Abstract
Lyme borreliosis (LB) group spirochetes, collectively known as Borrelia burgdorferi sensu lato, are distributed worldwide. Wild rodents are acknowledged as the most important reservoir hosts. Ixodes scapularis is the primary vector of B. burgdorferi sensu lato in the eastern United States, and in the southeastern United States, the larvae and nymphs mostly parasitize certain species of lizards. The primary aim of the present study was to determine whether wild lizards in the southeastern United States are naturally infected with Lyme borreliae. Blood samples obtained from lizards in Florida and South Carolina were tested for the presence of LB spirochetes primarily by using B. burgdorferi sensu lato-specific PCR assays that amplify portions of the flagellin (flaB), outer surface protein A (ospA), and 66-kDa protein (p66) genes. Attempts to isolate spirochetes from a small number of PCR-positive lizards failed. However, PCR amplification and sequence analysis of partial flaB, ospA, and p66 gene fragments confirmed numerous strains of B. burgdorferi sensu lato, including Borrelia andersonii, Borrelia bissettii, and B. burgdorferi sensu stricto, in blood from lizards from both states. B. burgdorferi sensu lato DNA was identified in 86 of 160 (54%) lizards representing nine species and six genera. The high infection prevalence and broad distribution of infection among different lizard species at different sites and at different times of the year suggest that LB spirochetes are established in lizards in the southeastern United States.
Lyme borreliosis, the most frequently reported arthropod-borne infection in the United States (6), is caused by several species within the Borrelia burgdorferi sensu lato genogroup (38). B. burgdorferi sensu lato includes at least 11 genospecies worldwide, three of which are present in North America (Borrelia andersonii, Borrelia bissettii, and B. burgdorferi sensu stricto) (16, 28, 33). Thus far, only B. burgdorferi sensu stricto has been proven to cause human disease in the United States.
In the northeastern United States, the spirochetes are transmitted to humans by the blacklegged tick, Ixodes scapularis (5), and maintained in nature primarily by small rodents (4, 23, 31). In the southeastern and western United States, immature stages of the vector ticks feed primarily on lizards (2, 10, 35, 43). Although B. burgdorferi sensu lato has been isolated from birds, rodents, and ticks in southern and western states (9, 31, 33), the organism has never been isolated from wild lizards. Indeed, several studies have shown that strains of two B. burgdorferi sensu lato species do not survive in the blood of two lizard species found in California (19, 21, 43), leading to a widely held belief that lizards do not serve as reservoirs of the bacteria. However, a different study (22) showed in laboratory experiments that two common lizards in the southeastern United States, green anoles and southeastern five-lined skinks, were reservoir competent for one strain of B. burgdorferi sensu stricto. In the present study, we sought to determine whether lizards in the southeastern United States are naturally infected with B. burgdorferi sensu lato by attempting to isolate spirochetes and by using DNA amplification methods to genetically characterize strains present in lizards and to conduct initial experiments to determine if I. scapularis ticks could acquire B. burgdorferi sensu lato from feeding on naturally infected lizards collected in the wild. Here we present the first reported evidence of B. burgdorferi sensu lato among naturally infected wild lizards; these findings demonstrate a broad geographic distribution, three B. burgdorferi sensu lato species, and high infection prevalence among multiple lizard species in two southeastern states.
MATERIALS AND METHODS
Sample collection.
Lizards were captured and sampled from national forests and state parks in northern and central Florida and southeastern South Carolina from March 2003 through May 2004. The primary habitats at the collection localities are mixed pine and oak uplands, mixed pine flatwoods, and bay swamps. Prescribed burning of vegetation is conducted regularly at some sites as part of ongoing habitat management programs. Lizards were obtained by stalking and capturing either by hand or via noosing. Attached ticks were immediately removed with forceps and preserved in ethanol. A sample (∼50 to 100 μl) of blood was obtained via tail fracture and blotted onto filter paper strips for DNA extraction. The blood from most lizards was obtained by removing the distal portion of the tail by hand, as the tails of most common lizards collected in the study fracture readily, providing a few drops of blood without harming the animals (tail fracturing is a natural escape mechanism for the animals). Most animals were returned to their site of capture immediately after examination and blood collection. Twelve broad-headed skinks (Eumeces laticeps) were euthanized humanely to harvest organs and tissues for Borrelia isolation attempts. An additional six PCR-positive E. laticeps skinks were kept in the laboratory for several months for transmission experiments.
DNA extraction and PCR testing.
DNA was extracted from dried filter paper blood samples, tick pools, and cultures using a commercially available kit (MasterPure; Epicentre, Madison, WI) with optimized modifications of the manufacturer's protocols for each starting material. The starting template for filter paper blood samples was an approximately 5- by 5-mm square piece of blood-soaked paper. Culture aliquots of 200 μl were taken from approximately the middle of each conical tube of 4 ml of media suspension in attempts to avoid obtaining dead spirochetes that would presumably settle to the bottom of the tubes. The resulting DNA pellets for all extracts were rehydrated in 100 μl of Tris-EDTA buffer. Negative control samples free of any template were included in each round of DNA extractions. Extracts were first screened by nested PCR targeting a 389-bp portion of the conserved 41-kDa chromosomal flagellin gene (flaB) of B. burgdorferi sensu lato using slight modifications of published primers (Table 1) (15, 32, 42). All flaB-positive samples were further tested with a nested PCR assay targeting a 236-bp portion of the chromosomal 66-kDa protein (p66) using a combination of published primers (36) and one that we modified (Table 1). The same subset of flaB-positive samples was tested with primers (13) targeting a 352-bp portion of the genetically diverse outer surface protein A (ospA) gene of B. burgdorferi sensu lato.
TABLE 1.
