Abstract
Food-borne illness caused by Salmonella enterica has been linked traditionally to poultry products but is associated increasingly with fresh fruits and vegetables. We have investigated the role of the production of autoinducer 2 (AI-2) in the ability of S. enterica serovar Thompson to colonize the chicken intestine and the cilantro phyllosphere. A mutant of S. enterica serovar Thompson that is defective in AI-2 production was constructed by insertional mutagenesis of luxS. The population size of the S. enterica serovar Thompson parental strain was significantly higher than that of its LuxS− mutant in the intestine, spleen, and droppings of chicks 12 days after their oral inoculation with the strains in a ratio of 1:1. In contrast, no significant difference in the population dynamics of the parental and LuxS− strain was observed after their inoculation singly or in mixtures onto cilantro plants. Digital image analysis revealed that 54% of S. enterica serovar Thompson cells were present in large aggregates on cilantro leaves but that the frequency distributions of the size of aggregates formed by the parental strain and the LuxS− mutant were not significantly different. Carbon utilization profiles indicated that the AI-2-producing strain utilized a variety of amino and organic acids more efficiently than its LuxS− mutant but that most sugars were utilized similarly in both strains. Thus, inherent differences in the nutrients available to S. enterica in the phyllosphere and in the chicken intestine may underlie the differential contribution of AI-2 synthesis to the fitness of S. enterica in these environments.
Quorum sensing is a process by which small molecules released by bacteria increase in concentration and provide signals to the bacterial cells about the density of their neighboring microbial population. This cell-cell communication controls bacterial behaviors such as virulence, conjugation, bioluminescence, sporulation, and biofilm formation and occurs in a wide range of species that include marine bacteria, epiphytic and plant-pathogenic bacteria, and human pathogenic bacteria. At least three different common signaling systems mediate quorum sensing in gram-negative bacteria: the acyl-homoserine lactone (AHL) signal-producing pathway, the autoinducer 2 (AI-2) pathway (28), and the AI-3 pathway, which has been demonstrated in Escherichia coli (24).
Salmonella enterica serovar Typhimurium does not produce any known AHL, although the expression of the sdiA virulence gene, a luxR homolog, has been suggested to be regulated by AHLs synthesized by other bacterial species (25). In contrast, production of the AI-2 signal in S. enterica serovar Typhimurium has been well characterized. It proceeds via pfs and luxS, is increased in culture under conditions of high glucose and osmotic stress, and regulates the lsr operon, which is involved in the transport of AI-2 into the cell (23, 27-29). Unlike several other species, in which production of AI-2 is involved in virulence, motility, or biofilm formation (28), the role of AI-2 signaling in S. enterica serovar Typhimurium has not been well defined. With the exception of a requirement of LuxS for the development of complete biofilms on gallstone surfaces (22), the significance of AI-2 synthesis to the ecology of S. enterica serovar Typhimurium remains to be elucidated.
S. enterica is prevalent in chickens, where it can reach high population densities in the intestinal tract, but also survives well in nonhost environments, such as soil and water (32). This high adaptability to nonhost habitats likely contributes to its contamination cycle in the environment. In the last decade, outbreaks of salmonellosis have been linked increasingly to fresh fruits and vegetables (1). Additionally, S. enterica contaminated ca. 3.5% of the domestic and imported fresh produce sampled in a recent survey by the U.S. Food and Drug Administration (http://www.cfsan.fda.gov). The observation that S. enterica was present on fresh produce prior to retail to consumers suggests that preharvest contamination with this pathogen is probable. We have shown previously that a strain of S. enterica serovar Thompson that was linked to an outbreak associated with cilantro reached significant population densities on cilantro plants under warm and moist conditions, recovered from dry conditions on plants to achieve high population sizes under subsequent humid conditions, and formed aggregates singly or with other species on leaf surfaces (6).
