Abstract
Cell lines and tumors with defined genetic alterations provide ideal systems in which to test the molecular mechanisms of tumor sensitivity to pathway-targeted therapy. We have generated mouse ovarian epithelial tumor cell lines that contain various combinations of genetic alterations in the p53, c-myc, K-ras and Akt genes. Using both in vitro and in vivo approaches, we investigated the effect of rapamycin on cell proliferation, tumor growth, and the accumulation of peritoneal ascites. We demonstrated that rapamycin effectively inhibits the growth of tumors that rely on Akt signaling for proliferation, whereas tumors in which Akt signaling is not the driving force in proliferation are resistant to rapamycin. The introduction of activated Akt to the rapamycin-resistant cells does not render the cells susceptible to rapamycin if they can use alternative pathways for survival and proliferation. Accordingly, the rapamycin-sensitive tumors develop resistance to rapamycin when presented with alternative survival pathways, such as the mitogen-activated extracellular kinase signaling pathway. The combination of rapamycin and the mitogen-activated extracellular kinase inhibitor PD98059 is required to diminish proliferation in these cell lines. Our results indicate that mammalian target of rapamycin inhibitors may be effective in a subset of tumors that depend on Akt activity for survival but not effective in all tumors that exhibit Akt activation. Tumors with alternative survival pathways may require the inactivation of multiple individual pathways for successful treatment.
Keywords: mouse model, ovarian cancer, rapamycin
The etiology of ovarian cancer is not well understood because the majority of ovarian cancer patients are diagnosed at late stages of the disease (1). Advanced ovarian cancers typically show numerous genetic alterations and chromosomal abnormalities (2). Mutation of the p53 tumor suppressor gene is the most frequently identified genetic alteration in ovarian carcinomas. In addition, proto-oncogenes such as Her-2, c-myc, K-ras, and Akt are often amplified or mutated in ovarian cancer (3). However, it is difficult to discern which of these alterations are required for the maintenance of ovarian cancers and, thus, could be used for targeted therapy.
The mammalian target of rapamycin (mTOR) has recently gained attention as a therapeutic target because of its key regulatory function downstream of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. The inhibition of mTOR with immunosuppressive macrolide rapamycin blocks oncogenic transformation induced by either PI3K or Akt, indicating that the mTOR function is required for the oncogenic effects of PI3K or Akt (4, 5). Rapamycin and its derivatives CCI-779 and RAD001 have demonstrated drastic antiproliferative and anti-angiogenic effects in preclinical models (6–9). Previous studies have shown that the antitumor effects of mTOR inhibitors correlate with heightened Akt activity (10, 11) and that cells from PTEN knockout mice and human tumor cell lines lacking PTEN are more sensitive than wild-type cells to the growth-inhibitory effects of rapamycin derivatives (12–14). Similarly, transfection of the CCI-779 nonresponsive human multiple myeloma cell line U266 with constitutively active Akt significantly increased sensitivity to CCI-779 in a xenograft mouse model (10), indicating that the sole activation of the Akt pathway might induce hypersensitivity of tumor cells to mTOR inhibition. However, several recent studies revealed the necessity of combined therapeutic approaches in the treatment of tumors with an activated Akt pathway (15–17).
Genetically defined cell lines and tumors provide ideal systems in which to determine the functional contributions of individual pathways that are necessary for tumor maintenance and to test the molecular mechanisms of tumor sensitivity to pathway-targeted therapy. Here, we dissect the molecular requirements for rapamycin inhibition and resistance in mouse ovarian cell lines and tumors with defined combinations of genetic alterations in the p53, c-myc, K-ras, and Akt genes.
Materials and Methods
Production of Viruses. The plasmid forms of RCAS vectors (18) were transfected into DF-1 cells, which is an immortalized line of chicken cells (19, 20) used for the production of viruses. The viral supernatant was prepared by growing the confluent virus-producing DF-1 cells in low-serum OPTI-MEM medium (GIBCO) overnight. The viruses were concentrated from viral supernatant by centrifugation at 25,000 × g for 1.5 h. The pBabe-puro-H-rasV12 vector was used to transfect the amphotropic packaging cell line LinX-A. The viral supernatant was collected 24 h after the addition of fresh medium, filtered, and stored at –80°C until needed.
Generation of Genetically Defined Ovarian Cancer Cell Lines. For the generation of the C1, C2, and C3 cell lines, ovarian explants from K5-TVA/p53–/– mice (18) were infected with different combinations of RCAS viruses carrying human c-myc, mouse K-rasG12D, and mouse myristoylated Akt1 oncogenes. The T1, T2, and T3 cell lines were derived by isolating cells from tumors that were generated by i.p. injection of C1, C2, and C3 cells into nude mice, respectively. The mC1+Akt cell line was derived from tumors that were generated by i.p. injection of C1 cells into nude mice and subsequent in vivo superinfection with RCAS-Akt viruses. The T2+K-ras and T2+Her-2 cell lines were generated by in vitro superinfection of T2 cells with RCAS-K-ras and RCAS-Her-2 viruses, respectively. The T2+H-ras cell line was generated by infecting T2 cells with pBabe-puro-H-rasV12 and selecting the cells for 10 days in a medium containing 2.5 μg/ml of puromycin.
