Highlights
-
•
L3HYPDH is induced by IFN-Ι in a cell type-dependent manner.
-
•
L3HYPDH inhibits EV71 replication.
-
•
L3HYPDH inhibits viral translation by targeting the 5′UTR.
Keywords: Interferon-stimulated gene, Enterovirus 71, L3HYPDH, Antiviral activity
Abstract
Enterovirus 71 (EV71) is the common causative agent of hand-foot-mouth disease (HFMD). Despite evidence in mice model suggested that the interferon (IFN) signaling pathways play a role in defending against this virus, knowledge on the IFN-mediated antiviral response is still limited. Here we identified an IFN-stimulated gene (ISG) called L3HYPDH, whose expression inhibits EV71 replication. Mapping assay indicated that amino acids 61–120 and 295–354 are critical for its optimal antiviral activity. Mechanismly, L3HYPDH specifically inhibits protein translation mediated by EV71 internal ribosome entry site (IRES). Our data thus uncovered a new mechanism utilized by the host cell to restrict EV71 replication.
1. Introduction
Hand-foot-mouth disease (HFMD) affects infants and children across the Asian-Pacific region, characterized by fever, rash, and occasionally severe neurological symptoms (Esposito and Principi, 2018). Its major causative agent is enterovirus 71 (EV71), an enterovirus belonging to the Picornaviridae family. RNA is used as the genomic material, single and positive-stranded, with a length of about 7400 nt. The genomic RNA encodes only one open reading frame (ORF) flanked with a 5′-untranslated region (5′-UTR) and a 3′-UTR (Brown et al., 1999; Swain et al., 2022). The 5′-UTR contains a cloverleaf structure and an internal ribosome entry site (IRES), responsible for viral RNA replication and protein translation, respectively (Martínez-Salas et al., 2015). The life cycle of EV71 starts with attachment to the host cell surface by recognition of specific receptors, followed by endocytosis and release of viral RNA into the cytoplasm (Dang et al., 2014). The identified receptors for EV71 infection include the widely expressed intracellular receptor SCARB2 (Scavenger Receptor Class B Member 2).Then, EV71 IRES initiates viral protein translation by recruiting translation machineries. The polyproteins are processed into structural and non-structural proteins by the protease 2A and 3C embedded. When viral proteins accumulate, viral protein 3CD binds to the cloverleaf structure of 5′-UTR to initiate viral RNA replication. The newly synthesized plus-stranded RNAs in turn direct viral protein synthesis in large quantities. With the assembly of viral RNAs and proteins into virions, the host cell lyses and progeny viruses are released for a new round of infection (Lai et al., 2020).
EV71 is capable to suppress type I interferon (IFN-I) production and its signaling activation (Sarry et al., 2022; Lei et al., 2010; Lei et al., 2011; Lu et al., 2012). Nonetheless, cells infected with EV71 can still respond to type I IFN treatment and display an enhanced antiviral state. For example, in vitro studies showed that some type I IFNs, including IFN-α4, IFN-α6, IFN-α14 and IFN-α16, significantly reduced cytopathic effect (CPE) induced by EV71 infection (Yi et al., 2011). An IFN-α2b aerosol therapy has been used topically to treat HFMD (Lin et al., 2016). We recently reported an IFN-induced gene TMEM106A encodes proteins to inhibit EV71 infection through restricting viral binding onto host cells (Guo et al., 2022). However, how IFNs suppress EV71 infection is still largely unknown.
Many new interferon-stimulated genes (ISGs) were identified from human immune cell lines after treatment with interferon (Zhang et al., 2018), but many of them are unclear of the antiviral activities. C14orf149 was one of the newly identified ISGs (Zhang et al., 2018). Using a fluorescent activated cell sorting-based strategy for screening, we identified several ISGs with anti-EV71 efficacy (data not shown). C14orf149 was one of them, this gene was first identified as a gene encoding a trans-3-hydroxy-L-proline dehydratase and then renamed L3HYPDH (Visser et al., 2012). This enzyme may function to degrade dietary proteins that contain trans-3-hydroxy-L-proline as well as other proteins such as collagen IV. The encoded protein can be converted to an epimerase by changing a threonine to a cysteine at a catalytic site (Visser et al., 2012). Little is known about its antiviral activity. We found that L3HYPDH possesses antiviral activity against EV71, and its mechanism of action was investigated with a series of biochemical and genetic assays.
2. Materials and methods
2.1. Plasmids construction
pCAG-DsRed, a red fluorescent protein-expressing plasmid, has been described previously (Matsuda and Cepko, 2004). pWSK-EV71-GFP is an infectious EV71-GFP cDNA clone, with a GFP-coding sequence inserted downstream of EV71 5′UTR and in frame fusion with the downstream VP4, and expression of EV71-GFP is driven by a T7 promoter (Zhang et al., 2017). pcDNA3.1-T7RNP expresses T7 RNA polymerase. These plasmids were kindly provided by Dr. Liguo Zhang at the Institute of Biophysics, Chinese Academy of Sciences (IBP, CAS). A siRNA targeting the coding sequence of L3HYPDH from position 791 to 811 was designed according to the recommendation of Sigma-Aldrich (https://www.sigmaaldrich.com/catalog/genes) and named shRNA-L3HYPDH. A pair of complementary oligonucleotides 5′-GATCCCCCAGATGAACAGGTTGACAGAATTCAAGAGATTCTGTCAACCTGTTCATCTGTTTTTA-3′ (sense) and 5′-AGCTTAAAAACAGATGAA CAGGTTGACAGAATCTCTTGAATTCTGTCAACCTGTTCATCTGGGG-3′ (antisense) were synthesized with 5′ ends being BglII and HindIII restriction site overhangs. For each oligonucleotide, the target sequence was sense followed by antisense orientations separated by a nine-nucleotide spacer. Oligonucleotides were annealed and then cloned into the BglII and HindIII sites of pSUPER.retro.neo+gfp (Oligoengine, herein abbreviated for pSUPER-GFP) to generate pSUPER-GFP-shRNA-L3HYPDH. L3HYPDH wild type (WT) and deletion mutants as indicated in Fig. 3 were amplified with PCR using pLPCX-C14orf149 (L3HYPDH) (Zhang et al., 2018), kindly provided by Dr. Guangxia Gao at IBP, CAS, as the template. PCR products of L3HYPDH WT and deleted mutants were digested with BamHI & NotI and KpnI & XbaI, respectively, and inserted into similarly digested pcDNA4-To/myc-His B (Invitrogen), resulting in pcDNA4-L3HYPDH, pcDNA4-L3HYPDHΔN1, pcDNA4-L3HYPDHΔN2, pcDNA4-L3HYPDHΔN3, pcDNA4-L3HYPDHΔC1, pcDNA4-L3HYPDHΔC2, and pcDNA4-L3HYPDHΔC3. psiCHECK2-M was a modified form of psiCHECK-2 (Promega) with deletion of the HSV-TK promoter (Fig. 5(A)). Inverse PCR was performed with high-fidelity DNA polymerase Phusion (ThermoFisher) and a pair of back-to-back primers to amplify the whole plasmid except the HSV-TK promoter sequence. PCR products were self-ligated and resulted in psiCHECK2-M; meanwhile, a SalI and a NotI sites within the back-to-back primers were introduced into the plasmid. EV71-5′UTR and HCV (Hepatitis C virus)-5′UTR were amplified from pWSK-EV71-GFP and pNL4-3RL-HCV-FL (Zhu et al., 2012) by PCR, respectively. After digestion with SalI and NotI, the PCR products were linked into the similarly digested psiCHECK2-M and resulted in psiCHECK2-M-EV71-5′UTR and psiCHECK2-M-HCV-5′UTR. All primers used are listed in Table S1.