The objective of this study was to investigate the role of AI-2 production in S. enterica serovar Thompson in host and nonhost environments, namely, in chicken and on plant surfaces, respectively. More specifically, we tested the competitive ability of S. enterica serovar Thompson and its LuxS− mutant, which is defective in the synthesis of AI-2, to colonize the chicken intestine and the cilantro phyllosphere. These competitive population studies were complemented by carbon utilization profiling of the LuxS− mutant and its parental strain. In addition, we investigated the formation of aggregates by the parental and the AI-2-deficient strain on the cilantro leaf surface using epifluorescence microscopy combined with digital image analysis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Thompson strain RM1987 is a clinical isolate that was linked to an outbreak from cilantro and was described previously (6). S. enterica serovar Thompson is commonly isolated from chicken (see http://www.fsis.usda.gov/OPHS/haccp/sero1yr and http://www.cdc.gov/ncidod/dbmb/phlisdata/samtab/2000) and thus was a relevant strain to use in our comparative study. A spontaneous nalidixic acid-resistant mutant of S. enterica serovar Thompson strain RM1987 was isolated by streaking the wild-type strain onto a Luria-Bertani (LB) agar (Becton Dickinson, Sparks, MD) plate containing 50 μg/ml nalidixic acid. The nalidixic acid-resistant mutant was designated S. enterica serovar Thompson strain RM1987N. Strain RM1987N grew at the same rate as the wild-type strain in chicken and on cilantro plants (data not shown). All strains were grown at 28°C (for plant inoculations) or 37°C (for all other experiments) in Luria-Bertani liquid broth amended with nalidixic acid at 50 μg/ml and kanamycin at 50 μg/ml or gentamicin at 15 μg/ml, when appropriate.
Construction of LuxS− mutant.
All plasmids and strains used in this study are described in Table 1. To clone luxS, primers LuxS-up (5′-TTAGATAGCTTCGCAGTCGATCATA-3′) and LuxS-low (5′-TTATTGCTGTTCACGCGCACATCAC-3′), which were derived from the luxS gene sequence of S. enterica serovar Typhimurium LT2 (GenBank accession number AE008828) (14), were used to amplify an internal fragment of the luxS gene of S. enterica serovar Thompson RM1987 from total genomic DNA by PCR. The resulting 490-nucleotide fragment was cloned into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, CA) to generate pCRLuxST and sequenced to confirm its homology to the luxS gene of S. enterica serovar Typhimurium. The luxS internal clone was ligated into the EcoRI site of pUC18 to generate pUCLuxST. A kanamycin resistance cassette was then inserted into the luxS fragment on pUCLuxST by ligation of the HincII-restricted AphI-encoding fragment from pUC4K (30) into the unique EcoRV restriction site of the luxS subclone. The entire luxS-aphI fusion fragment was then restricted out from the latter plasmid with EcoRI and ligated into the unique EcoRI site of the broad-host-range and low-copy-number plasmid pLAFR3 (26) to generate pLAFLuxSTK. This plasmid was introduced into S. enterica serovar Thompson RM1987N by electroporation, and its luxS gene was interrupted by homologous recombination of the aphI insertional derivative of the luxS subclone into the chromosome to generate strain RM1987NLUX.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica serovar Thompson | ||
| RM1987 | Wild type | 6 |
| RM1987N | Spontaneous Nalr mutant of RM1987 | This study |
| RM1987NLUX | RM1987N luxS::aphI (LuxS− Kmr); NaIr | This study |
| V. harveyi | ||
| BB170 | luxN::Tn5 (AI-2 reporter strain) | 3 |
| Plasmids | ||
| pPROBE-GT | Promoter trap::gfp plasmid; Gentr | 18 |
| pGT-KAN | pPROBE-GT with Pkan-gfp; Gentr | This study |
| pCRLuxST | Truncated luxS in pCR2.1 | This study |
| pLAFLuxSTK | Truncated luxS::aphI in pLAFR3 | This study |
| pCRLuxST2 | luxS in pCR2.1 | This study |
| pBBR1MCS-5 | Cloning vector; Gentr | 12 |
| pBBRLuxST2 | luxS in pBBR1MCS-5 | This study |
Subsequently, the full luxS gene of S. enterica serovar Thompson RM1987 was cloned in order to carry out complementation studies. This was done by amplifying the entire LuxS-coding region by PCR using primers derived from the sequence flanking the luxS of S. enterica serovar Typhimurium: LuxS-F (5′-AGTTGGTCGCCTGCTTTG-3′) and LuxS-R (5′-TCGTGAGTTATCCGCCTTTT-3′). The resulting 1.2-kb PCR fragment was cloned into the TA cloning vector pCR2.1 to generate pCRLuxST2 and sequenced to confirm its identity.