Treatment with Rapamycin and PD98059. Equal numbers of cells were plated into six-well dishes in DMEM/F12 media with 10% FCS. After 2 days of growth in culture, the medium was replaced with a medium that contained either 100 ng/ml rapamycin (LC Laboratories, Woburn, MA) dissolved in ethanol or ethanol alone (vehicle). The cells were treated with rapamycin or vehicle for 8 days, during which time the medium with rapamycin or vehicle was changed every second day. For combinatorial treatment with rapamycin and PD98059, the cells were treated as described above with a medium that contained one of the following: 100 ng/ml rapamycin dissolved in ethanol and 50 μM PD98059 dissolved in DMSO, 100 ng/ml rapamycin dissolved in ethanol and DMSO, 50 μM PD98059 dissolved in DMSO and ethanol, or ethanol and DMSO.
Determining Cell Proliferation in Vitro and the Presence of Apoptosis and Senescence. To monitor cell proliferation, drug- and vehicle-treated cells were harvested every second day and cell proliferation was determined by direct cell counting or by using crystal-violet dye as described in ref. 18. Apoptosis and senescence were determined by using the ApopTag Plus Peroxidase in Situ Apoptosis Detection Kit (Chemicon International, Temecula, CA) and the Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, Beverly, MA), respectively.
Rapamycin Treatment in Vitro. Rapamycin was initially dissolved in 100% ethanol at a concentration of 50 mg/ml and stored at –20°C. The working solution was further diluted in an aqueous phase of 5.2% Tween 80 and 5.2% polyethylene glycol 400 and prepared immediately before use. Female nude or FVB mice were injected s.c. or i.p. with 5 × 106 cells in PBS. After the formation of 0.2 cm3 to 0.5 cm3 s.c. tumor nodules, the mice were treated with 100 μl of rapamycin solution (5 mg/kg i.p.) every day for 2 weeks. For i.p. tumors, 7 days after the initial injection of cells, the mice were treated with 100 μl of rapamycin solution (5 mg/kg i.p.) every second day for 2–3 weeks.
Western Blotting. Western blotting was performed as described in ref. 18 by using antibodies against hemagglutinin (HA) (Santa Cruz Biotechnology), Phospho-p70 S6 Kinase (S6K) Thr-389 (Cell Signaling Technology), and α-tubulin (Sigma).
Vascular Endothelial Growth Factor (VEGF) ELISA. Equal numbers of cells were seeded in six-well dishes for VEGF secretion experiments. After cell attachment, the medium was replaced with low-serum OPTI-MEM medium treated with 100 ng/ml rapamycin dissolved in ethanol or ethanol alone. Cell culture supernatants were collected 48 h after treatment and subjected to ELISA for mouse VEGF by following the manufacturer's instructions (R & D Systems).
Results and Discussion
Generation of Mouse Ovarian Cancer Cell Lines and Tumors with Defined Genetic Alterations. Ovarian surface epithelial cells are thought to be the precursor tissue for ovarian carcinoma, but the molecular mechanism of their transformation is unknown. We have developed a system in which multiple defined genetic alterations can be introduced into mouse ovarian surface epithelial cells through RCAS retroviral delivery (18). To generate ovarian surface epithelial cell lines with defined genetic alterations, we introduced coding sequences for human c-myc, mouse K-rasG12D, and HA-tagged mouse myristoylated Akt1 oncogenes into ovaries isolated from K5-TVA/p53–/– mice. Confirming our previous findings (18), we demonstrated that c-myc, K-ras, and Akt cannot individually transform p53-null mouse ovarian surface epithelial cells, but a combination of at least two of these three oncogenes is necessary and sufficient for cell transformation (Table 1, which is published as supporting information on the PNAS web site).
The mouse ovarian cancer cell lines were generated as follows: C1 (genotype: p53–/–, c-myc, K-ras), C2 (genotype: p53–/–, c-myc, Akt), and C3 (genotype: p53–/–, K-ras, Akt). i.p. injection of C1, C2, and C3 cell lines into nude mice consistently resulted in carcinomatosis that closely resembled peritoneal metastases in women with stage III ovarian carcinoma. Similar to human ovarian cancer, tumors were rarely found on the liver or spleen and were confined to the peritoneal cavity where they were loosely adhering to the peritoneal surfaces, intestinal mesentery, and reproductive organs (Fig. 5, which is published as supporting information on the PNAS web site). Histologically, the i.p. tumors derived from C1 and C2 cells resembled ovarian papillary serous carcinoma, which is the most common type of ovarian carcinoma in women (Fig. 5D). Tumors derived from C3 cells resembled poorly differentiated ovarian carcinoma (data not shown). C1-, C2-, and C3-derived tumor nodules were explanted in culture to generate T1, T2, and T3 tumor cell lines (Table 2, which is published as supporting information on the PNAS web site).
