 5 ITFs from the 20 ITFs identified in stage 1.
Acknowledgments
We thank Celine Barlier for the processing of ChIP-Atlas raw data. We thank Moustapha Cheick, Valentin Grouès, Miroslav Kratochvil, Hesam Korki, François Ancien, and Jacek Lebioda for the development of the web interface. We thank Sascha Jung for giving us valuable feedback for developing the computational method. S.B. and S.O. are supported by a CORE grant from Fonds National de la Recherche Luxembourg (project number: R-AGR-3676-10).
Author contributions
A.d.S. conceived the overall study. B.S. and S.O. developed the computational method. S.B. performed the analysis, created SinCMatDB, and implemented the visualizations. S.B., S.O., and A.d.S. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.12.006.
Supplemental information
GO IDs were collected from the Cell Ontology Database with the keyword search “capable of” and complemented with prior literature knowledge for well known functions. 18 and 16 cell types are present in the Mouse Cell Atlas and Tabula Sapiens, respectively.
SinCMatDB is a manually curated database resuming known maturing TFs. TF evidence was obtained from PubMed search with the use of a list of keywords such as “mature”, “maturation”, “function”, “functional” associated with “transcription factors” and names of cell types. In total, SinCMatDB contains 950 TF evidences among 66 cell types. Literature evidence is classified by cell types.
SinCMat was applied to the MCA and Tabula Sapiens datasets. Sheet 1 contains predictions for MCA and Sheet 2 contains predictions for TS. Experimentally validated TFs are marked in bold. See also Table S2 for literature evidence.
SinCMat’s performance was assessed with cell types for which at least 5 TFs had literature evidence (Table S3). Given the decreasing number of co-target functional genes of predicted pairs, which on average falls below 50% at the 6th pair, we benchmarked SinCMat with the 5 top pairs (Figure S4). Fisher’s exact test was used to assess the enrichment of SinCMat’s predictions in validated TFs. Sheet 1 contains Fisher’s exact test results for predictions performed in mouse cell types, sheet 2 contains results for human predictions.
This table contains de entire set of predicted maturation TFs for dopaminergic neurons (sheet1), LSCs (sheet2) and CECs (sheet3). As input were used the scRNA-seq data published by Kamath et al. (2022) and the Tabula Sapiens dataset for dopaminergic neurons and LSCs/CECs respectively. For each predicted pair, the table provides information regarding the Jaccard coefficient index, the number of functional co-targets, the summed Pearson’s correlation score and the Functional Score.
The table compares ranked lists of predicted TFs for examples where predictions from all methods are available. The retrieved TFs are denoted with a yellow background (see Table S2 for literature references). Sheet 1 presents the comparison of SinCMat with d’Alessio et al. Sheet 2 presents the comparison with Mogrify and CellNet. Both sheets provide detailed information regarding the TF predictions and the number of retrieved well-known TFs for each method. Shet3 summarizes the sources cell types for all predictions. 10TFs were used for D’Alessio et al., CellNet and SinCMat. As only 8 TFs are available for Mogrify, to avoid any bias linked to the number of TFs, the numbers of validated TFs from Mogrify are multiplied by a factor of 1.25. Sheet4 summarizes which TFs among predictions of SinCMat, Mogrify, CellNet and d’Alessio et al. can target cardiomyocyte maturation markers genes listed in the first column based on prior TF-Target information. This table contains the gene names and references for cardiomyocyte maturation markers. Sheet 5 presents the comparison of SinCMat with FateCompass, The retrieved TFs are denoted with a yellow background. Sheet 5 provides also all the information about the parameters used for the FateCompass analysis (components, n_neighbors, tolerance, variability threshold and z-score threshold).
This table contains the classification ITF/STF used in this study.