To complement the luxS mutation in strain RM1987N, the fragment containing the S. enterica serovar Thompson luxS gene and its flanking regions was cleaved out of pCRLuxST2 with EcoRI and ligated into the unique EcoRI site of plasmid pBBR1MCS-5 (12) to generate pBBRLuxST2. In this plasmid, luxS is transcribed in the opposite orientation of lacZ transcription. Plasmids pBBR1MCS-5 and pBBRLuxST2 were introduced into strains RM1987N and RM1987NLUX by electroporation.
Construction of pGT-KAN.
To construct plasmid pGT-KAN, the Tn903 kanamycin resistance gene promoter first was amplified from the plasmid pUT mini-Tn5Km (9). The 131-bp amplified fragment, designed to contain a HindIII site at the 5′ end and a BamHI site at the 3′ end, was ligated into HindIII/BamHI-digested pPROBE-GT (18) to yield plasmid pGT-KAN. pPROBE-GT is a broad-host-range plasmid that confers gentamicin resistance; the gfp allele in this plasmid contains both the S65T and F64L mutations that increase green fluorescent protein (GFP) solubility (7, 10). GFP is constitutively expressed from the kanamycin resistance gene promoter in pGT-KAN. The presence of the kanamycin resistance gene promoter in pGT-KAN was confirmed by restriction analysis.
S. enterica serovar Thompson strain RM1987N and strain RM1987NLUX were labeled intrinsically with GFP by transformation with pGT-KAN. This plasmid was stably maintained over multiple generations in S. enterica serovar Thompson. The GFP-labeled strains were used for observation under the epifluorescence microscope and image analysis to study aggregate formation on the cilantro leaf surface. The introduction of plasmid-encoded GFP into S. enterica serovar Thompson did not affect its fitness on cilantro leaves relative to the wild-type strain (data not shown).
AI-2 assay.
Cell-free culture supernatants of S. enterica serovar Thompson RM1987N and its LuxS− mutant, strain RM1987NLUX, were obtained from early-, mid-, and late-log-phase cultures grown at 37°C in LB broth with 0.5% glucose and appropriate antibiotics. The supernatants were then assayed for AI-2 production by induction of light in Vibrio harveyi strain BB170 (3) as described previously by Surette and Bassler (27). Bioluminescence was measured with a Luminoskan Ascent luminometer (GMI Inc., Ramsey, MN). Light production induced in V. harveyi strain BB170 by the addition of bacterial culture supernatants was normalized for that induced by the addition of uninoculated sterile culture medium, which served as a control. Complementation of the LuxS− mutant with the full luxS clone was tested by assaying culture supernatants of strains RM1987N pBBR1MCS-5, RM1987NLUX pBBR1MCS-5, and RM1987NLUX pBBRLuxST2 for the presence of AI-2, as described above.
Chicken inoculation and bacterial enumeration.
Newly hatched female white leghorn chickens were obtained from an experimental farm at the University of California (Genetic Resources Conservation Program, Davis, CA). At 14 days of age, the chickens were inoculated orally with 1 ml of a suspension containing both the parental strain RM1987N and the LuxS− mutant strain RM1987NLUX, each at 5 × 106 cells ml−1 phosphate-buffered saline (PBS) (10 mM, pH 7.0). The chickens were held at 29°C and fed with Lab Chick Chow S-G feed (Purina Mills, Richmond, IN). Twelve days after inoculation, the chickens were euthanized with CO2 gas. Eight and 28 chickens were used in two independent replicate experiments.
