Abstract
We report the presence of three new O1 ElTor vibriophages named AS1, AS2 and AS3, isolated from the sewage and pond waters of the outskirts of Kolkata. A few phages, named AS4, with hexagonal heads and abnormally long tails with typical curly projections were also found in the water samples.
Keywords: Vibrio, phage, Electron microscopy
Vibrio cholerae, the causative agent of cholera in humans, is classified into two serotypes: O1 and nonO1 [1]. The O1 strains are divided into two biotypes: Classical and ElTor. Before 1961 most epidemics had been caused by the classical biotype. But with the passage of time the classical biotype disappeared from the scenario and the ElTor emerged as the major biotype causing the Vibrio cholerae in humans. In 1993, Vibrio cholerae serogroup O139 made an explosive appearance and caused a severe epidemic in the Indian continent [2]. The disease cholera spreads rapidly to far off places from the epicenter of its emergence. From the epidemiological point of view it is important to track down the spread of the disease. Phage typing is a widely accepted method for tracking down cholera epidemic [3]. The international phage-typing scheme of Basu and Mukerjee [3] includes five phages (I, II, III, IV and V). But in course of time this typing scheme proved inadequate as a large number of Vibrio cholerae strains were found to be untypeable using this scheme. In order to overcome this problem a new typing scheme for ElTor strains was proposed in 1993 [4]. In the recent times vibriophages are found to occur in amazingly in large numbers in the environment around the globe [5-8] which, prompted us to search for new cholera phages from the environmental resources.
Sewage and pond water were collected from different places from the outskirts of Kolkata. During the study period the recorded temperature was about 32–38°C and pH ranged from 7.8 to 10. The sample waters were processed for phage isolation as described previously [9] using Vibrio cholerae O1 ElTor (MAK 757) as the propagating strain. The procedure was repeated on nutrient agar or the appearance of plaques. The phages were purified from a single discrete plaque by the soft agar (0.9 %) overlay method. Phage lysates were prepared in nutrient broth. A few drops of chloroform were added to the freshly prepared phage lysate to remove bacterial content in it. The phage lysates (nearly 108 – 109phages/ml) was subjected to ultracentrifugation at 35,000 r.p.m. for 90 minutes in a Sorval T 865 rotor and phage pellets were obtained. The phage pellets were resuspended in 1 ml of 50 mM; Tris-HCl pH 7.5, 20 mM, MgCl2 (TM buffer) to concentrate and the phage was stored at 4°C. The phages was purified on a sucrose step gradient of 10% to 40% as described previously [6] using a Sorval TW 668 swing-out rotor at revolution speed of 35,000 r.p.m. for 75 minutes. The purified phage pellets were resuspended in 1 ml of TM buffer and stored at 4°C. Five microliters of the purified suspensions were deposited directly on Pioloform coated 300-mesh Nickel grids, stabilized with a thin layer of carbon, allowed to adsorb for two minutes and the excess liquid was blotted out. The samples were stained with 2% aqueous uranyl acetate (pH 4.5). Grids were examined in FEI Tecnai 12 Biotwin transmission electron microscope. Measurements of the dimensions of the head (distances between the opposite apices), length and thickness of the tail were done using 'analySIS' software (SIS GmbH, Germany). Calibration was done using catalase crystal with alternate lattice plane spacing of 8.75 nm and 6.85 nm (Agar Scientific Ltd. England). Several enteropathogens were included to test the susceptibility against these phages. Cultures of these enteropathogens were grown to their mid log phases and were plated as lawn on (0.5% NaCl) nutrient agar plate. The lawns are spotted with about 5–7 μl of the lysates. Table 1 shows the result of the phage sensitivity to the different pathogens. It is seen that phages are only sensitive to Vibrio cholerae O1 and are resistant to rest of the species.
Table 1.
Table showing the sensitivity of the newly isolated phages to the different species of enteropathogens
| Species | Phage AS1 | Phage AS2 | Phage AS3 |
| Vibrio cholerae O1 | Sensitive | Sensitive | Sensitive |
| Vibrio cholerae O139 | Resistant | Resistant | Resistant |
| Vibrio cholerae non-O1 | Resistant | Resistant | Resistant |
| non-O139 | |||
| Vibrio parahaemolyticus | Resistant | Resistant | Resistant |
| Escherichia coli | Resistant | Resistant | Resistant |
| Salmonella enterica | Resistant | Resistant | Resistant |
| serovar Typhimurium |
We have not included the phage AS4 for the sensitivity study as their number is very small which, may give erroneous results.