Rapamycin only Inhibits Proliferation of Cells and Tumors That Are Dependent on the Akt Pathway for Survival and Proliferation. The PI3K/Akt/mTOR pathway is commonly activated in human ovarian carcinomas (11, 21–26), underscoring the rationale for the use of mTOR inhibitors in ovarian cancer treatment. To determine whether rapamycin inhibits proliferation of mouse ovarian cancer cell lines in vitro, we treated C1, T1, C2, T2, C3, and T3 cells with 100 ng/ml rapamycin or vehicle every second day for up to 8 days. Cell proliferation was monitored by harvesting cells every second day and measuring light absorbance of cell-associated crystal-violet dye or by direct cell counting. We determined that rapamycin is not efficient in inhibiting proliferation of C1 and T1 cells but efficiently inhibits proliferation of C2 and T2 cells and C3 and T3 cells (Fig. 1 and Table 2). The rapamycin-treated C2, T2, C3, and T3 cells exhibited apoptosis and drastic changes in cell morphology that resembled cell senescence. The “senescent-like” cells were large and flat but did not express β-galactosidase. Apoptosis was more common in C2 and T2 cells, whereas the “senescence-like” phenotype was more common in C3 and T3 cells (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Rapamycin inhibits proliferation of cell lines with genetic alterations in p53, c-myc, and Akt (C2 and T2) and p53, K-ras, and Akt (C3 and T3) but does not inhibit proliferation of cell lines with genetic alterations in p53, c-myc, and K-ras (C1 and T1). (A–C) C1, T1, C2, T2, C3, and T3 cells were seeded in equal amounts into six-well dishes. Two days after the cells were seeded, the medium was replaced with medium containing 100 ng/ml rapamycin dissolved in ethanol or ethanol alone (vehicle). The culture medium with rapamycin or vehicle was changed every second day. To monitor cell proliferation, rapamycin- and vehicle-treated cells were harvested every second day and stained with crystal-violet dye. The numbers on the y axis represent relative absorbance of cell-associated dye at 590 nm or the cell number. The error bars indicate standard deviation in triplicate cultures. (D) Representative images of nude mice (n = 8) that were injected i.p. with T1 cells. Seven days after injection, the mice were randomized for i.p. injections of a 5 mg/kg dose of rapamycin (n = 4) or vehicle (n = 4) every second day for 20 days. The tumor burden and extensive ascites accumulation were equivalent in rapamycin-treated and vehicle-treated mice. (E) Representative images of nude mice (n = 10) that were injected i.p. with T2 cells. Seven days after injection, the mice were randomized for i.p. injections ofa5mg/kg dose of rapamycin (n = 5) or vehicle (n = 5) every second day for 17 days. Rapamycin-treated mice were free of tumors and ascites, whereas vehicle-treated mice displayed i.p. carcinomatosis with ascites.
In vivo inhibition by rapamycin was tested in nude mice that were injected with C1, T1, C2, and T2 cell lines at s.c. or i.p. sites (Table 3, which is published as supporting information on the PNAS web site). The mice were randomized for treatment with i.p. injection of 5 mg/kg rapamycin or vehicle. When the s.c. tumors reached a volume of 0.2 cm3 to 0.5 cm3, the mice were treated every day until the tumors reached ≈1cm3. For i.p. tumors, the treatment started 7 days after cell injection and was delivered every second day until the rapamycin-treated or vehicle-treated mice developed abundant ascites. Consistent with in vitro data, rapamycin was not efficient in inhibiting the growth of tumors that were derived from C1 and T1 cells (Fig. 1D and Table 3) but was highly efficient in inhibiting the growth of tumors that were derived from the C2 and T2 cells (Fig. 1E and Table 3).
Together, these results demonstrate that cell lines with genetic alterations in p53, c-myc, and K-ras (C1 and T1) are largely resistant to rapamycin inhibition. However, rapamycin efficiently inhibits the proliferation of cell lines with genetic alterations in p53, c-myc, and Akt (C2 and T2) and p53, K-ras, and Akt (C3 and T3). Therefore, the presence of activated Akt in the C2, T2, C3, and T3 cell lines is required not only for transformation (Table 1) but also for continuous cell proliferation and tumor maintenance (Tables 2 and 3). The C1 and T1 cell lines are resistant to rapamycin, presumably because the proliferation of these cells does not depend on the mTOR pathway.
Introduction of Activated Akt into Rapamycin-Resistant Cells Does Not Render the Cells Sensitive to Rapamycin. It has been demonstrated that the effects of mTOR inhibitors correlate with heightened Akt activity (10, 11) and that constitutive activation of Akt in an mTOR inhibitor nonresponsive human multiple myeloma cell line increases sensitivity to the mTOR inhibitor (10). Therefore, we investigated whether the introduction of constitutively activated Akt into the rapamycin-resistant C1 cells would affect their sensitivity to rapamycin. To generate a cell line that expresses a high level of Akt, we first injected nude mice with C1 cells i.p. and subsequently super-infected the cells in vivo by i.p. injection of RCAS-Akt. Although the addition of Akt is not necessary for the proliferation of C1 cells, the Akt-expressing C1 cells were positively selected during in vivo tumor formation. A high expression level of HA-tagged Akt was confirmed in tumor tissues (data not shown), and the tumors were explanted in culture to generate cell lines. The cell line with the highest level of Akt-HA was selected and named the mC1+Akt cell line (genotype: p53–/–, c-myc, K-ras, and Akt). The mice that were injected with mC1+Akt cells developed tumors and accumulated hemorrhagic ascites ≈1 week earlier than the mice that were injected with the original C1 cells, indicating possible “gain of function.”