The ratio of each strain in the inoculum was estimated immediately before inoculation by dilution plating individually six subsamples from the inoculum suspension onto growth agar as described below. For enumeration of S. enterica serovar Thompson strains in the chicken entera, the intestine (consisting of the duodenum, cecum, and cloaca) of each chicken was excised 12 days after inoculation. Each intestine was weighed, chopped, added to PBS at a proportion of 1 g per 10 ml, and homogenized in a Stomacher Lab Blender 400 (Seward Medical, London, England) at medium speed for 2 min. Three subsamples of each homogenate were diluted individually and serially in PBS and plated with an automated plater (Autoplate 4000; Spiral Biotech Inc., Norwood, MA) onto SS agar (Becton Dickinson, Sparks, MD) containing nalidixic acid for estimation of the total S. enterica serovar Thompson population size (parental and mutant strain) in the tissue. This was followed by replica plating of the SS agar plates onto LB agar containing kanamycin to assess LuxS− mutant strain population sizes; the parental strain population sizes were then estimated by the difference between the total S. enterica serovar Thompson population and that of the mutant strain. The counts from the three subsamples were pooled for each of the parental and mutant strains and used to calculate a single ratio for each chicken.
The chicken feces were sampled at regular time intervals at three separate random locations in the chicken cages. For enumeration of the S. enterica serovar Thompson parental and mutant population sizes in the feces, 1 g of each sample was homogenized in 10 ml PBS and dilution plated as described above.
Plant inoculation and incubation.
S. enterica serovar Thompson cells in stationary phase of growth were centrifuged and washed twice in potassium phosphate buffer (KP buffer) (10 mM, pH 7.0). The inoculum suspensions were prepared in 0.5 mM KP buffer at 1 × 104 cells ml−1 for inoculations with the parental or LuxS− mutant strain separately and 104 and 3 × 104 cells ml−1 of the parental and mutant strains, respectively, for coinoculations at a ratio of 1:3.
Cilantro plants (Coriandrum sativum cv. Leisure) grown to the four- to six-true-leaf stage were inoculated by immersion of the upper plant part in the bacterial suspension for 2 seconds. Six replicate pots of eight plants each were used per treatment. Immediately after inoculation, 18 leaves were removed at random from the eight pots for each treatment and assessed for initial S. enterica serovar Thompson population size as described below. The plants were then placed under fluorescent lights at 28°C in a randomized design in a humid chamber that allowed for the presence of visible free water on part of the cilantro leaf surface. For subsequent exposure of the bacteria to water stress on plants, the plants were then incubated in a growth chamber at 50% relative humidity at 28°C for 24 h to trigger low water availability on the leaf surface. The plants were then returned to the humid chamber to allow the bacteria to recover from water stress under conditions where free water was present on the leaves. Each experiment was repeated twice independently.
To determine the effect of AI-2 production on aggregate formation, the upper part of cilantro plants was inoculated with either the GFP-labeled parental strain (RM1987N pGT-KAN) or the LuxS- mutant strain (RM1987NLux pGT-KAN) at 105 cells ml−1 in 0.5 mM KP buffer, as described above. Five replicate pots of eight plants each were used per treatment (strain). The plants were placed in a randomized design at 28°C in a humid chamber under conditions described above. The plants were incubated for 3 days, with a 12-h period per day of illumination under fluorescent lights, before the leaves were sampled at random from each treatment for microscopy and image analysis.
Bacterial recovery from plants and enumeration.
For estimation of bacterial population sizes on cilantro leaves, 18 leaves were removed at each sampling time from the cilantro plants at random from the replicate pots and placed in 10 ml of KP buffer. The leaves were then sonicated in a sonicator bath for 75 s and vortexed vigorously for 30 s to dislodge the bacterial cells from the leaf surface and to break up bacterial aggregates. Homogenization of the leaves in buffer prior to plating did not result in a significantly higher number of cells recovered from the cilantro leaves than by the above-described method. The suspensions obtained from the leaf washings were dilution plated with an automated plater (Autoplate 4000; Spiral Biotech Inc., Norwood, MA) onto LB agar containing nalidixic acid. Enumeration of S. enterica serovar Thompson colonies from leaves inoculated with a single strain was performed by automated counts (QCount; Spiral Biotech Inc., Norwood, MA). Enumeration of the parental and LuxS− mutant strains on coinoculated leaves was done manually as described above for enumeration of these strains from chicken tissue samples, with the exception that the suspensions obtained from the leaf washings were plated onto LB agar containing nalidixic acid. Each leaf was weighed to allow for the normalization of bacterial population size to the weight of leaf tissue.
