Abstract
α-Deuterated amino acids are valuable building blocks for developing deuterated drugs, and are important tools for studying biological systems. Biocatalytic deuteration represent an attractive strategy to directly access enantiopure α-deuterated amino acids. Here, we show that a PLP-dependent Mannich cyclase, LolT, involved in the biosynthesis of loline alkaloids, is capable of deuterating a diverse range of l-amino acids, including basic and acidic, nonpolar and polar, aliphatic and aromatic amino acids. Furthermore, complete deuteration of many amino acids can be achieved within minutes with exquisite control on the site- and stereoselectivity. During the course of this investigation, we also unexpectedly discovered that LolT exhibits β-elimination activity with l-cystine and O-acetyl-l-serine, confirming our previous hypothesis based on structural and phylogenetic analysis that LolT, a Cα-C bond forming enzyme, is evolved from a primordial Cβ-S lyase family. Overall, our study demonstrates that LolT is an extreme versatile biocatalyst, and can be used for not only heterocyclic quaternary amino acid biosynthesis, but also biocatalytic amino acid deuteration.
Keywords: Enzyme, PLP, Deuteration, Amino Acids
Graphical Abstract

A PLP-dependent Mannich cyclase LolT is repurposed for α-deuteration of amino acids. Complete deuteration can be achieved with diverse l-amino acids bearing assorted functional groups, e.g. from lysine to glutamic acid. The broad substrate scope and strict stereoselectivity make LolT a powerful biocatalyst for preparation of α-deuterated l-amino acids.
Introduction
Deuterated compounds have very different chemical and physical properties from their nondeuterated counterparts, and have found wide applications in chemistry, biology, and medicine.[1] Deuterated amino acids can help to improve signals in protein NMR spectroscopy,[2] and are essential tools for protein neutron crystallography.[3] Due to the kinetic isotope effect, deuterated drugs often have slower metabolism and extended half-lives in vivo, maintaining potency but allowing for lower doses.[4] As a result, deuterium incorporation plays a very important role in drug discovery: to date two deuterated drugs have been approved by FDA (e.g. deutetrabenazine[5] and deucravacitinib[6]) while many others are currently under clinical trials.[7]
The importance of deuterated compounds has hence spurred great interests to develop synthetic methods to incorporate deuterium into common building blocks. Although a few chemical approaches have been developed to achieve direct stereoselective α-deuteration on unprotected α-amino acids, such as organocatalysis,[8] and metal-dependent C-H bond activation,[9] these methods often have limited functional group tolerance, incomplete deuteration, and sometimes modest site- and/or stereoselectivity. In comparison, biocatalytic deuteration represents an attractive strategy and offers several advantages, including excellent selectivity, green and sustainable reaction conditions.[10] In particular, enzyme catalyzed facile H/D exchange can efficiently incorporate deuterium from D2O, an inexpensive deuterium source.
Pyridoxal 5’-phosphate (PLP)-dependent enzymes are such enzymes. They are known to catalyze reversible deprotonation of amino acids with help from the PLP cofactor, which acts as an electron sink to stabilize corresponding carbanion intermediates.[11] Several PLP-dependent enzymes have been utilized for biocatalytic preparation of deuterated amino acids. For example, l-allo-isoleucine epimerase DsaD/DsaE system was recently used to prepare α/β-deuterated hydrophobic l-amino acids;[12] and an α-oxoamine synthase from saxitoxin biosynthetic pathway was capable of producing few α-deuterated l-amino acids and a relatively broad panel of α-amino acid methyl esters.[13] However, practical applications of these enzymes are still limited to deuteration of amino acids whose structures resemble enzymes’ native substrates.[14] Although PLP-dependent transaminases and racemases have broader substrate tolerance,[15] and have been shown to catalyze amino acids deuteration,[16] unfortunately these enzymes rarely have control over site- and stereoselectivity. Therefore, there is still an unmet need to expand the current toolbox of deuteration biocatalysts.