Oligonucleotide primers used in this study
| B. burgdorferi sensu lato target gene | Primer | Primer sequence (5′ — 3′)a | Base position | Annealing temp (°C) | Amplicon size (bp) | Source or reference |
|---|---|---|---|---|---|---|
| flaB | Outer 1 | AAR-GAA-TTG-GCA-GTT-CAA-TC | 271-290 | 52 | This study | |
| Outer 2 | GCA-TTT-TCW-ATT-TTA-GCA-AGT-GAT-G | 743-767 | 497 | This study | ||
| Inner 1 | ACA-TAT-TCA-GAT-GCA-GAC-AGA-GGT-TCT-A | 301-328 | 55 | This study | ||
| Inner 2 | GAA-GGT-GCT-GTA-GCA-GGT-GCT-GGC-TGT | 663-689 | 389 | 15 | ||
| ospA | N1 | GAG-CTT-AAA-GGA-ACT-TCT-GAT-AA | 334-356 | 42 | 13 | |
| C1 | GTA-TTG-TTG-TAC-TGT-AAT-TGT | 894-874 | 561 | 13 | ||
| N2 | ATG-GAT-CTG-GAG-TAC-TTG-AA | 362-381 | 42 | 13 | ||
| C2 | CTT-AAA-GTA-ACA-GTT-CCT-TCT | 713-693 | 352 | 13 | ||
| p66 | Outer 1 | CGA-AGA-TAC-TAA-ATC-TGT | 147-158 | 50 | 36 | |
| Outer 2 | GCT-GCT-TTT-GAG-ATG-TGT-CC | 442-461 | 315 | This study | ||
| Inner 1 | TGC-AGA-AAC-ACC-TTT-TGA-AT | 168-187 | 50 | 36 | ||
| Inner 2 | AAT-CAG-TTC-CCA-TTT-GCA | 385-403 | 236 | 36 |
R in a primer sequence signifies A or G; W signifies A or T.
First-round amplifications contained between 2.5 and 5 μl of DNA extract per individual sample in a total reaction volume of 50 μl. All reactions utilized a hot start master mix (HotMasterMix; Brinkmann-Eppendorf, Westbury, NY), resulting in a final concentration of 1.0 U of Taq DNA polymerase, 45 mM KCl, 2.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, and a 0.5 μM concentration of each primer, and were carried out in an automated DNA thermal cycler (PTC 200; MJ Research, Watertown, MA). Outer reaction PCRs consisted of initial denaturation at 95°C for 1 min followed by 40 cycles of 94°C for 30 s, primer annealing at the temperature listed in Table 1 for 30 s, and extension at 68°C for 45 s. Nested reactions included between 1 and 2.5 μl of outer reaction product as template for another 30 cycles with the same parameters and annealing temperature profile as described above and in Table 1.
PCRs were set up in a separate area within a PCR clean cabinet (CleanSpot Workstation; Coy Laboratory Products, Grass Lake, MI) equipped with a germicidal UV lamp. Other precautions to prevent carryover contamination of amplified DNA included different sets of pipettes dedicated for DNA extraction, PCR setup, and postamplification activities; the use of aerosol barrier filter pipette tips; and exposing PCR tubes, pipettes, and tips to UV light prior to PCR setup. Each outer PCR included a negative control sample with sterile water as template and a positive control sample from B. burgdorferi sensu stricto strain B31 culture extract. Negative control samples included in each round of extractions were also screened to detect any possible extraction contamination. A portion of the positive and negative control outer reaction samples was carried over as template in each nested reaction, just as for experimental samples. PCR products were electrophoresed in 2% agarose gels, which were stained with ethidium bromide, and visualized and recorded with a digital gel documentation unit.
DNA sequence analysis.
PCR products were purified using the MinElute PCR purification kit (QIAGEN, Valencia, CA). DNA templates were sequenced using the fluorescent dideoxy terminator method of cycle sequencing on either a Perkin-Elmer, Applied Biosystems, Inc. 373A or 377 automated DNA sequencer following Applied Biosystems protocols (29). Sequences were generated using the Sequencher Software (Gene Codes Corporation, Ann Arbor, MI). Investigator-derived sequences were compared with those obtained by searching the GenBank database (National Center for Biotechnology Information) using the Basic Local Alignment Search Tool (BLAST) (1) and aligned using Clustal X (40). Phylogenetic trees were constructed using the neighbor-joining (NJ) and maximum parsimony (MP) methods (37, 39) with the tree-building program MEGA version 2.1 (18). Tree topologies and genetic relationships obtained with the two methods were compared for consensus. To estimate node reliability of trees obtained with each method, bootstrap values (11) based on an analysis of either 100 (MP) or 1,000 (NJ) replicates were determined. Pairwise distances were computed by the Kimura two-parameter model (17).
Borrelia culture isolation.
We chose 12 flaB PCR-positive broad-headed skinks (nine males and three females) for attempts to isolate spirochetes. Within 1 week of testing PCR positive, the animals were euthanized humanely and dissected with sterile technique inside a biosafety cabinet. Whole blood (∼100 μl), heart, and liver samples (and testes from males; ∼50 mg of each tissue type per sample) were inoculated into tubes containing 4 ml of modified Barbour-Stoenner-Kelly (BSK-H) culture medium supplemented with antibiotics (Sigma, St. Louis, MO) for isolating Borrelia (3), incubated at 32°C, and examined weekly for spirochetes by dark-field microscopy for 8 weeks. Samples from B. burgdorferi sensu lato reference strains (B. andersonii MOK-1c; B. bissettii 25015; B. burgdorferi sensu stricto B31, JD1, NC92, and WI90; and Borrelia sp. strain SCW-30h) were also inoculated and maintained in culture to ensure the ability of the medium to support spirochete growth.
Tick-feeding experiments.
Six additional broad-headed skinks (three each from Florida and South Carolina) that tested positive for B. burgdorferi sensu lato in initial PCR tests were chosen for a tick-feeding and infection experiment. Individual animals were placed inside a ∼1.5-in.-diameter tube constructed of 1/4-in.-mesh hardware cloth, with PVC end caps modified from published designs (14, 22). Approximately 100 I. scapularis tick larvae from a laboratory-reared colony at Colorado State University were introduced to each lizard's tube. Feeding tubes were suspended over white plastic photo trays with wet paper towel substrate in separate aquaria. The rims of the trays and aquaria were coated with petroleum jelly to prevent escape of ticks that dropped off. Trays were checked daily for detached ticks. Lizards were fed one or two crickets every other day by removing the PVC end cap on one end of the tube. Water was provided via a moistened cotton ball kept inside the tube at all times. After blood-fed ticks dropped off, they were rinsed in a 5% bleach solution, followed by a rinse in sterile distilled water, and then maintained in glass vials with cloth mesh lids in a glass desiccator jar at ∼97% rH and 82°F until they molted to nymphs (∼1 month later). The resultant nymphs were tested by PCR to determine if they acquired B. burgdorferi sensu lato and maintained infection through the molt. All procedures involving capturing, maintaining, and testing lizards were conducted in accordance with protocols approved by the University of North Florida Institutional Animal Care and Use Committee.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the B. burgdorferi sensu lato flaB gene sequences generated in the present study are AY662999 to AY663008, AY663010 to AY663017, AY823241, AY823242, and AY823244 to AY823249. The B. burgdorferi sensu lato ospA gene sequences reported here are AY663018 to AY663021, AY663023, AY663024, AY823229, AY823231, and AY823232. The B. burgdorferi sensu lato p66 gene sequences reported here are AY823233 to AY823240. The sequences used for comparison are listed in Table 2.