Epifluorescence microscopy.
For each strain, four first true leaves were sampled at random from five replicate pots containing cilantro plants that were inoculated with the parental strain RM1987N pGT-KAN or the LuxS− strain RM1987Nlux pGT-KAN. Three leaf disks that were 10 mm in diameter were cut out of each leaf. Each disk was mounted onto a microscope slide with Aqua Poly/Mount (Polysciences, Warrington, PA). The GFP-labeled S. enterica serovar Thompson cells were visualized on the leaf surface under a Leica DMRB microscope fitted with a 20×/0.5 HC PL FLUOTAR objective and GFP filter set 513847 (Leica Microsystems, Wetzlar, Germany). The fluorescence images were captured with a Hamamatsu (Bridgewater, NJ) Orca C4742-95 cooled charge-coupled-device camera and acquired with the software Openlab, version 2.1 (Improvision, Lexington, MA). Ten different random fields of view were imaged per leaf disk.
GFP was a suitable marker for viable S. enterica serovar Thompson cells in our study, since we observed that after application of various stresses in the laboratory, only a minor fraction of S. enterica serovar Thompson cells had both green and red fluorescence following staining with the viability stain propidium iodide; all other cells exhibited distinct green or red fluorescence, suggesting that most S. enterica serovar Thompson cells retaining GFP were viable (data not shown). Lowder et al. (13) reported similar observations for GFP-labeled Pseudomonas fluorescens.
Digital image analysis.
The frequency distribution of the size of aggregates formed by the parental strain or the LuxS− mutant on cilantro leaves was obtained by digital image analysis using the software IPLab Spectrum, version 3.5.5, for Macintosh (Scanalytics, Fairfax, VA). The size of each S. enterica serovar Thompson aggregate was estimated as follows. Each GFP-S. enterica serovar Thompson single cell or aggregate in the image was identified by thresholding on the bright pixels originating from the GFP-labeled cells, which were of higher intensity than the background pixels originating from the leaf surface. This thresholding (segmentation) yielded objects (segments) that consisted of a single or a group of GFP-S. enterica serovar Thompson cells. After manual deletion of all segments in the image that originated from bright fluorescent debris on the leaf surface or out-of-focus GFP cells, the area of each segment (single cell or aggregate) was measured automatically by the software as the sum of the individual pixels that formed the segment. The size of each aggregate was then estimated as the total number of GFP-labeled cells by dividing the area of each segment by the average number of pixels forming one bacterial cell (six pixels). This procedure may have underestimated the number of cells in multidimensional large aggregates because of its assumption that most aggregates would be formed of a single layer of cells. The measurements were pooled for all three disks for each leaf. The distribution of the aggregate sizes on all four leaves was then analyzed statistically for each bacterial strain.
Carbon source utilization.
The S. enterica serovar Thompson parental, mutant, and complemented strains were assayed for respiration induced by carbon substrates in BIOLOG plates (Biolog Inc., Hayward, CA). The S. enterica serovar Thompson strains were grown to mid-log phase of growth (optical density at 600 nm [OD600], 1.0) in LB broth with 0.5% glucose at 28°C. Production of AI-2 by S. enterica serovar Thompson parental cells was determined to be the highest at that growth phase; LuxS− mutant cells did not produce any detectable level of AI-2 at any growth phase of the culture (data not shown). The S. enterica serovar Thompson cells were centrifuged and washed in KP buffer twice and resuspended in Biolog GN/GP inoculation fluid to an absorbance of 0.65, as measured by densitometry (Biolog absorbance meter), and the suspension was added to GN2 Microplates (Biolog Inc., Hayward, CA) containing substrates. The microtiter plates were incubated for 48 h at 37°C. Three independent replicate assays were performed for each strain. The color change resulting from the reduction of tetrazolium violet by metabolic respiration was measured by optical density in a Biolog spectrophotometer according to the manufacturer's instructions (Biolog Inc., Hayward, CA).