As was the case with the original C1 cell line (Fig. 1 A), rapamycin was not very efficient in inhibiting proliferation of the mC1+Akt cells in vitro (Fig. 2A). The in vivo effect of rapamycin treatment on mC1+Akt cells was assessed by i.p. injection of mC1+Akt cells into nude mice and subsequent treatment with rapamycin. Eighteen days after the injection of the mC1+Akt cells, all vehicle-treated mice developed i.p. tumors with ascites. None of the rapamycin-treated mice showed signs of significant ascites formation (Fig. 2 B and C). However, the i.p. tumor burden and pattern of metastatic spread were similar in vehicle-treated and rapamycin-treated mice (data not shown). Therefore, rapamycin was effective in inhibiting ascites formation induced by the injection of mC1+Akt cells but not effective in inhibiting tumor proliferation (Table 3).
Fig. 2.
Rapamycin inhibits ascites production but does not inhibit the proliferation of rapamycin-resistant C1 cells with constitutively activated Akt. (A) Effect of rapamycin on in vitro proliferation of mC1+Akt cells. Two days after an equal number of cells were seeded into six-well dishes, the medium was replaced with medium containing 100 ng/ml rapamycin dissolved in ethanol or ethanol alone (vehicle). The culture medium with rapamycin or vehicle was changed every second day. To monitor cell proliferation, rapamycin- and vehicle-treated cells were harvested every second day and stained with crystal-violet dye. The numbers on the y axis represent relative absorbance of the cell-associated dye at 590 nm. The error bars indicate standard deviation in triplicate cultures. (B) Representative images of nude mice (n = 15) that were injected i.p. with mC1+Akt cells. Seven days after injection, the mice were randomized for treatment with a 5 mg/kg dose of rapamycin or vehicle every second day for 11 days. Rapamycin-treated mice (n = 8) and vehicle-treated mice (n = 7) displayed i.p. carcinomatosis. However, ascitic fluid was only present in the vehicle-treated mice. (C) The volume of ascites isolated from mice in B. The error bars indicate standard deviation. *, P < 0.01. (D) Levels of VEGF secreted in cell culture supernatant by mC1+Akt cells treated with rapamycin or vehicle. Equal numbers of cells were seeded in six-well dishes for VEGF secretion experiments. After cell attachment, the medium was replaced with a low-serum medium containing 100 ng/ml rapamycin dissolved in ethanol or ethanol alone. Cell culture supernatants were collected 48 h after treatment and subjected to ELISA for mouse VEGF. The error bars indicate standard deviation in six cultures. *, P < 0.01. (E) Western blot showing effective inhibition of the mTOR pathway in rapamycin-resistant and rapamycin-sensitive cell lines. C1, C2, and mC1+Akt cell lines were grown in vitro for 2 days, after which they were treated with 100 ng/ml rapamycin or vehicle for 1 day. Total cell lysates were immunoblotted by using an antibody against the hemagglutinin (HA) tag to determine the levels of the HA-tagged Akt protein. The activity of the mTOR pathway was assessed by comparing the protein levels of phosphorylated p70S6K in vehicle- and rapamycin-treated cells. Phosphorylated p70S6K (Thr-389) was detected with an antibody that recognizes phospho-p70S6K and phospho-p85S6K.
VEGF is commonly overexpressed in the malignant ovarian epithelium (27) and is thought to be a potent mediator of ascites accumulation (28–32). Studies on human ovarian cancer cell lines have implicated the PI3K/Akt/mTOR pathway in VEGF transcriptional activation and demonstrated that rapamycin treatment decreases VEGF protein levels (33, 34). To determine whether rapamycin treatment affects the level of VEGF secreted by mC1+Akt cells, we subjected cell culture supernatants from rapamycin-treated and vehicle-treated cells to mouse VEGF ELISA. The level of VEGF in supernatants from rapamycin-treated cells was three times lower than in supernatants from vehicle-treated cells (Fig. 2D), indicating that rapamycin treatment may be effective in reducing the level of VEGF secreted by ovarian cancer cells. Thus, the introduction of Akt into the rapamycin-resistant C1 cell line did not sensitize the cells to proliferative inhibition by rapamycin. Rapamycin treatment reverted the Akt-induced ascites accumulation but did not impact the growth of i.p. tumors.