TABLE 2.
Borrelia species reference strains used in this study
| Species | Strain | Biological source | Geographic origin | GenBank sequence accession number
|
||
|---|---|---|---|---|---|---|
| flaB sequence | ospA sequence | p66 sequence | ||||
| B. andersonii | 19857 | Cottontail rabbit | NY | D83762 | A24008 | |
| 21038 | Ixodes dentatus | NY | D83763 | |||
| MOK-1c | Ixodes dentatus | MO | AY654915 | AY654919 | AY654939 | |
| SI-10 | Ixodes scapularis | GA | AF264883 | |||
| B. bissettii | 25015 | Ixodes scapularis | NY | L29245 | U65802 | AY654938 |
| DN127 | Ixodes pacificus | CA | D82857 | Y10897 | ||
| FL18 | Cotton mouse | FL | AY654922 | |||
| FL27 | Cotton rat | FL | AY654935 | |||
| FL35 | Cotton rat | FL | AY654923 | AY654937 | ||
| FL42 | Cotton rat | FL | AY654924 | |||
| IA1 | Ixodes affinis | FL | AY654925 | |||
| IS-19 | Ixodes spinipalpis | CO | U96243 | |||
| MI8 | Cotton rat | FL | AF264892 | |||
| SCGT8a | Ixodes minor | SC | AF264894 | |||
| B. burgdorferi sensu lato | SCW-30h | Ixodes minor | SC | AF264886 | AY654920 | AY654940 |
| B. burgdorferi sensu stricto | AA4Pool | Amblyomma americanum | FL | AY654928 | ||
| AA15Pool | Amblyomma americanum | FL | AY654929 | |||
| B31 | Ixodes scapularis | NY | X15661 | AY030279 | AE001161 | |
| CA8 | Ixodes pacificus | CA | L23144 | |||
| FLCL3 | Ixodes affinis | FL | AY654933 | |||
| JD1 | Ixodes scapularis | MA | AF369944 | U96240 | ||
| KC9 | Golden mouse | FL | AY654934 | |||
| MI2 | Cotton rat | FL | AF264889 | |||
| SCI2 | Cotton mouse | GA | AF264885 | |||
| Tr293 | Ixodes ricinus | Turkey | AB091813 | |||
| Borrelia afzelii | ACA1 | Human subject | Sweden | X75202 | AY090473 | |
| VS461 | Ixodes ricinus | Switzerland | Z29087 | |||
| Borrelia garinii | Ip90 | Ixodes persulcatus | Russia | X75203 | X87727 | |
| PBi | Human subject | Germany | X80257 | |||
| Borrelia japonica | HO14 | Ixodes ovatus | Japan | D82852 | ||
| IKA2 | Ixodes ovatus | Japan | Y10892 | |||
| Borrelia lusitaniae | PotiB2 | Ixodes ricinus | Portugal | D82856 | Y10838 | |
| Borrelia valaisiana | VS116 | Ixodes ricinus | Switzerland | D82854 | Y10840 | |
| Borrelia sinica | CMN3 | White-bellied rat | China | AB022138 | ||
| Borrelia hermsii | HS1 | Ornithodoros hermsii | United States | AF016408 | ||
| Borrelia lonestari | TX | Amblyomma americanum | TX | U26704 | ||
RESULTS
B. burgdorferi sensu lato-specific PCR.
We captured and obtained blood samples from 160 lizards representing seven genera and 10 species from sites in Florida and South Carolina. Most (77%) of the animals were captured in March and April of 2003 and 2004, which is before most I. scapularis larvae and nymphs become active, or during the period of initial activity in the spring (35). Most (93%) animals were free of ticks when they were captured. Only 18 I. scapularis larvae and 30 nymphs were observed and removed (all during April and May), and all came from three lizard species: broad-headed and southeastern five-lined skinks and eastern glass lizards.
To determine if the lizards' blood contained B. burgdorferi sensu lato DNA, we first screened blood sample extracts with the B. burgdorferi sensu lato-specific nested flaB PCR assay. Eighty-six (53.8%) of the samples tested positive, including those from nine lizard species (six genera) and nine sites in Florida and South Carolina (Table 3); positive blood samples were obtained from juvenile, subadult, and adult animals. None of the negative control extracts tested positive. The infection prevalence among different positive lizard species ranged from 18 to 100%. The prevalence among animals from South Carolina (72%) was higher than for those from Florida (40%), but a greater variety of species from Florida was tested. The only species for which no positives were obtained was the Mediterranean gecko, which, along with two brown anoles that did test positive, came from suburban residences. Positive results were obtained for several species that are not commonly, if ever, parasitized by I. scapularis: brown and green anoles, Florida scrub lizards, and six-lined racerunners (2, 10). flaB PCR-positive samples were obtained from lizards in February through May, September, and December, which includes months (February and December) when I. scapularis larvae and nymphs do not typically parasitize hosts (35).
TABLE 3.
Prevalence of B. burgdorferi sensu lato flagellin (flaB) gene DNA among lizards from Florida and South Carolina
| Location | Number of PCR-positive animals/number of each species tested (%)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Broad-headed skink | Brown anole | Fence lizard | Glass lizard | Scrub lizard | Green anole | Ground skink | Gecko | Race- runner | Five-lined skink | Total | |
| Florida | 8/18 (44) | 2/4 (50) | 3/9 (33) | 1/1 (100) | 6/14 (43) | 7/17 (41) | 5/7 (71) | 0/3 (0) | 2/11 (18) | 3/8 (38) | 37/92 (40) |
| South Carolina | 13/18 (72) | NTb | NT | 1/1 (100) | NT | 22/33 (67) | 1/1 (100) | NT | NT | 12/15 (80) | 49/68 (72) |
| Total | 21/36 (58) | 2/4 (50) | 3/9 (33) | 2/2 (100) | 6/14 (43) | 29/50 (58) | 6/8 (75) | 0/3 (0) | 2/11 (18) | 15/23 (65) | 86/160 (53.8) |
Species tested were the broad-headed skink (Eumeces laticeps), brown anole (Anolis sagrei), eastern fence lizard (Sceloporus undulatus), eastern glass lizard (Ophisaurus ventralis), Florida scrub lizard (Sceloporus woodi), green anole (Anolis carolinensis), ground skink (Scincella lateralis), Mediterranean gecko (Hemidactylus turcicus), six-lined racerunner (Cnemidophorus sexlineatus), and southeastern five-lined skink (Eumeces inexpectatus).