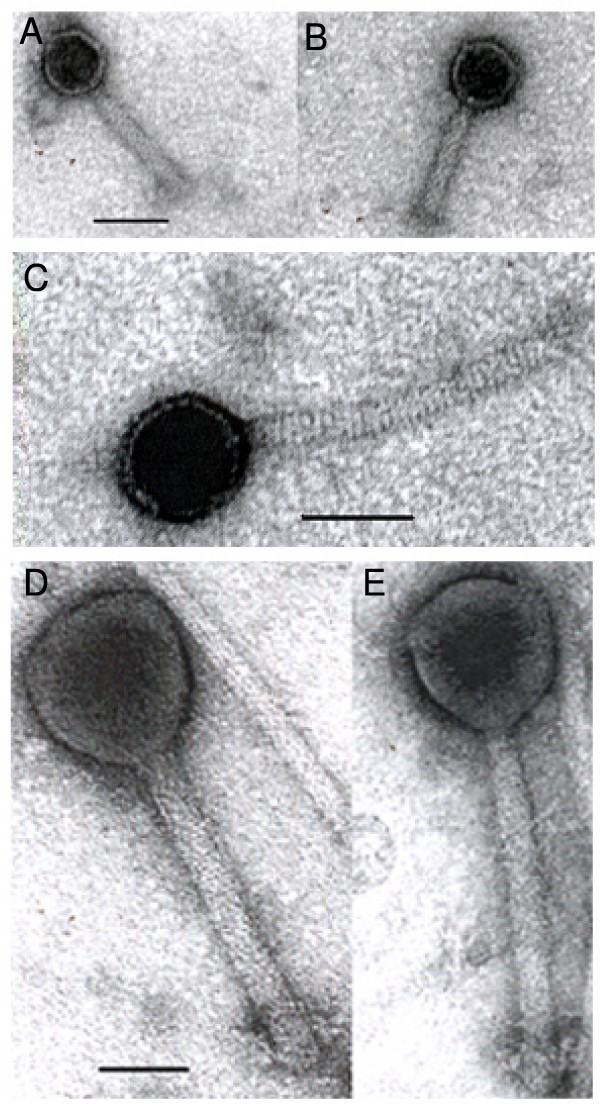
Three different types of phages, named AS1, AS2 and AS3 were found. All the three phages have hexagonal heads with long tail (figure 1). Phages AS1 and AS3 have hexagonal heads and contractile tails and falls in the family of Myoviridae while phage AS2 has a hexagonal head with non-contractile tail and falls within the family Siphoviridae (according to International Committee for the Taxonomy of Viruses; 1982). The dimensions of the head (distances between the opposite apices), length and thickness of the tail of these phages are summarized in table 1. Morphological comparisons of these three phages were made with several other important typing vibriophages that possess hexagonal heads and long tails have been made and are given in table 1.
Figure 1.
Vibrio cholerae O1 Biotype ElTor bacteriophages AS1-3. Panels A and B show the vibriophage AS1. They are contractile in nature and possess similar pattern as seen in the tail of another O1 ElTor typing vibriophage D10 (Chakrabarti et al., (1993). Panels A and B are shown at the same magnification. Panel C show the vibriophage AS2. The tails are non-contractile in nature. Panels D and E show the vibriophage AS3. Panels D and E are shown at the same magnification. The bars in Panels A, C and D: 50 nm.
From table 1 we find that phages AS1, AS2 and AS3 are morphologically different from the other typing vibriophages. While studying these phages we came across few phages (extremely small in number), named AS4, that have the head diameter of nearly equal to 65.24 ± 3.1 nm, straight-tail length nearly 460.20 ± 11.2 nm long and typical curly projection of length 230.20 ± 12.4 nm attached to the free end of the tail. In fact each of the curly projection has a constant contour length of 38.8 ± 5.72 nm. The thickness of the tails is about 10.52 ± 0.86 nm (figure 2). To best of our knowledge, till date, no vibriophage with such abnormally long tails are reported. However, Ackermann and DuBow [10] reported two non-cultivated rumen bacteriophages with such long tails but they do not have any curly projections as seen in AS4.
Figure 2.

Vibrio cholerae O1 Biotype ElTor bacteriophage AS4. The tails are enormously long and non contractile in nature. Typical curly projections are seen at the end of the tails. The number of such phages is extremely rare in the water samples. Bar: 50 nm.
Isolation of these new cholera phages from the sewage and pond waters collected from the outskirts of Kolkata, a high cholera-endemic region, (where a cholera outbreak took place nearly two years back) carries additional significance. Detailed physiochemical studies like host specificity, immunological analysis, study of structural proteins, thermal and light inactivation, growth characteristics and extensive study of their genomes of these newly isolated vibriophages will prove helpful in modifying the present phage typing scheme of Vibrio cholerae O1 biotype ElTor untypeable strains in future as it was needed for the old Basu and Mukerjee [3] O1 biotype ElTor typing scheme almost a decade back.
Table 2.