Recently our group reported the discovery of a C-C bond forming PLP-dependent enzyme, LolT, involved in the biosynthesis of loline alkaloids.[17] We showed that LolT catalyzes Mannich-type 5-endo-trig cyclization transforming an iminium amino acid 1 to a pyrrolizidine quaternary amino acid 2 (Figure 1a). Besides, we also demonstrated that LolT is a highly versatile cyclase in that it can also catalyze two-component Mannich cyclization reactions using various diamino acids and aldehydes as substrates, resulting in the biosynthetic formation of a variety of pyrrolidine and piperidine-based quaternary amino acids (e.g, 3 and 4, Figure 1b). Structural and mechanistic study of LolT predicts that in the absence of an imine or iminium electrophile, LolT should catalyze reversible proton exchange at Cα of amino acid substrates in a stereoretentive manner (Figure 1c). Indeed, LolT was shown to stereoselectively α-deuterate l-diamino acids of varying chain-lengths, including l-lysine, l-ornithine, l-2,3,-diaminobutanoic acid (l-Dab), and l-diaminopropanoic acid (l-Dap).[17] However, the promiscuous potential of LolT-catalyzed amino acid deuteration has not yet been revealed.
Figure 1.
LolT is a PLP-dependent Mannich cyclase. a) LolT’s natural function and its role in loline biosynthesis. b) LolT catalyzes two-component Mannich cyclization reactions. c) Proposed stereoretentive Cα H/D exchange catalyzed by LolT in the absence of electrophile. Residue Lys236 was proposed as the general base (abbreviated as B) for reversible deprotonation of l-amino acid substrate through the PLP si face.
Motivated by the continuous need of new amino acids deuteration biocatalysts with broader or complementary substrate scope while maintaining excellent selectivity, and encouraged by the promising results from LolT-catalyzed α-deuteration of l-diamino acids, we set out to test the possibility of repurposing LolT to deuterate a broad range of substrates beyond diamino acids. Here, we show that LolT has impressively relaxed substrate specificity as it is able to catalyze α-deuteration on almost all types of l-amino acids. More importantly, rapid and complete deuteration can be achieved with exquisite site- and stereoselectivity. The remarkable substrate promiscuity and high fidelity on stereochemical outcomes render LolT as one of the most promising amino acids deuteration biocatalyst.
Results and Discussion
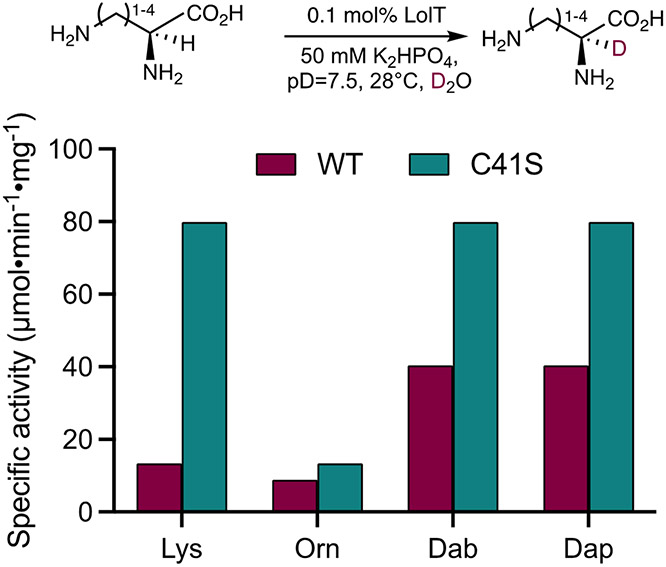
Before we tested the substrate flexibility of LolT-catalyzed amino acid deuteration, we first sought to engineer LolT to improve its stability, since in the previous investigation precipitated proteins were often observed if purified LolT was left at room temperature for overnight. Analysis of the crystal structure of holo-LolT (PDB entry 8DL5) reveals a cysteine residue (Cys41) partially buried at the dimer interface forming a hydrogen bond with the side chain from Ser67, a residue from the nearby monomer within the homodimer (Figure S1). We envisaged that an isosteric replacement of Cys41 with Ser could potentially strengthen this intermolecular hydrogen bond interaction due to increased electronegativity.[18] In addition, removing surface exposed cysteine residues can potentially facilitate protein crystallization and improve protein crystal quality,[19] which will simultaneously benefit our on-going crystallographic study of LolT. Accordingly, we expressed and purified LolT-C41S mutant using the previously described protocol.[17] No protein precipitates were observed with this mutant after overnight incubation at room temperature, suggesting C41S has improved protein stability as we predicted. More importantly, C41S was also found to exhibi higher α-deuteration activity with several l-diamino acids when compared to the wild-type (WT) protein (Figure 2). Because of its superior performance, all the subsequent characterizations were performed with C41S mutant instead of the WT protein.