NT, none tested.
All 86 flaB-positive samples were then tested with the p66 and ospA nested PCR assays. Thirty (34.9%) of these 86 tested positive with the p66 assay, and 17 (19.8%) were also positive with the ospA PCR assay. Only 10 of 86 (11.6%) flaB-positive samples tested positive with both p66 and ospA assays. Most PCR-amplified target gene fragments were visible in ethidium bromide-stained gels only in the nested reaction products, indicating generally low target copy number in the extracts. Nevertheless, many lizard blood extracts were tested several times for each of the three target gene fragments, with consistent results. Some samples were positive only with the flaB assay. Some were consistently flaB and p66 positive but ospA negative, and others were consistently flaB and ospA positive but p66 negative.
B. burgdorferi sensu lato flaB, p66, and ospA sequences.
We sequenced PCR-amplified flaB, ospA, and p66 gene fragments from many individual lizard samples and compared the sequences to those obtained via BLAST (1) searching the GenBank database. Most of the flaB and ospA DNAs analyzed in this study were sequenced in one direction only, with the forward primers used in the PCRs. A few of the flaB DNAs and all of the p66 DNAs were sequenced in both directions with the forward and reverse PCR primers. For a few samples, the initially derived sequences contained some unresolved nucleotides caused by multiple signal polymorphisms at those positions. When these were resequenced, or the sequences obtained with both forward and reverse primers were compared, the unresolved bases in all but a few of these samples could be determined. Based on the BLAST scores and phylogenetic comparisons we conducted, all flaB, ospA, and p66 DNAs obtained from lizards or I. scapularis ticks fed upon them in the lab belonged to B. burgdorferi sensu lato.
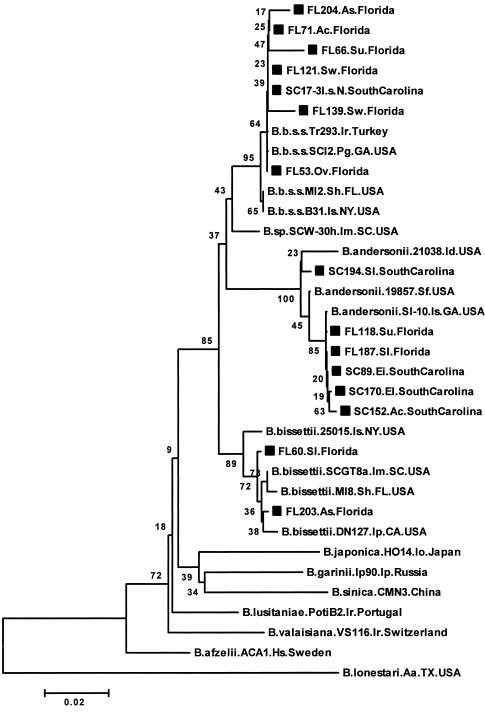
Figure 1 shows an NJ phylogenetic tree based on 362 bp of the flaB gene, which has been used to reliably differentiate B. burgdorferi sensu lato strains to the species level (12, 15, 32). flaB sequences derived from 28 lizards and four I. scapularis nymph pools (from larvae fed on infected lizards in the lab) clustered into three clades defined by reference strains of the B. burgdorferi sensu lato species previously identified in the United States; these included 19 B. burgdorferi sensu stricto strains in eight lizard species and the four I. scapularis nymph pools, 11 strains of B. andersonii in five lizard species, and 2 B. bissettii strains in two lizard species (Fig. 1; Table 4; not all data shown). Based on pairwise distances, the flaB sequences from lizards and ticks that clustered with B. burgdorferi sensu stricto strains were between 98.0 and 100% identical and between 98.6 and 100% similar to the B. burgdorferi sensu stricto reference strains included in the analysis. flaB sequences from several other lizards clustered with those from B. andersonii reference strains (Fig. 1). All except one (SC194) of these sequences from the lizards were nearly identical to that for strain SI-10, a B. andersonii isolate from a blacklegged tick in Georgia. The similarity among all of the B. andersonii flaB sequences ranged between 98.0 and 100%. The lizard flaB sequences from FL60 and FL203 were 99.7% identical and between 98.9 and 99.7% identical to B. bissettii reference strain sequences included in the comparison.
FIG. 1.
Unrooted neighbor-joining phylogenetic tree based on 362 bp of the flaB gene obtained from lizards in Florida and South Carolina and B. burgdorferi sensu lato reference strains. The relapsing fever group species Borrelia lonestari was included as an outgroup. Numbers at the branch nodes represent bootstrap values as percentages of 1,000 replications. All sequences obtained in this study are identified by a square symbol preceding the sequence name, and names end with the state, either Florida or South Carolina. Reference strain sequences end with the country name. The strain name for reference sequences follows the species name (e.g., B.bissettii.DN127). Letter symbols representing the specific host and/or source follow the sequence and species or strain name. Abbreviations: Aa, Amblyomma americanum; Ac, Anolis carolinensis; As, Anolis sagrei; B.b.s.s., B. burgdorferi sensu stricto; B.sp., Borrelia species; Ei, Eumeces inexpectatus; El, Eumeces laticeps; Hs, human subject; Id, Ixodes dentatus; Im, Ixodes minor; Io, Ixodes ovatus; Ip, Ixodes pacificus or Ixodes persulcatus; Ir, Ixodes ricinus; I.s.N., Ixodes scapularis nymphs; Ov, Ophisaurus ventralis; Pg, Peromyscus gossypinus; Sf, Sylvilagus floridanus; Sh, Sigmodon hispidus; Sl, Scincella lateralis; Su, Sceloporus undulatus; and Sw, Sceloporus woodi. Examining the tree from top to bottom, the first major clade, beginning with strain FL204 and ending with B.b.s.s.B31, delineates B. burgdorferi sensu stricto strains. Below this group, the next major clade groups B. andersonii strains. Below this is a clade grouping B. bissettii strains.
TABLE 4.