To confirm that rapamycin was effective in inhibiting the mTOR pathway in mouse ovarian cancer cell lines in vitro, we compared the protein levels of phosphorylated p70S6K, a downstream target of mTOR, in C1, C2, and mC1+Akt cell lines treated with rapamycin or vehicle (Fig. 2E). The cells were propagated in culture for 2 days and treated with 100 ng/ml rapamycin or vehicle for 1 day, after which the cells were lysed for protein analysis. The phosphorylation status of S6K was determined by immunoblotting cell lysates with the phospho-p70S6K (Thr-389) antibody that recognizes phospho-p70S6K and phospho-p85S6K. It has been shown that phosphorylation of Thr-389 most closely correlates with p70S6K activity in vivo (35). p70S6K was phosphorylated in all cell lines treated with vehicle, indicating that the mTOR pathway was active in these cells. The phosphorylation of p70S6K was completely inhibited in the C1, C2, and mC1+Akt cell lines treated with rapamycin (Fig. 2E), implying that biochemical inhibition of the mTOR pathway occurs efficiently in the three cell types. The level of myristoylated Akt protein in mC1+Akt cells was higher than in rapamycin-sensitive C2 cells (Fig. 2E), indicating that the level of activated Akt does not necessarily correlate with sensitivity to rapamycin. Although rapamycin effectively inhibited the mTOR pathway in C1 and mC1+Akt cells, cell proliferation was not inhibited by rapamycin. Therefore, C1 and mC1+Akt cell lines can proliferate in the absence of the mTOR signal.
Introduction of Alternative Survival Pathways to Rapamycin-Sensitive Cells Renders the Cells Resistant to Rapamycin. Our results indicate that the dependency of a cell on Akt signaling for proliferation determines whether the cell will be sensitive to rapamycin inhibition. If our hypothesis is correct, we should be able to convert rapamycin-sensitive cells into rapamycin-resistant cells by providing them with an alternative pathway for survival, such as Ras and Her-2, which activate the Akt pathway and several other survival pathways (reviewed in ref. 36).
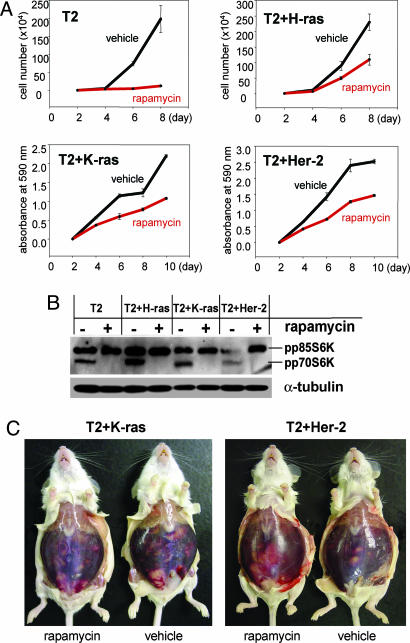
We superinfected rapamycin-sensitive T2 cells with the pBabe-puro-H-rasV12 vector or with an RCAS vector carrying K-ras or Her-2. The resulting cell lines were named T2+H-ras (genotype: p53–/–, c-myc, Akt, and H-ras), T2+K-ras (genotype: p53–/–, c-myc, Akt, and K-ras), and T2+Her-2 (genotype: p53–/–, c-myc, Akt, and Her-2). The T2 cell lines and the superinfected cell lines were subjected to in vitro treatment with rapamycin or vehicle. We found that rapamycin inhibits the growth of the T2+H-ras, T2+K-ras and T2+Her-2 cells by ≈50% (Fig. 3A), thus showing a significant increase in rapamycin resistance when compared with the original T2 cells (Figs. 1B and 3A). The phosphorylation of p70S6K was completely inhibited in all four cell lines treated with rapamycin (Fig. 3B), implying efficient biochemical inhibition of the mTOR pathway.
Fig. 3.
Introduction of alternative survival pathways into the rapamycin-sensitive ovarian cell lines induces resistance to rapamycin. (A) Equal numbers of T2, T2+H-ras, T2+K-ras, and T2+Her-2 cells were seeded into six-well dishes. After 2 days, the medium was replaced with medium containing 100 ng/ml rapamycin dissolved in ethanol or ethanol alone (vehicle). The culture medium with rapamycin or vehicle was changed every second day. To monitor cell proliferation, rapamycin- and vehicle-treated cells were harvested every second day and stained with crystal-violet dye. The numbers on the y axis represent relative absorbance of the cell-associated dye at 590 nm or the cell number. The error bars indicate standard deviation in triplicate cultures. (B) Western blot showing effective inhibition of the mTOR pathway in T2, T2+H-ras, T2+K-ras, and T2+Her-2 cell lines. The cells were grown in vitro for 2 days, after which they were treated with 100 ng/ml rapamycin or vehicle for 1 day. The activity of the mTOR pathway was assessed by comparing the protein levels of phosphorylated p70S6K in vehicle- and rapamycin-treated cells. Phosphorylated p70S6K (Thr-389) was detected with an antibody that recognizes phospho-p70S6K and phospho-p85S6K. (C) Representative images of immunocompetent FVB mice that were injected with T2+K-ras (n = 10) or T2+Her-2 (n = 8) cells and subjected to treatment with rapamycin or vehicle. Seven days after cell injection, the mice were randomly selected for treatment with 5 mg/kg of rapamycin or vehicle every second day. After 20 days of treatment, rapamycin- (n = 6) and vehicle-treated (n = 4) T2+K-ras-injected mice displayed i.p. carcinomatosis with an extensive accumulation of ascites. Similarly, rapamycin- (n = 4) and vehicle-treated (n = 4) T2+Her-2-injected mice displayed extensive i.p. carcinomatosis and ascites accumulation 20 days after treatment.