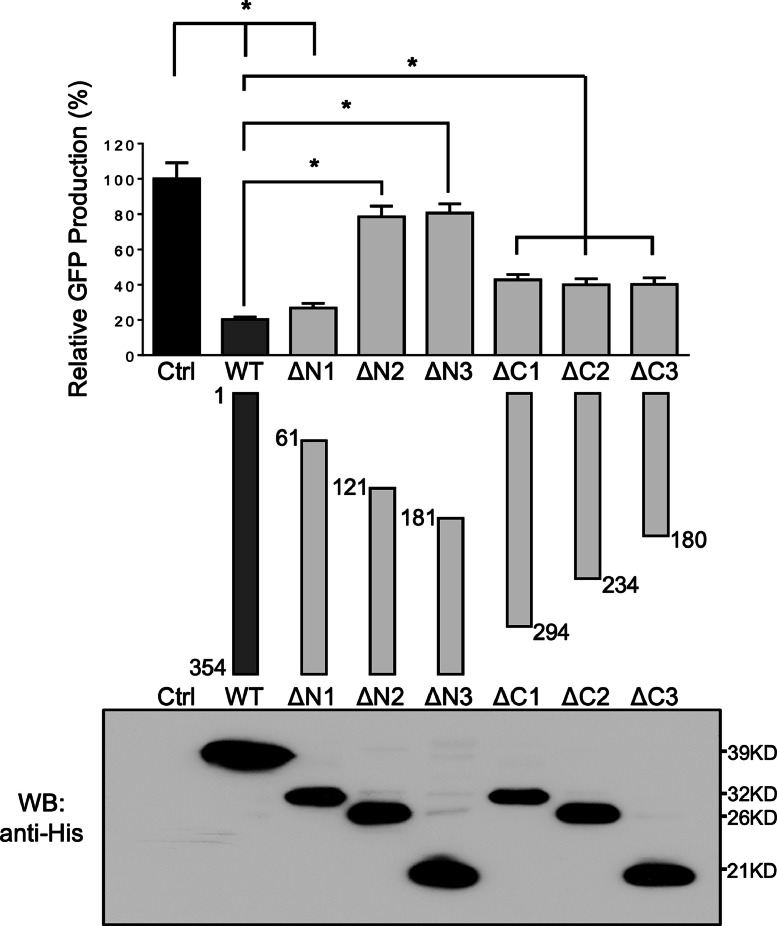
Fig. 3.
Mapping amino acid sequence required for anti-EV71 activity of L3HYPDH. Deletion mutants of L3HYPDH were schematically shown in the middle panel. Numbers indicate starting and ending amino acid. The plasmids expressing L3HYPDH WT or truncated mutants were individually transfected into 293A-SCARB2 cells together with pCAG-DsREd at a ratio of 3:1, followed by EV71-GFP infection. Same performance was done the empty vector used as a control (Ctrl). L3HYPDH WT proteins or deletion mutants were analyzed by Western blot with anti-6 × His mAb (lower panel). GFP production from EV71-GFP was detected using FACS and calculated as described in Fig. 1(C). The value from control was set as 100 %. Data are represented as mean ± SD of three independent experiments. *, P < 0.05.
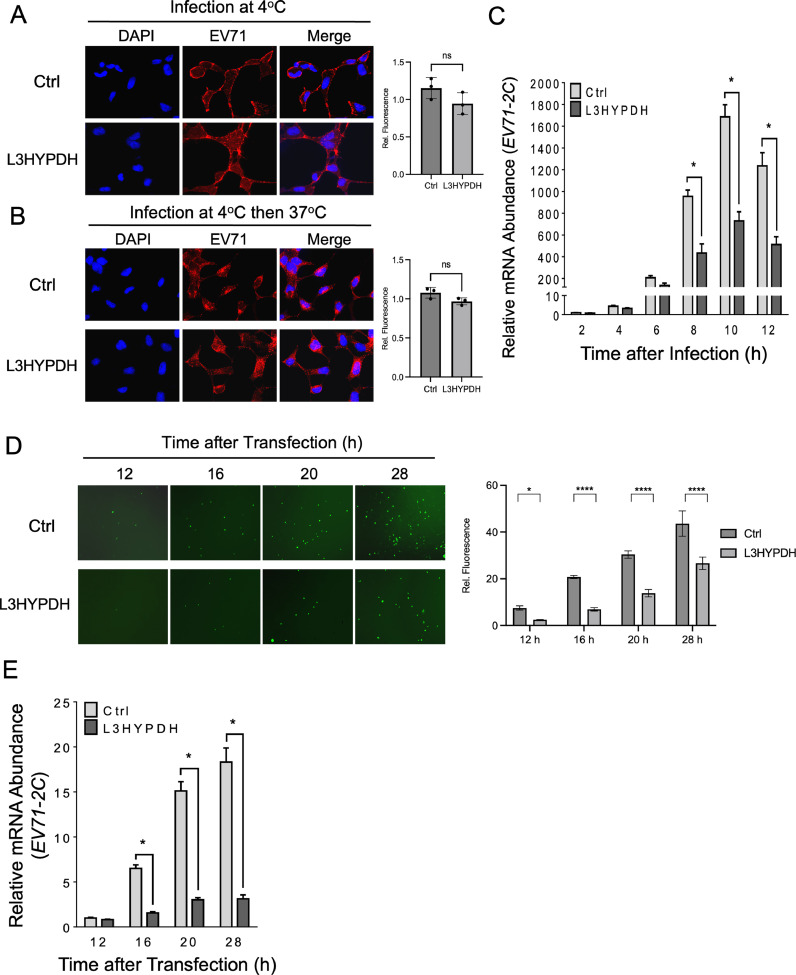
Fig. 5.