Representative Lyme borreliosis group spirochete strains identified in the present study based upon phylogenetic analysis of flaB, ospA, and p66 gene sequencesa
| Host ID | Host or tick species | Collection locality (site, county, state) | Genospeciesa |
|---|---|---|---|
| FL 53 | Eastern glass lizard | University of North Florida, Duval Co., FL | B. burgdorferi sensu stricto |
| FL 60 | Ground skink | Guana River State Park, St. Johns Co., FL | B. bissettii |
| FL 66 | Eastern fence lizard | University of North Florida, Duval Co., FL | B. burgdorferi sensu stricto |
| FL 71 | Green anole | Appalachicola National Forest, Leon/Wakulla Co., FL | B. burgdorferi sensu stricto |
| SC 87 | Green anole | Francis Marion National Forest, Berkley/Charleston Co., SC | B. andersonii |
| SC 107 | Broad-headed skink | Francis Marion National Forest, Berkley/Charleston Co., SC | B. andersonii |
| FL 121 | Florida scrub lizard | Ocala National Forest, Marion Co., FL | B. burgdorferi sensu stricto |
| FL 131 | Six-lined racerunner | Ocala National Forest, Marion Co., FL | B. burgdorferi sensu stricto |
| FL 203 | Brown anole | Fort Pierce Residence, St. Lucie Co., FL | B. bissettii |
| FL 204 | Brown anole | Atlantic Beach Residence, Duval Co., FL | B. burgdorferi sensu stricto |
| SC17-3I.s.Nb | Blacklegged tick | Colony-reared samples | B. burgdorferi sensu stricto |
See figures for phylogenetic trees based on flaB, ospA, and p66 sequences.
This strain was identified in DNA extracted from a pool of three nymph ticks fed as larvae in the laboratory on a broad-headed skink collected in Francis Marion National Forest, Berkley/Charleston Co., SC.
The ospA sequences (334 bp analyzed) derived from lizards clustered into three different clades defined by the previously described North American strains (Fig. 2). The sequences were heterogeneous, and their phylogenetic clustering did not agree with that obtained from the flaB sequence analysis for several individual lizard templates. For example, the flaB sequence from lizard samples SC89, SC167, and FL187 clustered with B. andersonii strains (Fig. 1), but the ospA sequences from these samples clustered with B. bissettii sequences (Fig. 2). The FL121 and FL126 flaB sequences both clustered with B. burgdorferi sensu stricto strains (Fig. 1; FL126 not shown), yet while the FL121 ospA sequence clustered along with strain SCW-30h most closely to B. burgdorferi sensu stricto reference strain sequences, the FL126 ospA sequence clustered with B. bissettii strains.
FIG. 2.
Unrooted maximum parsimony bootstrap consensus phylogenetic tree based on 334 bp of the ospA gene obtained from lizards in Florida and South Carolina and B. burgdorferi sensu lato reference strains. Numbers at the branch nodes represent bootstrap values as percentages of 100 replications. Sequences obtained in this study are identified by a square symbol preceding the sequence name. Ia, Ixodes affinis. See the legend to Fig. 1 for other abbreviations used in descriptive information for the sequences.
The ospA sequences from lizards FL126, FL187, SC89, SC167, and SC194 were between 99.1 and 100% identical and between 93.4 and 100% similar to all B. bissettii reference strain sequences included in the comparison and most similar to previously described B. bissettii strains from the southeastern United States. The ospA sequence from lizard FL121 was 99.7% similar to that for strain SCW-30h and between 93.6 and 95% similar to B. burgdorferi sensu stricto reference strains included in the analysis. The sequences from lizards SC87, SC106, and SC107 were 99.7 to 100% identical and between 97.2 and 98.7% similar to the B. andersonii reference strains.
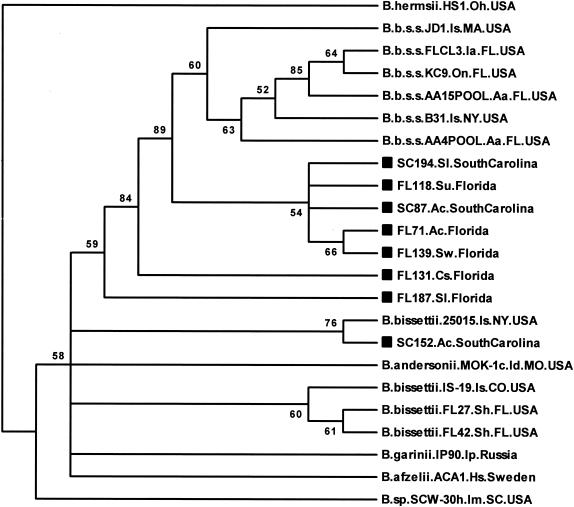
All of the p66 sequences derived from lizards were also very similar to B. burgdorferi sensu lato reference strains. The phylogenetic tree based on 233 bp of data showed that all but one of the lizard-derived p66 sequences analyzed in this study clustered most closely with B. burgdorferi sensu stricto reference strains (Fig. 3). The phylogenetic placement of some lizard p66 sequences also did not correlate with the B. burgdorferi sensu lato species clustering produced by the flaB sequence analysis for those lizard samples. The sequences from lizards FL71, FL118, FL131, FL139, FL187, SC87, and SC194 were 98.3 to 100% identical to each other as well as to the B. burgdorferi sensu stricto reference strains that they were most similar to. The p66 sequence from lizard SC152 was 99.6% identical to that from B. bissettii strain 25015, 98.3% identical to that from FL187, 97.6% identical to that from B. bissettii IS-19 from Colorado, and 96.8% identical to that from B. andersonii MOK-1c.
FIG. 3.
Unrooted neighbor-joining bootstrap consensus phylogenetic tree based on 233 bp of the p66 gene obtained from lizards in Florida and South Carolina and B. burgdorferi sensu lato reference strains. The relapsing fever group species Borrelia hermsii was included as an outgroup. Numbers at the branch nodes represent bootstrap values as percentages of 1,000 replications. Sequences obtained in this study are identified by a square symbol preceding the sequence name. Abbreviations: Cs, Cnemidophorus sexlineatus; Is, Ixodes scapularis or Ixodes spinipalpis; Oh, Ornithodoros hermsii; and On, Ochrotomys nuttalli. See the legend to Fig. 1 for other abbreviations used in descriptive information for the sequences.
Borrelia isolation.