To test the susceptibility of T2+K-ras and T2+Her-2 cells to in vivo treatment with rapamycin, the cells were injected i.p. into nude and the immunocompetent mice. The in vivo results showed complete resistance of the T2+K-ras and T2+Her-2 tumors to rapamycin inhibition. The rapamycin-treated and vehicle-treated nude mice and immunocompetent mice displayed the same levels of tumor burden and ascites accumulation, indicating that T2+K-ras and T2+Her-2 cells are resistant to rapamycin treatment in vivo (Fig. 3C). Cumulatively, these results show that rapamycin-sensitive T2 ovarian cancer cells become resistant to rapamycin upon introduction of activated H-ras, K-ras, or Her-2 (Table 3).
The mTOR Inhibitor Rapamycin and the Mitogen-Activated Extracellular Kinase (MEK) Inhibitor PD98059 Cooperate to Inhibit Proliferation of Ovarian Cancer Cell Lines with Multiple Survival Pathways. The most likely explanation for rapamycin resistance of cells with genetic alterations in p53, c-myc, and ras is that ras activates pathways other than the Akt/mTOR pathway, and these other pathways are sufficient for sustained cell proliferation. Several ras effectors are likely to cooperate with c-myc and Akt to sustain proliferation of transformed ovarian cancer cell lines. For example, the MEK/extracellular signal-regulated kinase pathway has been identified as a key mediator of ras-transforming activity in rodent cell lines (37, 38). Recently however, significant contributions of the PI3K (39) and Ral-GDS (40) pathways in the transformation of mammalian cells has been demonstrated.
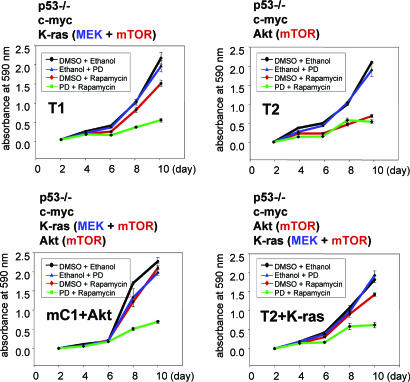
We wanted to test whether simultaneous inactivation of the MEK and mTOR pathways would be more effective in inhibiting proliferation of the rapamycin-resistant ovarian cancer cells. The T1, T2, mC1+Akt, and T2+K-ras cell lines were treated with rapamycin and the MEK inhibitor PD98059, individually and in combination. Cell proliferation was monitored by harvesting cells every second day for 10 days and measuring light absorbance of cell-associated crystal-violet dye (Fig. 4). Consistent with the notion that K-ras signals through the MEK and Akt/mTOR pathways and that c-myc can collaborate with either of these pathways to sustain cell proliferation, neither rapamycin nor PD98059 was effective in inhibiting the proliferation of the T1, mC1+Akt, and T2+K-ras cell lines (Fig. 4). However, in combination, rapamycin and PD98059 were significantly more effective in inhibiting cell proliferation (Fig. 4). Cell proliferation was not completely abolished by simultaneous treatment with rapamycin and PD98059 (Fig. 4), indicating that other K-ras effectors could be contributing to the proliferative signals. It is also possible that c-myc, K-ras, and Akt are connected through feedback mechanisms, which can be altered in the presence of rapamycin and PD98059. It has been demonstrated that the growth-promoting potential of myc and ras was strongly enhanced by rapamycin in transformed chicken fibroblasts (4).
Fig. 4.
Effect of rapamycin, PD98059, and combination treatment on the proliferation of ovarian cancer cell lines that contain multiple survival pathways. Ovarian cancer cell lines T1, T2, mC1+Akt, and T2+K-ras were seeded into six-well dishes in triplicate. After 2 days of growth in culture, the medium was removed and replaced with a medium that contained one of the following: 100 ng/ml rapamycin dissolved in ethanol and 50 μM PD98059 dissolved in DMSO, 100 ng/ml rapamycin with ethanol and DMSO, 50 μM PD98059 with ethanol and DMSO, or ethanol and DMSO. The cells were treated for 8 days, during which time the culture medium was changed every second day. To monitor cell proliferation, drug- and vehicle-treated cells were harvested every second day and stained with crystal-violet dye. The numbers on the y axis represent relative absorbance of the cell-associated dye at 590 nm. The error bars indicate standard deviation in triplicate cultures.