Stage assays for unveiling the mechanism of L3HYPDH against EV71. Effect of L3HYPDH on attachment (A) and endocytosis (B) of EV71 was examined using IFA. 293A-SCRB2-L3HYPDH and 293A-SCARB2-Ctrl cells were infected with EV71-MZ (MOI, 100). Nuclei were stained with DAPI. (C) Effect of L3HYPDH on viral RNA measured with RT-qPCR. 293A-SCARB2-L3HYPDH and control cell (Ctrl) were infected with EV71-MZ (MOI at 2 Lin and Huang, 2020). EV71 2C mRNA level was measured at indicated times and normalized to that of GAPDH, with the relative level in control cell at 2 h post infection set as 1. EV71-GFP RNAs were transfected into 293A-SCARB2-L3HYPDH and control cell (Ctrl). GFP signal and EV71 2C RNA level at different times post transfection were examined by fluorescent microscope (D) and RT-qPCR (E), respectively. Quantitative analysis of three independent imaging tests was shown on the right. All the RT-qPCR data are represented as means ± SD of three independent experiments. ns, not significant. *, P < 0.05. ****, P < 0.0001.
2.2. Cell culture and virus preparation
293A, 293A-SCARB2, RD, Vero, HeLa, and A549 cells were cultured in DMEM (Gibco) supplemented with 10 % fetal bovine serum (FBS, Gibco). 293A-SCARB2 (Kindly provided by Dr. Liguo Zhang at IBP, CAS), originated from a 293A cell line and constitutively expresses the main EV71 receptor scavenger receptor class B member 2 (SCARB2). To generate the cell line constitutively expressing tagged L3HYPDH, 293A-SCARB2 cells were transfected with pcDNA4-L3HYPDH as described below and selected with Zeocin (200 μg/ml). Resistant colonies were individually expanded and detected by Western blot. One positive clone was chosen and named 293A-SCARB2-L3HYPDH. This process was applied to the empty vector and resulted in control cell 293A-SCARB2-Ctrl.
EV71-MZ (GenBank accession no. KY582572), isolated from the throat swab of an ICU patient at Meizhou People's Hospital in 2014 (Zeng et al., 2019), was amplified by successive passages in RD cells until apparent CPE appeared. EV71-GFP was generated by co-transfecting pWSK-EV71-GFP and pcDNA3.1-T7RNAP into 293A-SCARB2 cells as described previously (Zhang et al., 2017). Viral supernatants were titrated using a plague assay, aliquoted, and then used for infection.
2.3. Transfection and infection
Depending on the experiments, cells were seeded into a 24-well or 6-well plate or 10 cm dish and were grown to approximately 80 % confluence prior to transfection or infection. All plasmid and RNA transfections were carried out by using Lipofectamine TM 2000 (Life Technology) according to the manufacturer's instructions. After incubation for the indicated time, cells were treated as required.
Viral infection was performed by incubating cells with EV71-GFP or EV71-MZ at a different multiplicity of infection (MOI) for 1 h, with shaking every 15 min, and then the unbound viruses were aspirated. Cells were washed with PBS, added fresh medium, and incubated for specific time, followed by FACS assay, RT-qPCR measurement, or supernatant titration.
2.4. Plaque assay
The plaque assay was performed as described previously (Tang et al., 2016). Briefly, RD cells were incubated with viral supernatants undiluted or diluted in 10-fold series for 1 h. Subsequently, the supernatants were aspirated, and cells were covered with DMEM containing 1 % methylcellulose (Sigma-Aldrich) and 2 % FBS. After incubation for 4 days, cells were fixed with 4 % paraformaldehyde (Sigma-Aldrich) and stained with 0.1 % crystal violet. Plaques were then quantified via visual scoring.
2.5. Fluorescent-activated cell sorting (FACS) assay
To measure GFP production from EV71-GFP, 1 × 106 infected cells were collected and fixed in 4 % paraformaldehyde for 15 min. After washing three times with PBS, cells were resuspended in 0.5 ml of PBS for flow cytometry (LSRFortessa, BD). To assess effects of L3HYPDH knockdown by RNAi, the cells were transfected with pSUPER-GFP-shRNA-L3HYPDH or pSUPER-GFP. After incubation for the indicated time, the cells were harvested and washed with PBS. GFP-positive cells were obtained through FACS, and then lysed for Western blot or seeded into a 24-well plate for EV71-GFP infection or reporter plasmid transfection as required.
2.6. IFN stimulation
Cells were treated with 1000 IU/ml of recombinant human IFN-α2b (Prospec) at different time, and then total RNAs were isolated and used to measure specific mRNA abundance by RT-qPCR.
2.7. In vitro transcription of EV71-GFP and microscope assay of GFP
pWSK-EV71-GFP was linearized XbaI and EV71-GFP RNAs were transcribed using the T7 RiboMax kit (Promega). After transfection into 293A-SCARB2-L3HYPDH and 293A-SCARB2-Ctrl cells, the GFP signal was observed under a fluorescence microscope (System Microscope BX63, Olympus) at the indicated times; total RNAs were isolated for RT-qPCR assay.
2.8. RNA isolation and RT-qPCR
Total RNAs were isolated from cells using TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. RT-qPCR was carried out as described previously (Zhou et al., 2015). Briefly, RNAs were treated with DNase using an RQ1 RNase-Free DNase Kit (Promega); cDNAs were synthesized using PrimeScript RT reagent Kit (Takara, Dalian) and then diluted and subjected to quantitative PCR using TransStart Green qPCR SuperMix (TransGen Biotch) in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Primers were shown in Table S1.
2.9. Immunofluorescence assay (IFA)
Subcellular localization of L3HYPDH proteins and the attachment and endocytosis of EV71 virions were detected using IFA as described previously (Li et al., 2017) with some modifications. Briefly, 293A-SCARB2-L3HYPDH and 293A-SCARB2-Crtl were individually seeded onto a coverslip. Polyclonal antibody (pAb) specific to c-myc (Sigma-Aldrich, 1:100) and Alexa Fluor 555-labeled anti-rabbit IgG (ThermoFisher, 1:100) were used as primary and secondary antibody, respectively, to localize the subcellular distribution of the tagged L3HYPDH proteins. Similarly, 293A-SCARB2-L3HYPDH and 293A-SCARB2-Crtl cells were infected with EV71-MZ (MOI, 100) at 4 °C for 1 h to allow viral attachment or incubated for an additional 30 min at 37 °C to allow viral endocytosis. The Anti-EV71 VP2 monoclonal antibody (mAb) (Millipore, 1:50) and Alex flour 555-labeled anti-mouse IgG (ThermoFisher, 1:100) were used as primary and secondary antibody, respectively, to visualize EV71 virions. Nuclei were stained with DAPI (Roche). Fluorescent images of cells were captured using a Zeiss LSM780 META confocal imaging system.
2.10. Western blot
Western blot was performed as described previously (Li et al., 2017) with some modifications. Briefly, 48 h after transfection, the cells were lysed with SDS-lysis buffer (30 mM SDS, 50 mM pH 6.8 Tris–HCL, 100 mM DTT and 20 mg/L bromophenol blue) directly and proteins were isolated with 10 % SDS-PAGE. The membrane was probed with anti-6 × His mAb (Abcam), anti-GAPDH pAb (Sangon), and anti-VP2 mAb (Millipore) followed by incubation with HRP-conjugated anti-mouse IgG and anti-rabbit IgG (Santa Cruz Biotechnology), respectively. Proteins were visualized with ECL.