Although all lizard blood and tissue cultures were examined weekly for spirochetes for 8 weeks, no spirochetes were detected microscopically. During that time, we extracted and tested DNA from cultures of different starting materials at different times postinoculation using the nested flaB PCR assay. At least one template type culture sample from each of the 12 animals tested positive via PCR. Target products were seen only in the nested reactions, indicating a very low copy number of target DNA template in the culture extracts. One of 12 whole-blood cultures and 1 of 12 liver tissue cultures tested at 1 week postinoculation were PCR positive. At 4 weeks postinoculation, 7 of 12 blood culture extracts tested positive. At 5 weeks, 5 of 12 heart tissue cultures and 3 of 9 testes cultures were positive. However, upon testing again at 8 weeks, only 2 of 12 new extracts from previously positive culture samples (seven blood samples, three heart samples, and two testes samples) were positive. Most of the lizard samples were visibly contaminated with other bacteria. PCR amplification of lizard blood and tissue culture DNA extracts using broad-range 16S rRNA gene primers and DNA sequencing confirmed the presence of Mycoplasma spp. Reference strain cultures of several B. burgdorferi sensu lato species grew extremely well and did not become contaminated. During the follow-up, flaB PCR-positive lizard culture samples were subcultured multiple times into fresh medium; however, the contaminating bacteria persisted and continued to grow in the medium.
Tick-feeding experiments.
In August 2003, blood sample extracts from 9 of 20 (45%) broad-headed and southeastern five-lined skinks that had been kept in the laboratory since being captured in April of that year tested positive via flaB PCR, and 6 of these (three broad-headed skinks each from Florida and South Carolina) were used in the tick-feeding and transmission experiment. Approximately 100 I. scapularis larvae were placed on each animal, and 393 attached and fed to repletion. The minimum and maximum numbers that fed on different lizards were 11 and 124, respectively, with an average of 66 per animal. Blood-fed ticks were kept in the lab until they molted to nymphs. Unfortunately, due to fungal growth in the tubes, only 28 ticks survived through the molt, which was deemed too few to attempt further transmission studies. However, DNA was extracted from the nymphs in six pools of three to six ticks per pool to aim for maximum sensitivity of detection and tested by nested flaB PCR to determine whether they acquired B. burgdorferi sensu lato and maintained detectable B. burgdorferi sensu lato DNA through the molt. All six pools tested positive, indicating a minimum estimated “infection” prevalence of 6 of 28 (21%); the actual number of infected individual ticks may have been higher. The flaB products were sequenced from four of the pools. They were not identical, but all were found to be >99% similar to lizard-derived and reference strain B. burgdorferi sensu stricto sequences as described above.
DISCUSSION
Studies have shown that two lizard species in California, the western fence lizard (Sceloporus occidentalis) and the southern alligator lizard (Elgaria multicarinata), have poor reservoir potential for some strains of B. burgdorferi sensu lato (19, 20, 21, 41, 43). Another study (22) showed that green anoles and southeastern five-lined skinks, both common in the southeastern United States, were reservoir competent under laboratory-controlled conditions for one strain of B. burgdorferi sensu stricto. In the present study, we were unable to obtain pure cultures of B. burgdorferi sensu lato from wild caught lizards, including those containing strains belonging to B. burgdorferi sensu lato species that have been cultured in BSK medium. We did, however, demonstrate the presence of B. burgdorferi sensu lato DNA in cultures of different sample types by PCR testing up to 8 weeks postinoculation.
These results may be explained by the fact that culturing in BSK medium is selective for specific genotypes of B. burgdorferi sensu lato (26, 30). Oliver et al. (31) found relatively low Borrelia culture isolation prevalence (6.5%) among 200 specimens of three reservoir-competent small mammal species sampled from several sites in Florida. Attempts to isolate spirochetes in BSK medium from a smaller number (n = 65) of small mammals from northeast Florida in a different study conducted by one of us (K. Clark) also failed, even though PCR testing showed the B. burgdorferi sensu lato prevalence to be very high (7). An alternative explanation for the failure to isolate B. burgdorferi sensu lato from lizards in the present study is that the number of spirochetes present in the lizard blood and tissues may have been below the sensitivity level of this detection method, especially if the spirochetes were competing with other bacteria that may have been present in the samples. Additional attempts at isolating spirochetes from more PCR-positive lizards are necessary to determine whether such strains are cultivable in BSK medium. These efforts could include filtering inoculated cultures in attempts to remove bacteria other than spirochetes that may be present in lizard blood and tissues. Attempting to isolate these Borrelia organisms in tick cell lines is another potentially successful strategy.
Enzootic cycles of B. burgdorferi sensu lato transmission in the southeastern United States are more complex than those in the northeastern United States due to the involvement of a greater number of vertebrate and tick species in the South (2, 8, 31). This ecologic diversity may explain the genetic heterogeneity among southern B. burgdorferi sensu lato strains and the variation between southern strains and those found elsewhere (24, 25). This heterogeneity may explain why the p66 and ospA primers we used did not amplify products from some of the flaB-positive samples we tested. Primer mismatch could have led to a complete lack of amplification or simply reduced the sensitivity of the PCR assays below the level needed for detection. Numerous strains of B. burgdorferi sensu lato were described previously from ticks and small mammals in Florida by using similar techniques (7). Many of the B. burgdorferi sensu stricto flaB-positive samples in that study also failed to test positive with primers designed to amplify portions of several genes, including p66 and ospA, indicating either low target gene copy number in the samples or primer mismatch. BSK culture attempts with human tissue and fluid samples also fail to isolate spirochetes from many samples that test positive via PCR assays. Interestingly, the p66- and ospA-positive prevalences among flaB-positive lizards in the present study (34.9 and 19.8%, respectively) are comparable to the p66 and ospA PCR-positive prevalences among human spinal fluid (39 and 23%, respectively) and urine (42 and 24%, respectively) samples from Lyme arthritis patients in one study (34).
Based on the findings of several studies (12, 15, 32), we considered the phylogenetic clustering of lizard-derived B. burgdorferi sensu lato strains from partial sequences of the highly conserved flaB gene as indicative of species-specific groupings. Our analyses showed much more variability in the partial ospA and p66 gene sequences derived from lizards. For some templates, the ospA and p66 sequences were very similar to those of reference strains of the same species. For other animals, however, the ospA and p66 sequences derived from them clustered with reference strains of a different species group. There are several possible explanations for this. There may be significant genetic variability in the ospA and p66 genes of southern B. burgdorferi sensu lato strains. This might include, for example, genetic exchange of the plasmid ospA gene among strains of different species, similar to that shown for ospC (27). It is also possible that multiple B. burgdorferi sensu lato species were present in the blood of many individual lizards, representing a relatively high rate of multiple infection. Additional testing of our samples with an adequately sensitive, PCR-based method that employs B. burgdorferi sensu lato species-specific DNA probes could determine this.