Morphology of different Vibriophages
| Phage | Host | Diameter of head (nm) | Length of tail (nm) | Thickness of tail (nm) | Nature of tail | Reference |
| AS1 | VC O1 ElTor MAK 757 | 43.60 ± 2.34 | 85.21 ± 3.80 | 13.54 ± 0.91 | Contractile tail | Present study |
| AS2 | VC O1 ElTor MAK 757 | 44.93 ± 1.35 | 123.88 ± 5.21 | 8.83 ± 0.43 | Non-contractile tail) | Present study |
| AS3 | VC O1 ElTor MAK 757 | 90.1 ± 2.21 | 193.5 ± 14.5 | 22.8 ± 1.25 | Contractile tail | Present study |
| M4 (O1 ElTor typing phage) | VC O1 ElTor MAK 757 | 97.7 ± 0.03 | 109.6 ± 0.2 | 18.2 ± 0.4 | Contractile tail | Chattopadhyay et al., 1993 |
| D10 (O1 ElTor typing phage) | VC O1 ElTor MAK 757 | 62.9 ± 0.06 | 101.4 ± 0.3 | 15.8 ± 0.4 | Contractile tail | Chattopadhyay et al., 1993 |
| MAD-5 O139 typing phage | VC O139 NPR-4 | 58.0 ± 2.7 | 141.2 ± 4.8 | 8.33 ± 0.4 | Contractile tail | Chakrabarti et al., 2000 |
| VE-2 O139 typing phage | VC O139 NPR-4 | 112.5 ± 1.8 | 204.0 ± 2.8 | 23.0 ± 0.2 | Contractile tail | Chakrabarti et al., 2000 |
| Group II classical typing phage | VC Classical 154 | 62.1 ± 3.1 & 65.5 ± 3.7 nm for the widest & narrowest sections | 81.0 ± 3.2 | 16.6 ± 2.0 | Contractile tail | Chatterjee and Maiti, 1984 |
| Group IV classical typing phage | VC Classical 154 | 73.8 ± 3.3 & 83.6 ± 4.0 for the widest & narrowest sections | 152.8 ± 8.2 | 10.7 ± 1.4 | Non-contractile tail | (Chatterjee and Maiti, 1984) |
| DR1 | VC O26 | 77.5 ± 0.3 | 100.0 ± 0.6 | 19.0 ± 0.4 | Contractile tail | (Sarkar et al., 2004) [11] |
| DR2 | VC O39 | 83.3 ± 0.3 | 111.0 ± 0.8 | 17.0 ± 0.5 | Contractile tail | (Sarkar et al., 2004) |
| ΦP15 | VC O1 ElTor Inaba | 52.9 ± 9.0 & 40.5 ± 9.5 for the widest & narrowest sections | 105.4 ± 3.2 | 22.5 nm | Contractile tail | (Talledo et al., 2003) |
(VC stands for Vibrio cholerae)
Acknowledgments
Acknowledgements
Authors are grateful to Dr. S. K. Bhattacharya, Director of the institute, for encouragement and kind cooperation.
Contributor Information
Anindito Sen, Email: sena@mail.nih.gov.
Amar N Ghosh, Email: anghosh@vsnl.net.
References
- Chatterjee SN, Maiti M. Vibriophages and Vibriocins: physical, chemical and biological properties. Adv Virus Res. 1984;29:263–312. doi: 10.1016/s0065-3527(08)60411-x. [DOI] [PubMed] [Google Scholar]
- Ramamurthy T, Garg R, Sharma R, Bhattacharya SK, Nair GB, Simada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;314:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- Basu S, Mukerjee S. Bacteriophage typing of Vibrio ElTor. Experimenta. 1968;24:299–300. doi: 10.1007/BF02152832. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay DJ, Sarkar BL, Ansari MQ, Chakrabarti BK, Roy MK, Ghosh AN, Pal SC. New phage typing scheme for Vibrio cholerae O1 biotype El Tor strains. J Clin Microbiol. 1993;31:1579–1585. doi: 10.1128/jcm.31.6.1579-1585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti AK, Ghosh AN, Nari GB, Niyogi SK, Bhattacharya SK, Sarkar BL. Development and evaluation of a phage-typing scheme for Vibrio choleare O139. J Clin Microbiol. 2000;38:44–49. doi: 10.1128/jcm.38.1.44-49.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti BK, Chattopadhyay DJ, Ghosh AN. Vibriophage D10 contains non-permutated DNA with cohesive ends. J Gen Virol. 1993;74:2749–2752. doi: 10.1099/0022-1317-74-12-2749. [DOI] [PubMed] [Google Scholar]
- Ghosh AN, Ansari MQ, Dutta GC. Isolation and morphological characterization of El Tor cholera phages. J Gen Virol. 1989;70:2241–2243. doi: 10.1099/0022-1317-70-8-2241. [DOI] [PubMed] [Google Scholar]
- Talledo M, Rivera ING, Lipp EK, Neale A, Karolis D, Huq A, Colwell R. Characterization of a Vibrio cholerae phage isolated from coastal water of Peru. Environ Microbiol. 2003;5:350–354. doi: 10.1046/j.1462-2920.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- Adams MH. Bacteriophages. Interscience publishers, Inc New York; 1959. [Google Scholar]
- Ackerman HW, DuBow MS. Viruses of Prokaryotes. I. CRC press, Inc Florida, USA; 1987. Vibriophages. [Google Scholar]
- Sarkar BL, Ghosh AN, Sen A, Rodrigues DP. Newly isolated Vibro cholerae non-O1 non-O139 phages. Emerg Infect Dis. 2003;10:754–756. doi: 10.3201/eid1004.030413. [DOI] [PMC free article] [PubMed] [Google Scholar]