References
- Alexander G.E. Biology of Parkinson’s disease: Pathogenesis and pathophysiology of a multisystem neurodegenerative disorder. Dialogues Clin. Neurosci. 2004;6:259–280. doi: 10.31887/DCNS.2004.6.3/galexander. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Dominguez J.R., Melton D.A. Cell maturation: Hallmarks, triggers, and manipulation. Cell. 2022;185:235–249. doi: 10.1016/j.cell.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S., Choi H., Kim J. Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 2014;1587:23–32. doi: 10.1016/j.brainres.2014.08.059. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Serror L., Nir E., Dhiraj D., Altshuler A., Khreish M., Tiosano B., Hasson P., Panman L., Luxenburg C., et al. SOX2 Regulates P63 and Stem/Progenitor Cell State in the Corneal Epithelium. Stem Cell. 2019;37:417–429. doi: 10.1002/stem.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans L.L., Soh H., Wood W.M., Nolan T.M., Mandoiu I.I., Yanagawa Y., Tzingounis A.V., Nishiyama A. Direct reprogramming of oligodendrocyte precursor cells into GABAergic inhibitory neurons by a single homeodomain transcription factor Dlx2. Sci. Rep. 2021;11:3552. doi: 10.1038/s41598-021-82931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou A.H., Darnell J.E. Transcription: Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- Bus C., Zizmare L., Feldkaemper M., Geisler S., Zarani M., Schaedler A., Klose F., Admard J., Mageean C.J., Arena G., et al. Human Dopaminergic Neurons Lacking PINK1 Exhibit Disrupted Dopamine Metabolism Related to Vitamin B6 Co-Factors. iScience. 2020;23:101797. doi: 10.1016/j.isci.2020.101797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Li H., Morris S.A., Lummertz Da Rocha E., Daley G.Q., Collins J.J. CellNet: Network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieślar-Pobuda A., Knoflach V., Ringh M.V., Stark J., Likus W., Siemianowicz K., Ghavami S., Hudecki A., Green J.L., Łos M.J. Transdifferentiation and reprogramming: Overview of the processes, their similarities and differences. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1359–1369. doi: 10.1016/j.bbamcr.2017.04.017. [DOI] [PubMed] [Google Scholar]
- Collin J., Queen R., Zerti D., Bojic S., Dorgau B., Moyse N., Molina M.M., Yang C., Dey S., Reynolds G., et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 2021;21:279–298. doi: 10.1016/j.jtos.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio A.C., Fan Z.P., Wert K.J., Baranov P., Cohen M.A., Saini J.S., Cohick E., Charniga C., Dadon D., Hannett N.M., et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015;5:763–775. doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Pellegrini G., Green H. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen. Med. 2006;1:45–57. doi: 10.2217/17460751.1.1.45. [DOI] [PubMed] [Google Scholar]
- de Lucas B., Pérez L.M., Gálvez B.G. Importance and regulation of adult stem cell migration. J. Cell Mol. Med. 2018;22:746–754. doi: 10.1111/jcmm.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Teixeira K.L., Sharifian Gh M., Romano J., Norouzi F., Laurie G.W. Autophagy in the normal and diseased cornea. Exp. Eye Res. 2022;225 doi: 10.1016/j.exer.2022.109274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl A.D., Meehan T.F., Bradford Y.M., Brush M.H., Dahdul W.M., Dougall D.S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S., et al. The cell ontology 2016: Enhanced content, modularization, and ontology interoperability. J. Biomed. Semant. 2016;7:44. doi: 10.1186/s13326-016-0088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhusseiny A.M., Soleimani M., Eleiwa T.K., ElSheikh R.H., Frank C.R., Naderan M., Yazdanpanah G., Rosenblatt M.I., Djalilian A.R. Current and Emerging Therapies for Limbal Stem Cell Deficiency. Stem Cells Transl. Med. 2022;11:259–268. doi: 10.1093/stcltm/szab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K., Park G., Iezaki T., Horie T., Kanayama T., Ozaki K., Onishi Y., Takahata Y., Yoneda Y., Takarada T., et al. ATF3 controls proliferation of osteoclast precursor and bone remodeling. Sci. Rep. 2016;6:30918. doi: 10.1038/srep30918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmozzi V., Gentile G., Riekschnitz D.A., Von Troyer M., Lavdas A.A., Kerschbamer E., Weichenberger C.X., Rosato-Siri M.D., Casarosa S., Conti L., et al. Generation of hiPSC-Derived Functional Dopaminergic Neurons in Alginate-Based 3D Culture. Front. Cell Dev. Biol. 2021;9:708389. doi: 10.3389/fcell.2021.708389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Hashizume K., Yamanaka M., Ueda S. POU3F2 participates in cognitive function and adult hippocampal neurogenesis via mammalian-characteristic amino acid repeats. Gene Brain Behav. 