Our DNA sequencing did produce some unresolved bases indicative of possible polymorphisms at some locations in the flaB and p66 sequences from a small number of animals. Most of these were resolved upon resequencing those templates with the same primers or the complementary primers used in the PCRs. However, in the flaB sequences for a couple of samples, polymorphisms were still evident, and these were located at positions where nucleotide variation occurs between B. andersonii and B. burgdorferi sensu stricto strains.
We took extensive measures to prevent and identify any contamination of our DNA samples. There was no evidence of contamination of our DNA extracts with reference strain DNA. We used a B. burgdorferi sensu stricto reference strain (B31) as a positive control in our PCR assays, and although we did identify several B. burgdorferi sensu stricto strains in lizards, none of their flaB, ospA, or p66 sequences were identical to those for B31. The significant genetic variability among the lizard-derived B. burgdorferi sensu lato strains, compared to B. burgdorferi sensu lato reference strains of B. andersonii, B. bissettii, and B. burgdorferi sensu stricto, therefore renders contamination from reference strains a very unlikely explanation of our findings.
In the present study, we confirmed the presence of DNA of three B. burgdorferi sensu lato species in over 50% of tested lizards representing several genera and nine species from Florida and South Carolina. We detected the spirochetes in some lizards sampled during months of the year when I. scapularis larvae and nymphs do not normally parasitize hosts, and the duration of the PCR-positive status of several lizards kept in the lab exceeded 4 months. Furthermore, I. scapularis nymphs from larvae that fed on PCR-positive lizards also tested positive for B. burgdorferi sensu lato via PCR, evidence that the ticks acquired the bacteria from the lizards and maintained them after molting to nymphs. All of these findings suggest, but do not prove, the presence of live spirochetes in these samples.
If lizards serve as reservoirs of B. burgdorferi sensu lato, two factors affecting their significance as such would be the duration of infection and their longevity. The duration of B. burgdorferi sensu lato infection of lizards in the wild is not known, and reliable estimates of lizard longevity in the wild are difficult to find in the published literature. However, one website that contains information provided by 234 institutions and 425 private collections (F. Slavens and K. Slavens, Reptiles and amphibians in captivity—longevity—home page. Last updated 20 March 2003. Retrieved 8 November 2004. http://www.pondturtle.com/longev.html) provides longevities of captive specimens of many species. The figures provided for representatives of the species included in the present study ranged from approximately 5 to 7 years for green anoles, 2 to 7 years for various Eumeces spp. skinks, 1 to 4 years for Cnemidophorus spp., 8 years for the Mediterranean gecko (Hemidactylus turcicus), 3 to 14 years for the eastern glass lizard (Ophisaurus ventralis), 1 to 8 years for the eastern fence lizard (Sceloporus undulatus) and other Sceloporus spp., and 2 years for the ground skink (Scincella lateralis). These figures agree with those provided at another site that contains detailed information about brown and green anoles (Under the leaves: complete anole care. Last updated 23 July 2003. Retrieved 8 November 2004. http://www.kingsnake.com/anolecare/5.htm). Although it is doubtful that most specimens live as long in the wild, it is possible that many wild specimens live for several years.
The minimal number of ticks removed from the lizards in this study, the distribution of infection among animals of different ages (including juveniles), and the diversity of infected lizard species, including some not commonly parasitized by ticks and a few individuals (brown anoles) from residential areas, all suggest the possibility of alternate transmission pathways besides the tick-borne route. Some other possible means of transmission to be investigated include transmission by mosquitoes, flies, or mites through blood feeding or mechanical transmission; by lizards ingesting infected arthropods; and from lizard to lizard vertically via transovarial transmission and horizontally during mating.
The present study's findings show that lizards in the southeastern United States are naturally infected with Lyme borreliae and suggest that they may play a role in the enzootic maintenance of B. burgdorferi sensu lato in the region. It remains to be conclusively shown whether the strains infecting lizards in the southeastern United States are cultivable in BSK medium and infectious or pathogenic to humans and whether ticks or other hematophagous arthropods acquire and maintain viable B. burgdorferi sensu lato spirochetes from feeding on lizards. Therefore, the relevance of this discovery to human disease risk in the region is not yet known.
Acknowledgments
We thank J. F. Piesman, Centers for Disease Control and Prevention, Fort Collins, CO, and J. H. Oliver, Georgia Southern University, Statesboro, GA, for providing reference strain culture samples. We also thank H. J. Hutcheson, Colorado State University, Fort Collins, CO, for providing colony ticks for the transmission experiments.
This work was supported in part by a grant from the American Lyme Disease Foundation, Somers, NY, and a University of North Florida College of Health Dean's professorship award to K.C. funded by the Brooks Health Foundation, Jacksonville, FL.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tools. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apperson, C. S., J. F. Levine, T. L. Evans, A. Braswell, and J. Heller. 1993. Relative utilization of reptiles and rodents as hosts by immature Ixodes scapularis (Acari: Ixodidae) in the coastal plain of North Carolina, USA. Exp. Appl. Acarol. 17:719-731. [DOI] [PubMed] [Google Scholar]
- 3.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, R. N., and R. S. Lane. 1996. Reservoir competence of four chaparral-dwelling rodents for Borrelia burgdorferi in California. Am. J. Trop. Med. Hyg. 54:84-91. [DOI] [PubMed] [Google Scholar]
- 5.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwadlt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 256:1439-1442. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2004. Lyme disease—United States, 2001-2002. Morb. Mortal. Wkly. Rep. 53:365-369. [PubMed] [Google Scholar]
- 7.Clark, K. 2004. Borrelia species in host-seeking ticks and small mammals in northern Florida. J. Clin. Microbiol. 42:5076-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, K. L., J. H. Oliver, Jr., J. M. Grego, A. M. James, L. A. Durden, and C. W. Banks. 2001. Host associations of ticks parasitizing rodents at Borrelia burgdorferi-enzootic sites in South Carolina. J. Parasitol. 87:1379-1386. [DOI] [PubMed] [Google Scholar]