2.11. Luciferase activity assay
Cell lysate was prepared by using passive lysis buffer (Promega). Firefly and renilla luciferase activities (Fluc and Rluc) were measured using a Dual Luciferase Assay kit (Promega) according to the manufacturer's instructions.
2.12. Statistical analysis
All the experiments involving counting or calculation were performed independently at least three times and data are means ± standard deviation (SD). A Student's two-tailed t-test was used for statistical analysis by using GraphPad Prism 6.1 (GraphPad 6 Software, San Diego, CA). P < 0.05 was considered statistically significant.
3. Results
3.1. Expression of L3HYPDH inhibits EV71-GFP replication
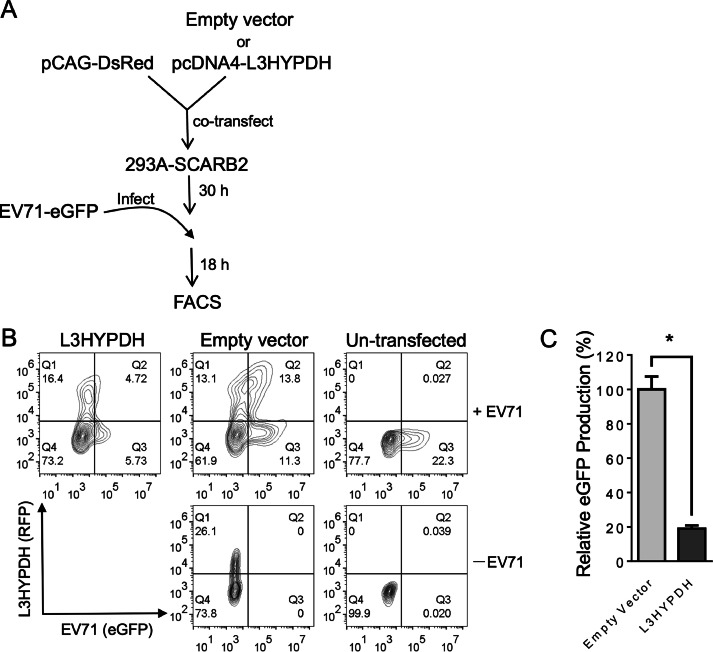
To study the activity of L3HYPDH, the recipient cell line is best to have little or no IFN signaling in response to EV71 infection. We thus used HEK293A cell line as it lacks the expression of many pattern recognition receptors (PRRs) (Hornung et al., 2002). To support the infection, the predominant receptor SCARB2 was stably introduced into the cells (293A-SCARB2) (Jiao et al., 2014). To check the transfection efficiency, we co-transfected pCAG-DsRed with L3HYPDH-expressing plasmids in a mass ratio of 1:3. Under this biased ratio, we assumed that cells bearing fluorescent signals would also have L3HYPDH expressed. The transfected cells were then infected with EV71-eGFP 30 h later. Viral replication in the DsRed-positive populations was quantified by FACS assay of GFP at 18 h post infection (Fig. 1(A)). Data showed that the GFP-positive population from empty vector-transfected cells was much larger than that from L3HYPDH-expressing cells (Fig. 1(B) and (C)), suggesting L3HYPDH acts as an inhibitory host factor against EV71.
Fig. 1.
FACS-based assay for antiviral activity of L3HYPDH against EV71-GFP replication. (A) Overview of the procedures detecting anti-EV71 activity of over-expressed L3HYPDH using FACS. (B) FACS plots of L3HYPDH inhibition of EV71-GFP in 293A-SCARB2 cells. Numerals represent percent of total cell counts. (C) GFP production, which was calculated by multiplying GFP and DsRed co-positive cell number by the mean value of GPF intensity. The value of the control cells transfected with empty vector was set as 100 %. Results are represented as means ± SD of three independent experiments. *, P < 0.05.
3.2. Expression of endogenous L3HYPDH suppresses EV71 replication
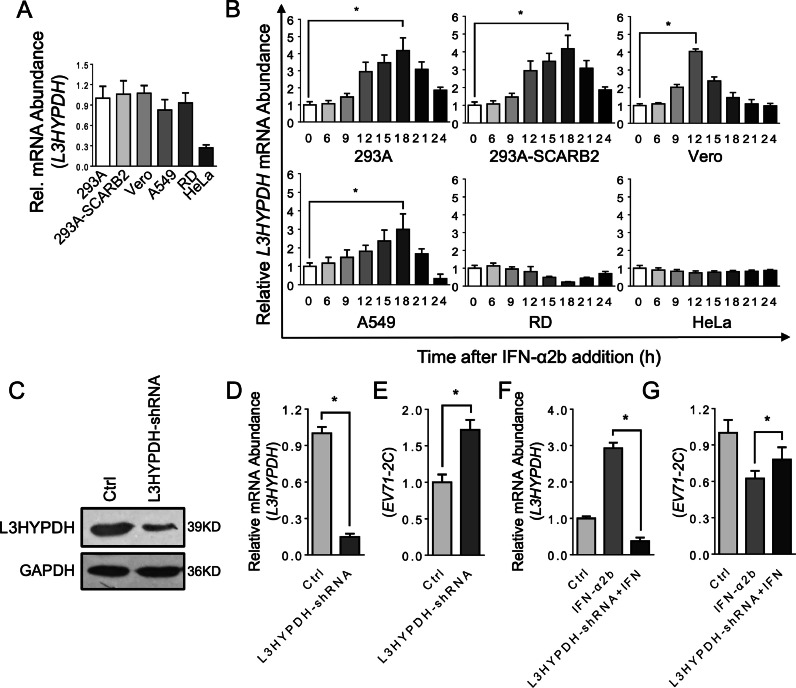
Next, we asked whether the endogenous L3HYPDH also inhibits EV71 infection. We set to test its expression pattern in commonly used cell lines. The RNA levels were measured by RT-qPCR. Data showed that HeLa cells express the least amount of L3HPDH while other tested cells have similar expression levels (Fig. 2(A)). As is mentioned previously, this gene was identified as an ISG in immune cell lines, we then set to test if its expression is also responsive to IFN treatment here. Upon exposure to IFN-α2b, the level of L3HYPDH mRNA was up-regulated and peaked at about 18 h in 293A, 293A-SCARB2 and A549 and at about 12 h in Vero cells (Fig. 2(B)). In contrast, the mRNA level changed little in HeLa cells and even decreased a little in RD cells (Fig. 2(B)). The data also showed that SCARB2 expression in 293A cells (293A-SCARB2) does not change L3HYPDH expression either in unstimulated state or IFN-stimulated state.
Fig. 2.