2018;17:118–125. doi: 10.1111/gbb.12408. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hounkpe B.W., Chenou F., de Lima F., de Paula E.V. HRT Atlas v1.0 database: Redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res. 2021;49:D947–D955. doi: 10.1093/nar/gkaa609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lu Y., Jiang X., Hu J., Li R., Liu Y., Zhu G., Rong X. Glucocorticoids induce osteoporosis mediated by glucocorticoid receptor-dependent and -independent pathways. Biomed. Pharmacother. 2020;125:109979. doi: 10.1016/j.biopha.2020.109979. [DOI] [PubMed] [Google Scholar]
- Jiménez S., Schreiber V., Mercier R., Gradwohl G., Molina N. Characterization of cell-fate decision landscapes by estimating transcription factor dynamics. Cell Rep. Methods. 2023;3:100512. doi: 10.1016/j.crmeth.2023.100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P., Harper W., et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376:6594. doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath T., Abdulraouf A., Burris S.J., Langlieb J., Gazestani V., Nadaf N.M., Balderrama K., Vanderburg C., Macosko E.Z. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease Nature Neuroscience. Springer US. 2022;25(Issue 5):588–595. doi: 10.1038/s41593-022-01061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Khang R., Ham S., Jeong G.R., Kim H., Jo M., Lee B.D., Lee Y.I., Jo A., Park C., et al. Activation of the ATF2/CREB-PGC-1α pathway by metformin leads to dopaminergic neuroprotection. Oncotarget. 2017;8:48603–48618. doi: 10.18632/oncotarget.18122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbassi E., Fenix A., Marchiano S., Muraoka N., Nakamura K., Yang X., Murry C.E. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020;17:341–359. doi: 10.1038/s41569-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T., Morizane A., Doi D., Magotani H., Onoe H., Hayashi T., Mizuma H., Takara S., Takahashi R., Inoue H., et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature. 2017;548:592–596. doi: 10.1038/nature23664. [DOI] [PubMed] [Google Scholar]
- Lebedeva O.S., Sharova E.I., Grekhnev D.A., Skorodumova L.O., Kopylova I.V., Vassina E.M., Oshkolova A., Novikova I.V., Krisanova A.V., Olekhnovich E.I., et al. An Efficient 2D Protocol for Differentiation of iPSCs into Mature Postmitotic Dopaminergic Neurons: Application for Modeling Parkinson’s Disease. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24087297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman E., Howard J.M., Garcia G.E., Miao Q., Ku A.T., Dekker J.D., Tucker H., Nguyen H. Foxp1 maintains hair follicle stem cell quiescence through regulation of Fgf18. Development (Camb.) 2013;140:3809–3818. doi: 10.1242/dev.097477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepack A.E., Werner C.T., Stewart A.F., Fulton S.L., Zhong P., Farrelly L.A., Smith A.C.W., Ramakrishnan A., Lyu Y., Bastle R.M., et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science. 2020;368:197–201. doi: 10.1126/science.aaw8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xu F., Zhu J., Krawczyk M., Zhang Y., Yuan J., Patel S., Wang Y., Lin Y., Zhang M., et al. Transcription factor PAX6 (paired box 6) controls limbal stem cell lineage in development and disease. J. Biol. Chem. 2015;290:20448–20454. doi: 10.1074/jbc.M115.662940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr K.M., Bernstein A.I., Stout K.A., Dunn A.R., Lazo C.R., Alter S.P., Wang M., Li Y., Fan X., Hess E.J., et al. Increased vesicular monoamine transporter enhances dopamine release and opposes Parkinson disease-related neurodegeneration in vivo. Proc. Natl. Acad. Sci. USA. 2014;111:9977–9982. doi: 10.1073/pnas.1402134111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajani S., Raina A., Fokken C., Kügler S., Bähr M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019;10:898. doi: 10.1038/s41419-019-2133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic J., Maisel M., Wegner F., Leuchtenberger J., Wenger R.H., Gerlach M., Storch A., Schwarz J. Lack of hypoxia-inducible factor-1α impairs midbrain neural precursor cells involving vascular endothelial growth factor signaling. J. Neurosci. 