In summary, we have designed mouse ovarian carcinoma cell lines and tumors with defined genetic alterations for the characterization of putative molecular pathways that can be therapeutically targeted. We dissected the susceptibility of ovarian cancer cell lines with different combinations of oncogenic alterations to the mTOR inhibitor rapamycin and demonstrated that the mTOR pathway is an excellent target for the treatment of cells that depend on Akt signaling for continuous proliferation and survival. However, tumors with alternative pathways for proliferation are independent of Akt signaling and cannot be effectively inhibited by solely targeting the mTOR pathway. When tumors acquire two redundant proliferation signals, such as Akt/mTOR or MEK, the inhibition of both pathways is necessary to suppress tumor growth. These results have significant implications for the use of pathway-targeted therapy in advanced human ovarian cancers, which typically display numerous genetic alterations that are likely to require impairment of multiple molecular pathways for successful treatment. Our results suggest that the interruption of multiple specific biochemical pathways may be a promising therapeutic strategy in ovarian carcinomas that exhibit resistance to an individual targeted therapy. We anticipate that mouse ovarian cancer cell lines with defined genetic alterations will be useful for the optimization of rationally designed cancer therapy that targets specific signaling pathways.
Supplementary Material
Acknowledgments
We thank Lynn Kim, Jeffrey Vainshtein, and Alex Yazhbin for mouse husbandry and genotyping; Yi Li, Brian Lewis, David Tuveson, and Gregory Hannon for viral plasmid constructs; Kristy Daniels for assistance in the preparation of the manuscript; Harold E. Varmus and Michael Seiden for support and guidance; Jeffrey Settleman for his critical reading of the manuscript; and the reviewers for their insightful comments. This work was supported by the National Cancer Institute Grant 1R01CA103924.
Author contributions: D.X. and S.O. designed research; D.X. performed research; D.X. and S.O. analyzed data; and S.O. wrote the paper.
Abbreviations: HA, hemagglutinin; mTOR, mammalian target of rapamycin; MEK, mitogen-activated extracellular kinase; PI3K, phosphatidylinositol 3-kinase; S6K, S6 kinase; VEGF, vascular endothelial growth factor.
References
- 1.Holschneider, C. H. & Berek, J. S. (2000) Semin. Surg. Oncol. 19, 3–10. [DOI] [PubMed] [Google Scholar]
- 2.Gray, J. W., Suzuki, S., Kuo, W. L., Polikoff, D., Deavers, M., Smith-McCune, K., Berchuck, A., Pinkel, D., Albertson, D. & Mills, G. B. (2003) Gynecol. Oncol. 88, S16–S21; discussion S22–S24. [DOI] [PubMed] [Google Scholar]
- 3.Aunoble, B., Sanches, R., Didier, E. & Bignon, Y. J. (2000) Int. J. Oncol. 16, 567–576. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, M., Blazek, E. & Vogt, P. K. (2001) Proc. Natl. Acad. Sci. USA 98, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debnath, J., Walker, S. J. & Brugge, J. S. (2003) J. Cell Biol. 163, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu, Y. & Sato, J. D. (1999) J. Cell Physiol. 178, 235–246. [DOI] [PubMed] [Google Scholar]
- 7.Guba, M., von Breitenbuch, P., Steinbauer, M., Koehl, G., Flegel, S., Hornung, M., Bruns, C. J., Zuelke, C., Farkas, S., Anthuber, M., et al. (2002) Nat. Med. 8, 128–135. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo, M. & Rowinsky, E. K. (2000) Oncogene 19, 6680–6686. [DOI] [PubMed] [Google Scholar]
- 9.Uhrbom, L., Nerio, E. & Holland, E. C. (2004) Nat. Med. 10, 1257–1260. [DOI] [PubMed] [Google Scholar]
- 10.Frost, P., Moatomed, F., Hoang, B., Shi, Y., Gera, J., Yan, H., Gibbons, J. & Lichtenstein, A. (2004) Blood 104, 4181–4187. [DOI] [PubMed] [Google Scholar]
- 11.Altomare, D. A., Wang, H. Q., Skele, K. L., De Rienzo, A., Klein-Szanto, A. J., Godwin, A. K. & Testa, J. R. (2004) Oncogene 23, 5853–5857. [DOI] [PubMed] [Google Scholar]
- 12.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills, G. B., Lu, Y. & Kohn, E. C. (2001) Proc. Natl. Acad. Sci. USA 98, 10031–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendel, H. G., De Stanchina, E., Fridman, J. S., Malina, A., Ray, S., Kogan, S., Cordon-Cardo, C., Pelletier, J. & Lowe, S. W. (2004) Nature 428, 332–337. [DOI] [PubMed] [Google Scholar]