Anti-EV71 activity of the endogenous L3HYPDH. (A) RT-qPCR assay of the endogenous L3HYPDH mRNA level in different cell lines, which was normalized to GAPDH mRNA level. (B) RT-qPCR assay of L3HYPDH expression with IFN-α2b (1000 IU/ml) treatment for indicated time in different cell lines. L3HYPDH mRNA level was normalized to that of GAPDH. The relative mRNA level from untreated cells (marked as 0 h) was set as 1. (C) Western blot of knockdown efficiency of shRNA-L3HYPDH. 293A-SCARB2 cells were transfected with pcDNA4-L3HYPDH together with shRNA-expressing plasmid or control plasmid (Ctrl) at a ratio of 1:3. The GFP-positive cells were sorted and then infected with EV71, followed by RT-qPCR analyses of L3HYPDH mRNA (D) and EV71 2C mRNA (E) after depression of L3HYPDH expression. RT-qPCR analyses of the L3HYPDH (F) and EV71-2C (G) mRNA in 293A-SCARB2 upon depression of L3HYPDH expression by shRNA in the presence of IFN-α2b. 293A-SCARB2 cells were seeded into a 10 cm dish and then transfected with 5 μg of pSUPER-GFP-shRNA-L3HYPDH or pSUPER-GFP. After incubation for 24 h, the GFP-positive cells were isolated using FACS and divided into two parts, one treated with 1000 IU/ml of IFN-α2b, the other mock-treated with water as control, followed by EV71-MZ infection (MOI, 0.1). Eighteen hours post-infection, total RNAs were isolated and used for RT-qPCR measurement. The target mRNA level was normalized to that of GAPDH, and the relative value from the control cells was set as 1. The results are represented as mean ± SD of three independent experiments. *, P < 0.05.
We then tested the function of L3HYPDH using shRNA-mediated gene knockdown. An shRNA specific to L3HYPDH was designed and transcribed from pSuper-GFP-shRNA-L3HYPDH. Its knocking down efficiency was detected in 293A-SCARB2 cells by co-transfecting pcDNA4-L3HYPDH together with pSUPER-GFP-shRNA-L3HYPDH or with pSUPER-GFP as a control. Western blot analysis showed that the myc-tagged L3HYPDH protein level decreased dramatically in the presence of shRNA-L3HYPDH (Fig. 2(C)), suggesting this shRNA is potent in down-regulating the expression level of L3HYPDH. To determine if the endogenous L3HYPDH could suppress EV71 replication, 293A-SCARB2 cells were transfected with pSUPER-GFP-shRNA-L3HYPDH, and the GFP-positive cells were sorted by FACS, followed by a clinical isolate of EV71, EV71-MZ infection at MOI of 0.1 according to established protocols (Guo et al., 2022; Tan et al., 2021). RT-qPCR assay revealed that L3HYPDH-shRNA reduced L3HYPDH mRNA level by more than 80 % (Fig. 2(D)) and increased EV71-transcribed 2C mRNA level from 1 to 1.7 (Fig. 2(E)), indicating that the expression of endogenous L3HYPDH impaired EV71 replication. Importantly, the L3HYPDH knockdown (Fig. 2(F)) relieved IFN-mediated EV71 replication impairment (Fig. 2(G)), suggesting both the basal level and IFN-stimulated expression of L3HYPDH encode antiviral activity against EV71.
3.3. Determination of the amino acid sequences essential for anti-EV71 activity of L3HYPDH
Next, we asked which domain is important for L3HYPDH antiviral activity. We mapped its regions using the FACS-based functional assay. Three N-terminal and three C-terminal serial deletions of L3HYPDH are schematically shown in Fig. 3 according to UniProt (middle panel), designated as ΔN1, ΔN2, ΔN3, ΔC1, ΔC2, and ΔC3. Their coding sequences were cloned in fusion with a myc-6 × His tag at the C-terminus as with the wild type (WT) L3HYPDH. The resulting plasmids were individually transfected into 293A-SCARB2 cells together with pCAG-DsRed followed by EV71-eGFP infection as described in Fig. 1(A). L3HYPDH protein expression was validated by western blotting (Fig. 3, lower panel) and virus infection was detected by eGFP measurement (Fig. 3, upper panel). FACS assay showed that L3HYPDH ΔN2 lacking the amino acids from position 1 to 120 significantly impaired the antiviral activity in comparison with WT, while L3HYPDH ΔN1 lacking the amino acids from position 1 to 60 only slightly weakened the antiviral activity. L3HYPDH ΔC1 lacking the C-terminal 60 amino acids from 295 to 354 also weakened the antiviral activity, while further deletion did not change this impairment. These results indicate that the amino acid sequences from position 61 to 120 and those from position 295 to 354 are both required for optimal anti-EV71 activity, and the N-terminus plays a more important role.
3.4. EV71 replication is suppressed in the cell line 293A-SCARB2-L3HYPDH expressing L3HYPDH constitutively
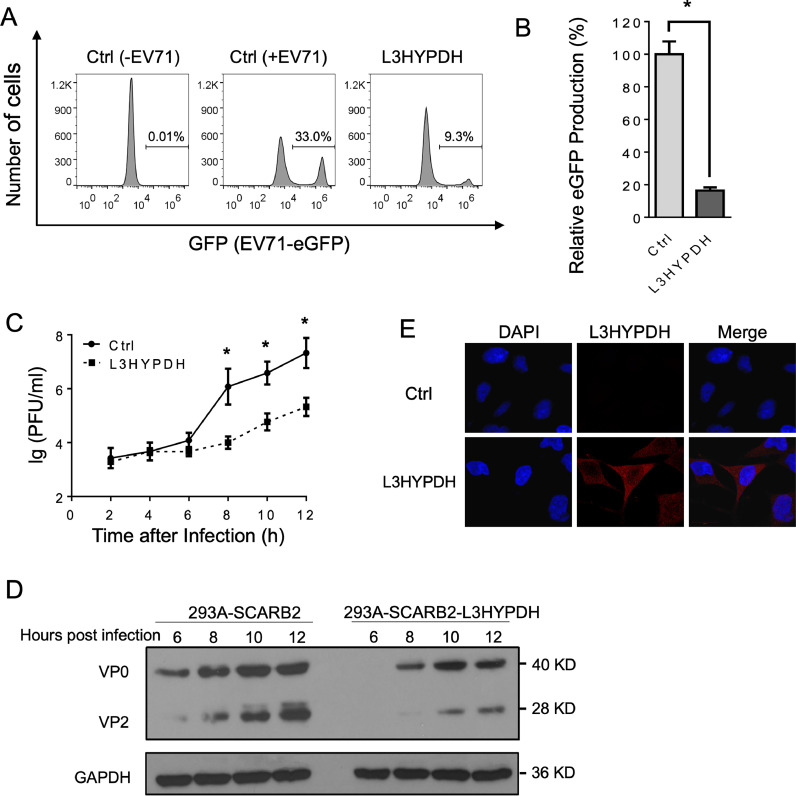
A cell line stably expressing L3HYPDH (293A-SCARB2-L3HYPDH) was generated to facilitate the study of its mechanism of action. They were infected with EV71-eGFP at an MOI of 0.1 (Guo et al., 2022; Tan et al., 2021). FACS assay showed that the eGFP production in 293A-SCARB2-L3HYPDH decreased significantly (Fig. 4(A) and (B)). Upon infection with EV71-MZ, there was significantly less viral multiplication in 293A-SCARB2- L3HYPDH than in the control cells (Fig. 4(C)). VP protein expression was consistent with viral titer (Fig. 4(D)). IFA showed that L3HYPDH proteins were mainly located in the cytoplasm (Fig. 4(E)), consistent with its anti-EV71 action. Therefore, the cell 293A-SCARB2-L3HYPDH displays remarkable anti-EV71 activity due to the ectopic expression of L3HYPDH, and thus can be exploited to uncover the underlying antiviral mechanism.