Figure 2.
LolT C41S mutant has improved catalytic activity compared to the WT.
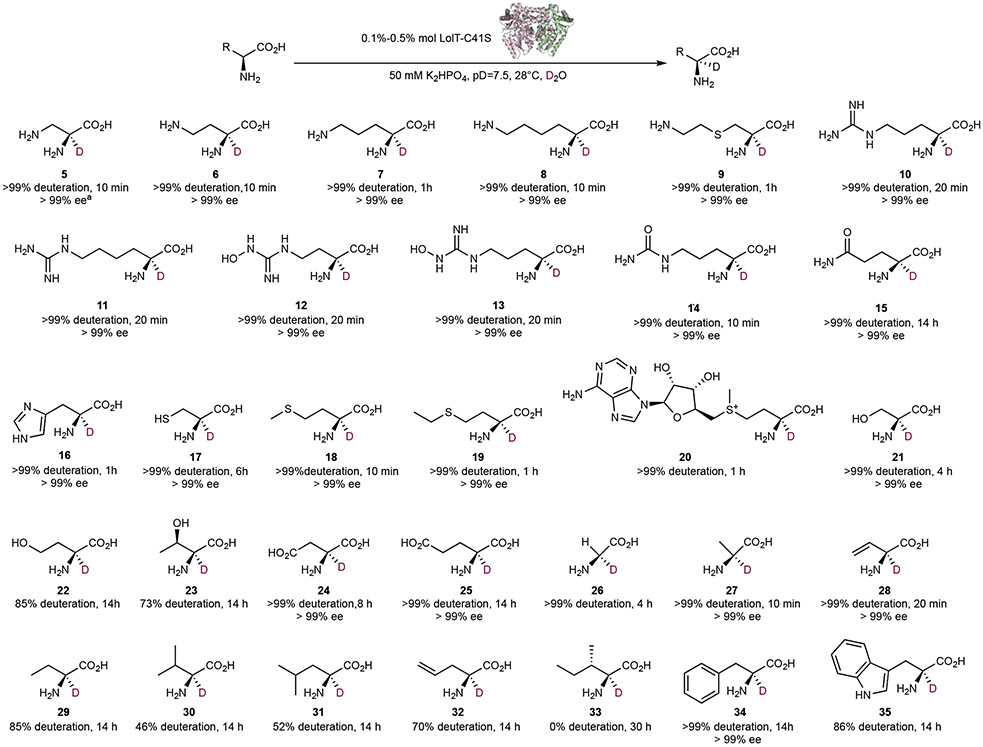
Next, we assayed the deuteration activity of LolT with a panel of amino acids. All enzymatic reactions were performed in D2O buffered with potassium phosphate. Reaction progresses were monitored by using NMR spectroscopy and LC-MS at 28 °C (see supporting information). As shown in Figure 3, most amino acid substrates showed modest to full level of deuteration. Not only does LolT recognizes supposedly preferred cationic amino acids, such as 5-11 and 20, but it can also efficiently deuterate amino acids whose physical properties are distinct from LolT’ s natural substrate 1. For example, neutral polar amino acids 12-17 and 21, nonpolar thioether-containing amino acids 18 and 19, as well as small and aliphatic amino acids 26-28, are all completely deuterated. Most of them can achieve complete deuteration within minutes. Surprisingly, even anionic amino acids 26 and 27, and aromatic amino acid 28, can be fully deuterated with extended incubation (i.e. 14h). Such unprecedented compatibility with both cationic and anionic substrates is consistent with the structural features revealed from previous docking study: there are minimal interactions between LolT and the amino acid substrate side chain.[17] Furthermore, the general preference to cationic substrates revealed here is also consistent with a cation-π interaction being operative in substrate recognition instead of ionic charge-charge interaction, since the latter would preclude binding of negatively charged substrates, e.g. 26 and 27.