- 16.Raje, N., Kumar, S., Hideshima, T., Ishitsuka, K., Chauhan, D., Mitsiades, C., Podar, K., Le Gouill, S., Richardson, P., Munshi, N. C., et al. (2004) Blood 104, 4188–4193. [DOI] [PubMed] [Google Scholar]
- 17.Majumder, P. K., Febbo, P. G., Bikoff, R., Berger, R., Xue, Q., McMahon, L. M., Manola, J., Brugarolas, J., McDonnell, T. J., Golub, T. R., et al. (2004) Nat. Med. 10, 594–601. [DOI] [PubMed] [Google Scholar]
- 18.Orsulic, S., Li, Y., Soslow, R. A., Vitale-Cross, L. A., Gutkind, J. S. & Varmus, H. E. (2002) Cancer Cell 1, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himly, M., Foster, D. N., Bottoli, I., Iacovoni, J. S. & Vogt, P. K. (1998) Virology 248, 295–304. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer-Klein, J., Givol, I., Barsov, E. V., Whitcomb, J. M., VanBrocklin, M., Foster, D. N., Federspiel, M. J. & Hughes, S. H. (1998) Virology 248, 305–311. [DOI] [PubMed] [Google Scholar]
- 21.Shayesteh, L., Lu, Y., Kuo, W. L., Baldocchi, R., Godfrey, T., Collins, C., Pinkel, D., Powell, B., Mills, G. B. & Gray, J. W. (1999) Nat. Genet. 21, 99–102. [DOI] [PubMed] [Google Scholar]
- 22.Kurose, K., Zhou, X. P., Araki, T., Cannistra, S. A., Maher, E. R. & Eng, C. (2001) Am. J. Pathol. 158, 2097–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellacosa, A., de Feo, D., Godwin, A. K., Bell, D. W., Cheng, J. Q., Altomare, D. A., Wan, M., Dubeau, L., Scambia, G., Masciullo, V. et al. (1995) Int. J. Cancer 64, 280–285. [DOI] [PubMed] [Google Scholar]
- 24.Cheng, J. Q., Godwin, A. K., Bellacosa, A., Taguchi, T., Franke, T. F., Hamilton, T. C., Tsichlis, P. N. & Testa, J. R. (1992) Proc. Natl. Acad. Sci. USA 89, 9267–9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philp, A. J., Campbell, I. G., Leet, C., Vincan, E., Rockman, S. P., Whitehead, R. H., Thomas, R. J. & Phillips, W. A. (2001) Cancer Res. 61, 7426–7429. [PubMed] [Google Scholar]
- 26.Yuan, Z. Q., Sun, M., Feldman, R. I., Wang, G., Ma, X., Jiang, C., Coppola, D., Nicosia, S. V. & Cheng, J. Q. (2000) Oncogene 19, 2324–2330. [DOI] [PubMed] [Google Scholar]
- 27.Olson, T. A., Mohanraj, D., Carson, L. F. & Ramakrishnan, S. (1994) Cancer Res. 54, 276–280. [PubMed] [Google Scholar]
- 28.Connolly, D. T., Olander, J. V., Heuvelman, D., Nelson, R., Monsell, R., Siegel, N., Haymore, B. L., Leimgruber, R. & Feder, J. (1989) J. Biol. Chem. 264, 20017–20024. [PubMed] [Google Scholar]
- 29.Nagy, J. A., Masse, E. M., Herzberg, K. T., Meyers, M. S., Yeo, K. T., Yeo, T. K., Sioussat, T. M. & Dvorak, H. F. (1995) Cancer Res. 55, 360–368. [PubMed] [Google Scholar]
- 30.Barton, D. P., Cai, A., Wendt, K., Young, M., Gamero, A. & De Cesare, S. (1997) Clin. Cancer Res. 3, 1579–1586. [PubMed] [Google Scholar]
- 31.Brustmann, H. & Naude, S. (2002) Gynecol. Oncol. 84, 47–52. [DOI] [PubMed] [Google Scholar]
- 32.Santin, A. D., Hermonat, P. L., Ravaggi, A., Cannon, M. J., Pecorelli, S. & Parham, G. P. (1999) Eur. J. Gynaecol. Oncol. 20, 177–181. [PubMed] [Google Scholar]
- 33.Skinner, H. D., Zheng, J. Z., Fang, J., Agani, F. & Jiang, B. H. (2004) J. Biol. Chem. 279, 45643–45651. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, X. S., Zheng, J. Z., Reed, E. & Jiang, B. H. (2004) Biochem. Biophys. Res. Commun. 324, 471–480. [DOI] [PubMed] [Google Scholar]
- 35.Weng, Q. P., Kozlowski, M., Belham, C., Zhang, A., Comb, M. J. & Avruch, J. (1998) J. Biol. Chem. 273, 16621–16629. [DOI] [PubMed] [Google Scholar]
- 36.Downward, J. (2003) Nat. Rev. Cancer 3, 11–22. [DOI] [PubMed] [Google Scholar]
- 37.Cowley, S., Paterson, H., Kemp, P. & Marshall, C. J. (1994) Cell 77, 841–852. [DOI] [PubMed] [Google Scholar]
- 38.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F. & Ahn, N. G. (1994) Science 265, 966–970. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, J. J., Gjoerup, O. V., Subramanian, R. R., Cheng, Y., Chen, W., Roberts, T. M. & Hahn, W. C. (2003) Cancer Cell 3, 483–495. [DOI] [PubMed] [Google Scholar]
- 40.Hamad, N. M., Elconin, J. H., Karnoub, A. E., Bai, W., Rich, J. N., Abraham, R. T., Der, C. J. & Counter, C. M. (2002) Genes Dev. 16, 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.