Fig. 4.
Evaluating antiviral activity of 293A-SCARB2- L3HYPDH cells. 293A-SCARB2-L3HYPDH and the control cell 293A-SCARB2-Ctrl were infected with EV71-GFP (MOI, 0.1). GFP production was detected using FACS (A) and calculated (B). Data are represented as mean ± SD of three independent experiments. *, P < 0.05. (C) Time-viral yield assay. 293A-SCARB2-L3HYPDH and the control cell were individually infected with EV71-MZ (MOI at 2 based on established protocols Lin and Huang, 2020). The culture supernatants were harvested at different time points as indicated and titrated by plaque assay. Data for each time point are means ± SD of three independent experiments, *, P < 0.05. (D) Detection of viral proteins by Western blotting. (E) Subcellular localization of tagged L3HYPDH proteins using IFA. Nuclear DNA was stained with DAPI.
3.5. L3HYPDH interferes with the synthesis of viral RNA and proteins
The effects of L3HYPDH on different life stages of EV71 replication were examined in 293A-SCARB2-L3HYPDH cells. Based on the knowledge that EV71 is only adsorbed on the cell surface and could not finish endocytosis at 4 ℃, 293A-SCARB2-L3HYPDH and the control cells were incubated with EV71-MZ at MOI of 100 (Guo et al., 2022; Yang et al., 2022) for 1 h at 4 ℃ followed by IFA with the antibody specific to EV71 VP2. As shown in Fig. 5(A), a massive number of virions distributed on the outer surfaces of both cell lines, showing no difference in numbers, indicating that L3HYPDH does not interfere with EV71 attachment. After attachment at 4 ℃, the viruses were further incubated with the cells for an additional 30 min at 37 ℃ to complete endocytosis. IFA showed that viruses entered both cell lines with little difference (Fig. 5(B)), indicating that L3HYPDH has no effect on EV71 endocytosis. In this way, these results demonstrate that L3HYPDH does not impede the viral attachment and endocytosis.
The effects of L3HYPDH on the synthesis of viral RNA and proteins were investigated by monitoring changes in their levels over time. 293A-SCARB2-L3HYPDH and 293A-SCARB2-Ctrl cells were infected with EV71-MZ. Total RNAs were isolated at different times post-infection and the viral RNA abundance was measured by RT-qPCR. The increase in EV71 RNA levels over time in 293A-SCARB2-L3HYPDH cells was much lower than that in the control cells (Fig. 5(C)). Due to the tight crosstalk between viral translation and viral RNA synthesis, we hypothesized that L3HYPDH might inhibit the synthesis of viral RNA, proteins, or both. To further confirm this assumption, EV71-GFP RNAs were transfected into 293A-SCARB2-L3HYPDH and the control cell, and then the viral RNA and GFP proteins were measured and compared at different times after transfection. Microscope and RT-qPCR analyses showed that both the number of GFP-positive cells and the viral RNA level were much lower in 293A-SCARB2-L3HYPDH cells than in the control cells (Fig. 5(D), (E)). These results suggested that L3HYPDH suppresses either EV71 RNA replication, viral protein synthesis, or both.
3.6. L3HYPDH impairs the translation mediated by EV71-5′UTR
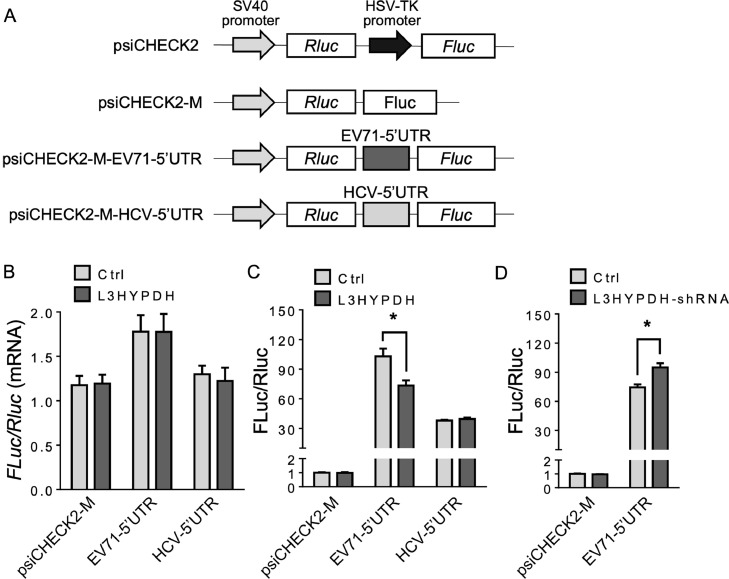
The repression of L3HYPDH on viral protein synthesis was investigated using a bicistronic reporter system. As shown in Fig. 6(A), psiCHECK-2-based reporter plasmids were constructed with the HSV-TK promoter deleted to generate the control (psiCHECK2-M) or replaced with EV71-5′UTR or HCV-5′UTR, which contains EV71 IRES or HCV IRES, respectively. pcDNA4-L3HYPDH or the empty vector was transfected into 293A cells together with one of the three reporter plasmids at a ratio of 3:1, and then the luciferase activity and mRNA level were measured after incubation for 48 h. For these reporters, the mRNA level ratio of Fluc/Rluc in L3HYPDH-overexpressed cells was equal to that in the empty vector-transfected cells as revealed by RT-qPCR (Fig. 6(B)). However, the luciferase activity ratio (Fluc/Rluc) showed variability (Fig. 6(C)). Whether L3HYPDH was over-expressed or not, the Fluc/Rluc ratio of the control reporter was extremely low due to the absence of IRES; the ratio of the EV71-5′UTR-containing reporter reduced by 29 % upon overexpression of L3HYPDH; while the ratio of the HCV-5′UTR-containing reporter changed little. We here proposed that L3HYPDH could specifically inhibit the reporter translation mediated by EV71 IRES. The knockdown assay further provided evidence for this speculation. 293A-SCARB2-L3HYPDH cells were transfected with the GFP-encoded L3HYPDH-shRNA-expressing plasmid or the empty vector. Then the GFP-positive cells were isolated and transfected with the reporter plasmids. Compared to the control cells, the Fluc/Rluc ratio of the EV71-5′UTR-containing reporter in 293A-SCARB2-L3HYPDH cells increased upon L3HYPDH knockdown (Fig. 6(D)). Altogether, these results indicate that L3HYPDH can specifically impair the translation initiated by EV71-5′UTR.
Fig. 6.