2007;27:412–421. doi: 10.1523/JNEUROSCI.2482-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Orlando D.A., Newman J.J., Lovén J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., Dekoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio D., Zhong Y., Li G., Xu Q., Yang Y., Tian Y., Chapkin R.S., Huang J.Z., Cai J.J. scTenifoldKnk: An efficient virtual knockout tool for gene function predictions via single-cell gene regulatory network perturbation. Patterns. 2022;3 doi: 10.1016/j.patter.2022.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. P63 Identifies Keratinocyte Stem Cells. Proc. Natl. Acad. Sci. USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S., Sun M., Mutoji K.N., Gesteira T.F., Coulson-Thomas V.J. Epithelial cell migration and proliferation patterns during initial wound closure in normal mice and an experimental model of limbal stem cell deficiency. Invest. Ophthalmol. Vis. Sci. 2020;61:27. doi: 10.1167/iovs.61.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puymirat J., Barret A., Picart R., Vigny A., Loudes C., Faivre-Bauman A., Tixier-Vidal A. Triiodothyronine enhances the morphological maturation of dopaminergic neurons from fetal mouse hypothalamus cultured in serum-free medium. Neuroscience. 1983;10:801–810. doi: 10.1016/0306-4522(83)90217-8. [DOI] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O.J.L., Firas J., Fang H., Oates M.E., Holmes M.L., Knaupp A.S., FANTOM Consortium. Suzuki H., Nefzger C.M., Daub C.O., et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016;48:331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- Ramaesh K., Ramaesh T., West J.D., Dhillon B. Immunolocalisation of leukaemia inhibitory factor in the cornea. Eye. 2004;18:1006–1009. doi: 10.1038/sj.eye.6701394. [DOI] [PubMed] [Google Scholar]
- Ribeiro M.M., Okawa S., del Sol A. TransSynW: A single-cell RNA-sequencing based web application to guide cell conversion experiments. Stem Cells Transl. Med. 2021;10:230–238. doi: 10.1002/sctm.20-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti M., Rama P., Bruscolini A., Lambiase A. Limbal stem cell transplantation: Clinical results, limits, and perspectives. Stem Cell. Int. 2018;2018:8086269. doi: 10.1155/2018/8086269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., Van Der Flier L.G., Cuppen E., Van Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.L., Hurley K., Villacorta-Martin C., Huang J., Hinds A., Gopalan K., Caballero I.S., Russo S.J., Kitzmiller J.A., Whitsett J.A., et al. Heterogeneity in human induced pluripotent stem cell–derived alveolar epithelial type II cells revealed with ABCA3/SFTPC reporters. Am. J. Respir. Cell Mol. Biol. 2021;65:442–460. doi: 10.1165/rcmb.2020-0259OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny S.S., Lachova J., Dupacova N., Kozmik Z. Multiple roles of Pax6 in postnatal cornea development. Dev. Biol. 2022;491:1–12. doi: 10.1016/j.ydbio.2022.08.006. [DOI] [PubMed] [Google Scholar]
- Tsukasaki M., Takayanagi H. Osteoclast biology in the single-cell era. Inflamm. Regen. 2022;42 doi: 10.1186/s41232-022-00213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaescusa J.C., Li B., Toledo E.M., Rivetti di Val Cervo P., Yang S., Stott S.R., Kaiser K., Islam S., Gyllborg D., Laguna-Goya R., et al. A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson’s disease. EMBO J. 2016;35:1963–1978. doi: 10.15252/embj.201593725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang Y., Liu J., Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 2021;22:410–424. doi: 10.1038/s41580-021-00335-z. Nature Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortham M., Sander M. Transcriptional mechanisms of pancreatic β-cell maturation and functional adaptation. Trends Endocrinol. Metabol. 2021;32:474–487. doi: 10.1016/j.tem.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Du Y., Deng H. Direct lineage reprogramming: Strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. Cell Press. [DOI] [PubMed] [Google Scholar]