Figure 3.
Substrate scope of LolT-catalyzed α-deuteration of amino acids. aenantiomeric excess (ee) was determined with Marfey derivatization.
By far, the only challenging substrates for LolT seem to be the hydrophobic branched-chain aliphatic amino acids. For example, l-valine 30 and l-leucine 31 showed modest deuteration after overnight incubation (14h), whereas l-isoleucine 33 showed no sign of deuteration even after prolonged incubation (up to 30h). Nevertheless, LolT is a highly promiscuous enzyme that can accommodate diverse amino acids, ranging from basic to acidic ones, from aliphatic to aromatic, and from the smallest amino acid glycine (26) to the bulky amino acid S-adenosyl-l-methionine (SAM, 20).
To assess the stereocontrol, all fully deuterated l-amino acids were subject to chiral resolution (see supporting information). High enantiopurity and complete stereoretentions were observed. In the cases where the corresponding d-enantiomers are commercially available, we also tested LolT’s deuteration activity with d-amino acids to examine its substrate enantioselectivity. In all cases tested, the respective d-enantiomers showed zero deuterium incorporation. Taken together, these results demonstrate that LolT shows strict stereochemical control, supporting the previous mechanistic proposal in which a general base residue must reside on the si face of PLP and is responsible for initial deprotonation and reprotonation, while the re face must be protected from solvent and other potential general acids to avoid stereoinversion during proton transfer steps, and to prevent activation of d-amino acids.
It is noteworthy to mention that developing a general approach for synthesizing enantiopure α-deuterated derivatives of serine 21, cysteine 17, and Dap 5 was considered a challenging task, and only a few chemical methodologies have been reported to access α-deuterated β-X-α-amino acids (X = N, O, S, or Se).[20] Our findings here demonstrate that LolT can efficiently deuterate these amino acids stereoselectively, which prompted us to further investigate whether LolT can deuterate their derivatives. Interestingly, when l-cystine 36 was incubated with LolT in D2O, enzymatic transformations in addition to α-deuteration took place (Figure 4a), as indicated by the unexpected loss of Cβ proton on the 1H-NMR spectra (Figure 4b). Such phenomenon is more consistent with an α,β-elimination reaction to give amino acrylate, which is unstable in aqueous solution and undergoes spontaneous tautomerization and hydrolysis to yield ammonium and pyruvate. Accordingly, we next derivatized the reaction mixtures with o-phenylenediamine (OPD) and confirmed the formation of the deamination product pyruvate (Figure 4c). Similar results were also observed with O-acetyl-l-serine (data not shown). These results suggest LolT may not be suitable for α-deuteration on amino acids with good leaving groups at Cβ. Determining the steady-state kinetics of cystine β-elimination shows a slow kcat of 0.53 ± 0.03 min−1 and a reasonable Km = 1.4 ± 0.3 mM (Figure 4d). Although the overall low catalytic efficiency (kcat/Km = 6.3 M−1s−1) indicates LolT is not optimized for this β-lyase reaction, it supports our previous postulate that LolT may share the last common ancestor with Cβ-S lyases,[17] and has evolved far away from Cβ-S lyases. However, it still remains a question how protein evolution reshaped LolT’s function to suppress unwanted β-elimination activity while favoring Cα-C bond Mannich cyclization activity.
Figure 4.
LolT-catalyzed β-elimination reaction with l-cystine. a) proposed mechanism. b) unexpected loss of both α and β-proton NMR signal. c) HPLC analysis confirms the end product as pyruvate. d) Steady-state kinetic analysis.
Conclusion
In summary, we have demonstrated that LolT, a PLP-dependent enzyme that natively catalyzes Cα-C bond forming cyclization reactions, can be repurposed for amino acids deuteration. The broad substrate scope and strict site- and stereoselectivity make LolT one of the best amino acids deuteration biocatalyst ever reported.
Supplementary Material
Acknowledgements
This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM151205. Y.H. acknowledges the University of California-Santa Barbara start-up funds for support of this work. C.Z. acknowledges the URCA grant from UCSB for financial support.