Bicistronic reporter assay to measure the effect of L3HYPDH on EV71 IRES mediated translation. (A) Schematic of bicistronic reporters. Rluc is translated in a cap-dependent manner and Fluc in an IRES-dependent manner. pcDNA4-L3HYPDH or empty vector was transfected into 293A cells together with psiCHECK2-M, psiCHECK2-EV71-5′UTR, or psiCHECK2-HCV-5′UTR. Effects of L3HYPDH on the reporter expression were estimated by RT-qPCR (B) and luciferase activity assay (C). Fluc/Rluc ratio was calculated, with the relative value from the cells transfected with empty vectors was set as 1. (D) RNAi assay of effect of L3HYPDH on reporter expression mediated by IRES. pSUPER-GFP-shRNA-L3HYPDH or pSUPER-GFP was transfected into 293A-SCARB2-L3HYPDH cell. The GFP positive cells were transfected with psiCHECK2-M or psiCHECK2-M-EV71-5′UTR. Luciferase activity was measured, with Fluc/Rluc ratio from cells co-transfected with psiCHECK2-M and pSUPER-GFP set as 1. Transfection with pcDNA4 was used as a control (Ctrl). Luciferase activities were measured. Fluc/Rluc ratio was calculated, with the relative value from the cells transfected with the empty vector set as 1. Data are means ± SD of three independent experiments. *, P < 0.05.
4. Discussion
In this work, we report that the ISG product L3HYPDH has antiviral activity against EV71 according to knockdown and over-expression experiments. Over-expression of L3HYPDH repressed eGFP production of EV71-eGFP (Fig. 1(B)) and caused significant inhibition of propagation of the clinical isolate EV71-MZ (Fig. 4(B)). Importantly, endogenous L3HYPDH is active in down-regulating EV71 mRNA in 293A-SCARB2 cells (Fig. 2(E)). Additionally, our data showed that IFN-α2b treatment was less effective against EV71 in cell culture when expression of L3HYPDH was depressed by shRNA (Fig. 2(F), (G)). Therefore, L3HYPDH contributes to the antiviral activity of IFN-α2b against EV71. The potential activity of L3HYPDH against other viruses is not known. Given that different viruses are usually targeted by unique sets of ISGs (Schoggins and Rice, 2011), an extensive investigation on L3HYPDH will help to further elucidate the mechanism of IFN-mediated innate immunity against invading viruses.
Our data show that L3HYPDH may interfere with EV71 replication at post-entry stage (Fig. 4). Bicistronic reporter assays confirmed that expression of L3HYPDH inhibited translation initiated by EV71 IRES (Fig. 6(B)), however, the reporter protein was less reduced than EV71 RNA and virus-carrying eGFP production during the first round of infection (Figs. 1(B), 4(A), 5(C)–(E)). These inconsistences suggest that L3HYPDH hampers EV71 replication at other steps as well. Although inhibition of viral RNA replication is likely, other potential effects on viral RNA stability, viral assembly and viral release cannot be excluded. Therefore, L3HYPDH inhibits EV71 replication at least at two levels, and these data agree with previous studies indicating that many ISGs block viral replication at multiple stages of the viral life cycle (Schoggins, 2019). Considering that a range of proteins are involved in the viral RNA replication and translation process, we performed co-immunoprecipitation and tandem affinity purification combination mass spectrometry to screen for proteins interacting with L3HYPDH. Neither viral nor host proteins were identified (data not shown). These results suggest that the association of L3HYPDH proteins with other proteins should be transient or weak. L3HYPDH might also function by binding to the viral RNA directly; however, no known RNA-binding domains were predicted with online software (data not shown).
Viral translation is in most cases host cell dependent. To maximize efficiency, different viruses evolved many strategies to facilitate selective translation of viral mRNAs over host transcripts (Swain et al., 2022; Lin et al., 2009). Among these, the IRES-mediated translation initiation is necessary for picornavirus and hepacivirus to replicate (Martínez-Salas et al., 2015; Honda et al., 1996). Reporter assays showed that expression of L3HYPDH impaired initiation of translation mediated by EV71 IRES but not HCV IRES (Fig. 6(B), (C)). These two tested IRES differ in nucleotide length and structure as well as in host factors required for translation initiation and regulation (Martinez-Salas et al., 2018). A potential target of L3HYPDH should be involved in EV71-5′UTR-mediated translation. Meanwhile, despite being present in all picornaviruses, IRES is also diverse in length and structure and requires different host factors to function (Andreev et al., 2007; Francisco-Velilla et al., 2022). Whether L3HYPDH can inhibit other genera of picornavirus by interfering with IRES-mediated translation is not clear is not clear, but this is worthy of study in the future.
L3HYPDH is a trans-3-hydroxy-L-proline dehydratase, and specifically catalyzes the dehydration of dietary trans-3-hydroxy-L-proline and from degradation of proteins such as collagen IV that contain it. This dehydratase contains two active sites, a Cys residue at the 104 position and a Thr residue at the 273 position (Visser et al., 2012). Interestingly, the region required for anti-EV71 activity was mainly mapped to the amino acid sequence from position 61 to 120 of L3HYPDH protein (Fig. 3), which contains the Cys104 active site. Whether this proline dehydratase activity is involved in the anti-EV71 activity is not known yet, but at least an extra region from the C-terminus is required for L3HYPDH antiviral activity. As virus often develops strategies to evade the antiviral defense, it would be very interesting to investigate whether EV-A71 evolved to antagonize L3HYPDH in the future work. The low expression of L3HYPDH in 293A-SCARB2 cells through the course of infection (data not shown) could be explained by the inability of IFN induction. The fact that L3HYPDH was not induced by IFN treatment in RD cells (Fig. 2(B)) suggested the expression pattern of L3HYPDH against EV-A71 is cell line-specific.
In sum, our current work uncovered a new anti-EV71 host factor whose expression is up-regulated by IFN-I treatment in certain cell lines, highlighting the protective role of IFN-I and ISGs upon viral infection. Understanding ISG products and antiviral spectra, as well as their mechanisms of action and biological function will help create novel therapeutics for HFMD in the future.
Funding
This work was supported by the grants to X.G from the Natural Science Foundation of Guangdong Province (2019A1515012133), the Science and Technology Program of Meizhou (2019B0202001, 2019B001), Key Scientific and Technological Project of Meizhou People's Hospital (PY-A2019003), Guangzhou Municipal Science and Technology Program (202206010114).
CRediT authorship contribution statement
Jian Liu: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Logen Liu: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Shinuan Zeng: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Xiaobin Meng: Conceptualization, Methodology, Formal analysis. Nanfeng Lei: Conceptualization, Methodology, Formal analysis. Hai Yang: Conceptualization, Methodology, Formal analysis. Runcai Li: Conceptualization, Methodology, Formal analysis. Xin Mu: Conceptualization, Formal analysis, Writing – review & editing. Xuemin Guo: Conceptualization, Formal analysis, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Liguo Zhang for providing the plasmids pCAG-DsRed, pWSK-EV71-GFP, pcDNA3.1-T7RNP and the cell line 293A-SCARB2. We also thank Dr. Guangxia Gao for providing the plasmids pLPCX-C14orf149 and pNL4-3RL-HCV-FL.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2024.199336.