- 9.Durden, L. A., R. G. McLean, J. H. Oliver, Jr., S. R. Ubico, and A. M. James. 1997. Ticks, Lyme disease spirochetes, trypanosomes, and antibody to encephalitis viruses in wild birds from coastal Georgia and South Carolina. J. Parasitol. 83:1178-1182. [PubMed] [Google Scholar]
- 10.Durden, L. A., J. H. Oliver, Jr., C. W. Banks, and G. N. Vogel. 2002. Parasitism of lizards by immature stages of the blacklegged tick, Ixodes scapularis (Acari: Ixodidae). Exp. Appl. Acarol. 26:257-266. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fukunaga, M., K. Okada, M. Nakao, T. Konishi, and Y. Sato. 1996. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int. J. Syst. Bacteriol. 46:898-905. [DOI] [PubMed] [Google Scholar]
- 13.Guy, E. C., and G. Stanek. 1991. Detection of Borrelia burgdorferi in patients with Lyme disease by the polymerase chain reaction. J. Clin. Pathol. 44:610-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James, A. M., and J. H. Oliver, Jr. 1990. Feeding and host preference of immature Ixodes dammini, I. scapularis, and I. pacificus (Acari: Ixodidae). J. Med. Entomol. 27:324-330. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, B. J. B., C. M. Happ, L. W. Mayer, and J. Piesman. 1992. Detection of Borrelia burgdorferi in ticks by species-specific amplification of the flagellin gene. Am. J. Trop. Med. Hyg. 47:730-741. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. C., G. P. Schmid, F. W. Hyde, A. G. Steigerwalt, and D. J. Brenner. 1984. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int. J. Syst. Bacteriol. 34:496-497. [Google Scholar]
- 17.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, M. M., R. S. Lane, and P. C. Giclas. 2000. A comparative study of mammalian and reptilian alternative pathway of complement-mediated killing of the Lyme disease spirochete (Borrelia burgdorferi). J. Parasitol. 86:1223-1228. [DOI] [PubMed] [Google Scholar]
- 20.Lane, R. S. 1990. Susceptibility of the western fence lizard (Sceloporus occidentalis) to the Lyme borreliosis spirochete (Borrelia burgdorferi). Am. J. Trop. Med. Hyg. 42:75-82. [DOI] [PubMed] [Google Scholar]
- 21.Lane, R. S., and G. B. Quistad. 1998. Borreliacidal factor in the blood of the western fence lizard (Sceloporus occidentalis). J. Parasitol. 84:29-34. [PubMed] [Google Scholar]
- 22.Levin, M., J. F. Levine, S. Yang, P. Howard, and C. S. Apperson. 1996. Reservoir competence of the southeastern five-lined skink (Eumeces inexpectatus) and the green anole (Anolis carolinensis) for Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 54:92-97. [DOI] [PubMed] [Google Scholar]
- 23.Levine, J. F., M. L. Wilson, and A. Spielman. 1985. Mice as reservoirs of the Lyme disease spirochete. Am. J. Trop. Med. Hyg. 34:355-360. [DOI] [PubMed] [Google Scholar]
- 24.Lin, T., J. H. Oliver, Jr., and L. Gao. 2003. Comparative analysis of Borrelia isolates from southeastern USA based on randomly amplified polymorphic DNA fingerprint and 16S ribosomal gene sequence analyses. FEMS Microbiol. Lett. 228:249-257. [DOI] [PubMed] [Google Scholar]
- 25.Lin, T., J. H. Oliver, Jr., L. Gao, T. M. Kollars, and K. L. Clark. 2001. Genetic heterogeneity of Borrelia burgdorferi sensu lato in the southern United States based on restriction fragment length polymorphism and sequence analysis. J. Clin. Microbiol. 39:2500-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowakowski, R. B. Nadelman, G. P. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livey, I., C. P. Gibbs, R. Schuster, and F. Dorner. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol. 18:257-269. [DOI] [PubMed] [Google Scholar]
- 28.Marconi, R. T., D. Liveris, and I. Schwartz. 1995. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J. Clin. Microbiol. 33:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCombie, W. R., C. Heiner, J. M. Kelly, M. G. Fitzgerald, and J. D. Gocayne. 1992. Rapid and reliable fluorescent cycle sequencing of double stranded templates. DNA Sequence 2:289-296. [DOI] [PubMed] [Google Scholar]
- 30.Norris, D. E., B. J. B. Johnson, J. Piesman, G. O. Maupin, J. L. Clark, and W. C. Black IV. 1997. Culturing selects for specific genotypes of Borrelia burgdorferi in an enzootic cycle in Colorado. J. Clin. Microbiol. 35:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver, J. H., Jr., T. Lin, L. Gao, K. L. Clark, C. W. Banks, L. A. Durden, A. M. James, and F. W. Chandler, Jr. 2003. An enzootic transmission cycle of Lyme borreliosis spirochetes in the southeastern United States. Proc. Natl. Acad. Sci. USA 100:11642-11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picken, R. N. 1992. Polymerase chain reaction primers and probes derived from flagellin gene sequences for specific detection of the agents of Lyme disease and North American relapsing fever. J. Clin. Microbiol. 30:99-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postic, D., N. Marti Ras, R. S. Lane, M. Hendson, and G. Baranton. 1998. Expanded diversity among Californian Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127). J. Clin. Microbiol. 36:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priem, S., M. G. Rittig, T. Kamradt, G. R. Burmeister, and A. Krause. 1997. An optimized PCR leads to rapid and highly sensitive detection of Borrelia burgdorferi in patients with Lyme borreliosis. J. Clin. Microbiol. 35:685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers, A. J. 1953. A study of the ixodid ticks of northern Florida, including the biology and life history of Ixodes scapularis Say (Ixodidae: acarina). Ph.D. thesis. University of Maryland, College Park, Md.
- 36.Rosa, P. A., D. Hogan, and T. G. Schwann. 1991. Polymerase chain reaction analyses identify two distinct classes of Borrelia burgdorferi. J. Clin. Microbiol. 29:524-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swofford, D. L., G. J. Olson, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics. Sinauer Associates, Sunderland, Mass.
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullmann, A. J., R. S. Lane, K. Kurtenbach, M. Miller, M. E. Schriefer, N. Zeidner, and J. Piesman. 2003. Bacteriolytic activity of selected vertebrate sera for Borrelia burgdorferi sensu stricto and Borrelia bissettii. J. Parasitol. 89:1256-1257. [DOI] [PubMed] [Google Scholar]
- 42.Wallich, R., S. E. Moter, M. M. Simon, K. Ebnet, A. Heiberger, and M. D. Kramer. 1990. The Borrelia burgdorferi flagellum-associated 41-kilodalton antigen (flagellin): molecular cloning, expression, and amplification of the gene. Infect. Immun. 58:1711-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, S. A., R. S. Lane, and J. R. Clover. 1998. Infestation of the southern alligator lizard (Squamata: Anguidae) by Ixodes pacificus (Acari: Ixodidae) and its susceptibility to Borrelia burgdorferi. J. Med. Entomol. 35:1044-1049. [DOI] [PubMed] [Google Scholar]