Footnotes
Supporting Information
The NMR spectra, mass spectra, and HPLC-traces were included in the Supporting Information.[30,
References
- [1].Atzrodt J, Derdau V, Kerr WJ, Reid M, Angewandte Chemie International Edition 2018, 57, 1758–1784. [DOI] [PubMed] [Google Scholar]
- [2].Sheppard D, Li D-W, Brüschweiler R, Tugarinov V, Journal of the American Chemical Society 2009, 131, 15853–15865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Meilleur F, The Biochemist 2020, 42, 16–20. [Google Scholar]
- [4].Timmins GS, Expert Opinion on Therapeutic Patents 2017, 27, 1353–1361. [DOI] [PubMed] [Google Scholar]
- [5].Schmidt C, Nature biotechnology 2017, 35, 493–495. [DOI] [PubMed] [Google Scholar]
- [6].Roskoski R Jr., Pharmacol Res 2023, 189, 106642. [DOI] [PubMed] [Google Scholar]
- [7].Di Martino RMC, Maxwell BD, Pirali T, Nature Reviews Drug Discovery 2023, 22, 562–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a Moozeh K, So SM, Chin J, Angewandte Chemie International Edition 2015, 54, 9381–9385; [DOI] [PubMed] [Google Scholar]; b Takeda R, Abe H, Shibata N, Moriwaki H, Izawa K, Soloshonok VA, Organic & Biomolecular Chemistry 2017, 15, 6978–6983. [DOI] [PubMed] [Google Scholar]
- [9].Taglang C, Martínez-Prieto LM, del Rosal I, Maron L, Poteau R, Philippot K, Chaudret B, Perato S, Sam Lone A, Puente C, Dugave C, Rousseau B, Pieters G, Angewandte Chemie International Edition 2015, 54, 10474–10477. [DOI] [PubMed] [Google Scholar]
- [10].Rowbotham JS, Ramirez MA, Lenz O, Reeve HA, Vincent KA, Nature Communications 2020, 11, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eliot AC, Kirsch JF, Annual review of biochemistry 2004, 73, 383–415. [DOI] [PubMed] [Google Scholar]
- [12].Doyon TJ, Buller AR, Journal of the American Chemical Society 2022, 144, 7327–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chun SW, Narayan ARH, ACS Catalysis 2020, 10, 7413–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a. Esaki N, Nakayama T, Sawada S, Tanaka H, Soda K, Biochemistry 1985, 24, 3857–3862; [DOI] [PubMed] [Google Scholar]; b. Winnicka E, Szymańska J, Kańska M, Isotopes Environ Health Stud 2016, 52, 231–238. [DOI] [PubMed] [Google Scholar]
- [15].Steffen-Munsberg F, Vickers C, Thontowi A, Schätzle S, Meinhardt T, Svedendahl Humble M, Land H, Berglund P, Bornscheuer UT, Höhne M, ChemCatChem 2013, 5, 154–157. [Google Scholar]
- [16].Babu UM, Johnston RB, Biochem Biophys Res Commun 1974, 58, 460–466; [DOI] [PubMed] [Google Scholar]; b Lim Y-H, Yoshimura T, Soda K, Esaki N, Journal of fermentation and bioengineering 1998, 86, 400–402. [Google Scholar]
- [17].Gao J, Liu S, Zhou C, Lara D, Zou Y, Hai Y, Nature Catalysis 2023, 6, 476–486. [Google Scholar]
- [18].Biswal HS, Shirhatti PR, Wategaonkar S, The Journal of Physical Chemistry A 2010, 114, 6944–6955. [DOI] [PubMed] [Google Scholar]
- [19].a. Ellis JM, Campbell ME, Kumar P, Geunes EP, Bingman CA, Buller AR, Nat. Catal 2022, 5, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]; b. Cai Y, Hai Y, Ohashi M, Jamieson CS, Garcia-Borras M, Houk KN, Zhou J, Tang Y, Nature Chemistry 2019, 11, 812–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Navo CD, Oroz P, Mazo N, Blanco M, Peregrina JM, Jiménez-Osés G, Organic Letters 2022, 24, 6810–6815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.