- Xu Q., Georgiou G., Frölich S., Van Der Sande M., Veenstra G.J.C., Zhou H., Van Heeringen S.J. ANANSE: An enhancer network-based computational approach for predicting key transcription factors in cell fate determination. Nucleic Acids Res. 2021;49:7966–7985. doi: 10.1093/nar/gkab598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yang Y., Huang J., Coppola G., Geschwind D.H., Bonni A. Sumoylated MEF2A coordinately eliminates orphan presynaptic sites and promotes maturation of presynaptic boutons. J. Neurosci. 2013;33:4726–4740. doi: 10.1523/JNEUROSCI.4191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.X., Glass C.K. Towards an understanding of cell-specific functions of signaldependent transcription factors. J. Mol. Endocrinol. 2013;51:T37–T50. doi: 10.1530/JME-13-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Guo T., Han Y., Huang H., Shi J., Hu J., Li H., Wang J., Saleem A., Zhou P., Lan F. Design of synthetic microenvironments to promote the maturation of human pluripotent stem cell derived cardiomyocytes. J. Biomed. Mater. Res. B Appl. Biomater. 2021;109(Issue 7):949–960. doi: 10.1002/jbm.b.34759. John Wiley and Sons Inc. [DOI] [PubMed] [Google Scholar]
- Zhu B., Carmichael R.E., Solabre Valois L., Wilkinson K.A., Henley J.M. The transcription factor MEF2A plays a key role in the differentiation/maturation of rat neural stem cells into neurons. Biochem. Biophys. Res. Commun. 2018;500:645–649. doi: 10.1016/j.bbrc.2018.04.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO IDs were collected from the Cell Ontology Database with the keyword search “capable of” and complemented with prior literature knowledge for well known functions. 18 and 16 cell types are present in the Mouse Cell Atlas and Tabula Sapiens, respectively.
SinCMatDB is a manually curated database resuming known maturing TFs. TF evidence was obtained from PubMed search with the use of a list of keywords such as “mature”, “maturation”, “function”, “functional” associated with “transcription factors” and names of cell types. In total, SinCMatDB contains 950 TF evidences among 66 cell types. Literature evidence is classified by cell types.
SinCMat was applied to the MCA and Tabula Sapiens datasets. Sheet 1 contains predictions for MCA and Sheet 2 contains predictions for TS. Experimentally validated TFs are marked in bold. See also Table S2 for literature evidence.
SinCMat’s performance was assessed with cell types for which at least 5 TFs had literature evidence (Table S3). Given the decreasing number of co-target functional genes of predicted pairs, which on average falls below 50% at the 6th pair, we benchmarked SinCMat with the 5 top pairs (Figure S4). Fisher’s exact test was used to assess the enrichment of SinCMat’s predictions in validated TFs. Sheet 1 contains Fisher’s exact test results for predictions performed in mouse cell types, sheet 2 contains results for human predictions.
This table contains de entire set of predicted maturation TFs for dopaminergic neurons (sheet1), LSCs (sheet2) and CECs (sheet3). As input were used the scRNA-seq data published by Kamath et al. (2022) and the Tabula Sapiens dataset for dopaminergic neurons and LSCs/CECs respectively. For each predicted pair, the table provides information regarding the Jaccard coefficient index, the number of functional co-targets, the summed Pearson’s correlation score and the Functional Score.
The table compares ranked lists of predicted TFs for examples where predictions from all methods are available. The retrieved TFs are denoted with a yellow background (see Table S2 for literature references). Sheet 1 presents the comparison of SinCMat with d’Alessio et al. Sheet 2 presents the comparison with Mogrify and CellNet. Both sheets provide detailed information regarding the TF predictions and the number of retrieved well-known TFs for each method. Shet3 summarizes the sources cell types for all predictions. 10TFs were used for D’Alessio et al., CellNet and SinCMat. As only 8 TFs are available for Mogrify, to avoid any bias linked to the number of TFs, the numbers of validated TFs from Mogrify are multiplied by a factor of 1.25. Sheet4 summarizes which TFs among predictions of SinCMat, Mogrify, CellNet and d’Alessio et al. can target cardiomyocyte maturation markers genes listed in the first column based on prior TF-Target information. This table contains the gene names and references for cardiomyocyte maturation markers. Sheet 5 presents the comparison of SinCMat with FateCompass, The retrieved TFs are denoted with a yellow background. Sheet 5 provides also all the information about the parameters used for the FateCompass analysis (components, n_neighbors, tolerance, variability threshold and z-score threshold).
This table contains the classification ITF/STF used in this study.
Data Availability Statement
SinCMat was implemented in R, and the code repository is available from GitLab (https://git-r3lab.uni.lu/CBG/sincmat). The web application was developed with the PAWS framework and is available at https://sincmat.lcsb.uni.lu/.