Contributor Information
Xin Mu, Email: xin_mu@tju.edu.cn.
Xuemin Guo, Email: guoxuemin@mzrmyy.com.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Andreev D.E., Fernandez-Miragall O., Ramajo J., Dmitriev S.E., Terenin I.M., Martinez-Salas E., Shatsky I.N. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Oberste M.S., Alexander J.P., Kennett M.L., Pallansch M.A. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 1999;73:9969–9975. doi: 10.1128/jvi.73.12.9969-9975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang M., Wang X., Wang Q., Wang Y., Lin J., Sun Y., Li X., Zhang L., Lou Z., Wang J., Rao Z. Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71. Protein Cell. 2014;5:692–703. doi: 10.1007/s13238-014-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Principi N. Hand, foot and mouth disease: current knowledge on clinical manifestations, epidemiology, aetiology and prevention. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:391–398. doi: 10.1007/s10096-018-3206-x. [DOI] [PubMed] [Google Scholar]
- Francisco-Velilla R., Embarc-Buh A., Abellan S., Martinez-Salas E. Picornavirus translation strategies. FEBS Open Bio. 2022;12:1125–1141. doi: 10.1002/2211-5463.13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zeng S., Ji X., Meng X., Lei N., Yang H., Mu X. Type I interferon-induced TMEM106A blocks attachment of EV-A71 virus by interacting with the membrane protein SCARB2. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.817835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Ping L.-H, Rijnbrand R.C.A., Amphlett E., Clarke B., Rowlands D., Lemon S.M. Structural Requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdörfer B., Giese T., Endres S., Hartmann G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- Jiao X.-Y., Guo L., Huang D.-Y, Chang X.-L., Qiu Q.-C. Distribution of EV71 receptors SCARB2 and PSGL-1 in human tissues. Virus Res. 2014;190:40–52. doi: 10.1016/j.virusres.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Lai M.-C., Chen H.-H., Xu P., Wang R.Y.L. Translation control of enterovirus A71 gene expression. J. Biomed. Sci. 2020;27:22. doi: 10.1186/s12929-019-0607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Liu X., Ma Y., Sun Z., Yang Y., Jin Q., He B., Wang J. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J. Virol. 2010;84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Sun Z., Liu X., Jin Q., He B., Wang J. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by toll-like receptor 3. J. Virol. 2011;85:8811–8818. doi: 10.1128/JVI.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Zhou S., Guo Q., Chen X., Lai D., Lun Z., Guo X. The eIF3 complex of Trypanosoma brucei : composition conservation does not imply the conservation of structural assembly and subunits function. RNA. 2017;23:333–345. doi: 10.1261/rna.058651.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Huang L., Zhou J., Lin K., Wang H., Xue X., Xia C. Efficacy and safety of interferon-α2b spray in the treatment of hand, foot, and mouth disease: a multicenter, randomized, double-blind trial. Arch. Virol. 2016;161:3073–3080. doi: 10.1007/s00705-016-3012-7. [DOI] [PubMed] [Google Scholar]
- Lin J.-Y., Chen T.-C., Weng K.-F., Chang S.-C., Chen L.-L, Shih S.-R. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Huang H.I. Autophagy is induced and supports virus replication in enterovirus A71-infected human primary neuronal cells. Sci. Rep. 2020;10(1):15234. doi: 10.1038/s41598-020-71970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Yi L., Zhao J., Yu J., Chen Y., Lin M.C., Kung H.-F., He M.-L. Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J. Virol. 2012;86:3767–3776. doi: 10.1128/JVI.06687-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Embarek A.M. Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol. 2018;8:2629. doi: 10.3389/fmicb.2017.02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 2015;206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarry M., Vitour D., Zientara S., Bakkali Kassimi L., Blaise-Boisseau S. Foot-and-mouth disease virus: molecular interplays with IFN response and the importance of the model. Viruses. 2022;14:2129. doi: 10.3390/v14102129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S.K., Panda S., Sahu B.P., Sarangi R. Activation of host cellular signaling and mechanism of enterovirus 71 viral proteins associated with hand, foot and mouth disease. Viruses. 2022;14:2190. doi: 10.3390/v14102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Yam W.K., Kooi R.J.W., Westman J., Arbrandt G., Chu J.J.H. Novel capsid binder and PI4KIIIbeta inhibitors for EV-A71 replication inhibition. Sci. Rep. 2021;11(1):9719. doi: 10.1038/s41598-021-89271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.-F., Huang R.-T., Chien K.-Y., Huang J.-Y., Lau K.-S., Jheng J.-R., Chiu C.-H., Wu T.-Y., Chen C.-Y., Horng J.-T. Host MicroRNA miR-197 plays a negative regulatory role in the enterovirus 71 infectious cycle by targeting the RAN protein. J. Virol. 2016;90:1424–1438. doi: 10.1128/JVI.02143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser W.F., Verhoeven-Duif N.M., de Koning T.J. Identification of a human trans-3-hydroxy-l-proline dehydratase, the first characterized member of a novel family of proline racemase-like enzymes. J. Biol. Chem. 2012;287:21654–21662. doi: 10.1074/jbc.M112.363218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen J., Lu Z., Huang S., Zhang S., Cai J., Zhou Y., Cao G., Yu J., Qin Z., Zhao W., Zhang B., Zhu L. Enterovirus A71 utilizes host cell lipid β-oxidation to promote its replication. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.961942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., He Y., Chen Y., Kung H.-F., He M.-L. Potent inhibition of human enterovirus 71 replication by type I interferon subtypes. Antivir. Ther. 2011;16:51–58. doi: 10.3851/IMP1720. [DOI] [PubMed] [Google Scholar]
- Zeng S., Meng X., Huang Q., Lei N., Zeng L., Jiang X., Guo X. Spiramycin and azithromycin, safe for administration to children, exert antiviral activity against enterovirus A71 in vitro and in vivo. Int. J. Antimicrob. Agents. 2019;53:362–369. doi: 10.1016/j.ijantimicag.2018.12.009. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yang P., Wang N., Zhang J., Li J., Guo H., Yin X., Rao Z., Wang X., Zhang L. The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection. Protein Cell. 2017;8:590–600. doi: 10.1007/s13238-017-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang W., Wang X., Zhang X., Tian H., Deng H., Zhang L., Gao G. Identification of new type I interferon-stimulated genes and investigation of their involvement in IFN-β activation. Protein Cell. 2018;9:799–807. doi: 10.1007/s13238-018-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Chen X., Meng X., Zhang G., Wang J., Zhou D., Guo X. ``Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci. Rep. 2015;5:8976. doi: 10.1038/srep08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang X., Goff S.P., Gao G. Translational repression precedes and is required for ZAP-mediated mRNA decay: ZAP-mediated translational repression versus mRNA decay. EMBO J. 2012;31:4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.