The same S. enterica serovar Thompson strains as those used in the Biolog assay were tested for their growth rate in minimal medium with single carbon sources. The cells were grown in 1× M9 minimal medium (Sigma-Aldrich, Inc.) with 10 mM d-glucose and 15 μg/ml gentamicin, washed twice in phosphate buffer (5 mM, pH 7.0), and inoculated at an initial concentration of 1 × 106 cells ml−1 into 1× M9 minimal medium containing a single carbon source. The concentration of glucose was 5, 10, or 25 mM, and that of the carbon sources listed in Table 2 was 10 mM. The cultures were incubated in two replicate tubes at 37°C and 250 rpm, and their OD600 was recorded at regular time intervals.
TABLE 2.
Carbon compounds utilized differentially by the S. enterica serovar Thompson parental and LuxS− strains in the Biolog assay and complementation with the luxS clone
| Carbon compound | Mutant/parentala | Complemented/parentala |
|---|---|---|
| M-inositol | 0.22 | 1.42 |
| Mono-methylsuccinate | 0.06 | 0.72 |
| d-Glucuronic acid | 0.19 | 0.65 |
| p-Hydroxyphenylacetic acid | 0.55 | 0.75 |
| Succinic acid | 0.06 | 0.91 |
| l-Alanine | 0.33 | 0.67 |
| l-Asparagine | 0.40 | 0.90 |
| l-Glutamic acid | 0.12 | 0.48 |
| l-Serine | 0.38 | 0.76 |
| Glycyl-l-aspartic acid | 0.53 | 1.04 |
In the Biolog assay, the color change resulting from the reduction of tetrazolium violet by bacterial respiration was measured by optical density. For each compound, the data are presented as the proportion of the optical density obtained with the mutant, or complemented mutant, to that obtained with the parental strain. Mutant strain, RM1987NLUX pBBR1MCS-5; parental strain, RM1987N pBBR1MCS-5; complemented strain, RM1987NLUX pBBRLuxST2.
Statistical methods.
Statistical calculations were performed with the program Prism, version 3.0 (GraphPad Software, Inc., San Diego, CA). Bacterial population sizes and population ratios were log transformed in order to obtain Gaussian distributions. All linear regression analyses were performed on the data from all replicate samples, rather than on the mean value, and the runs test was performed to verify that there was no significant departure from linearity.
Nucleotide sequence accession number.
The sequence of the luxS gene of S. enterica serovar Thompson strain RM1987 was deposited in the GenBank database under the accession number AY496970.
RESULTS
Construction and characterization of LuxS− mutant.
The luxS gene of S. enterica serovar Thompson strain RM1987 had 99% identity to that of S. enterica serovar Typhimurium strain LT2 at the deduced amino acid level (data not shown). The luxS gene of strain RM1987 was disrupted by marker exchange mutagenesis, resulting in the insertion of the kanamycin resistance gene aphI in the middle of the luxS open reading frame. Insertion of the kanamycin resistance cassette into luxS was confirmed by PCR amplification of the target region, as well as by Southern analysis, which revealed a single insertion of the cassette in the S. enterica serovar Thompson genome. No significant difference in the growth of the parental and mutant strains in LB broth at 28, 37, and 42°C and in M9 minimal medium with 0.5% glucose at 28 and 37°C was detected. The parental and mutant strains grew equally poorly in M9 minimal medium with 0.5% glucose at 42°C.
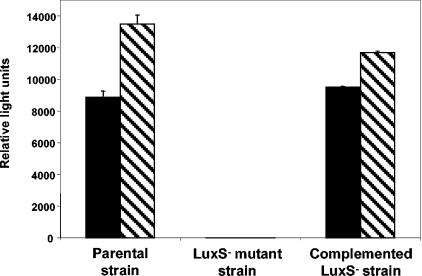
The synthesis of AI-2 in S. enterica serovar Thompson strains was assessed by induction of bioluminescence in the Vibrio harveyi system 2 reporter strain BB170. As reported for S. enterica serovar Typhimurium (27), production of AI-2 in the S. enterica serovar Thompson parental strain increased during the mid- to late log phase of growth in culture and decreased after entry into stationary phase (data not shown), and production of AI-2 was higher in LB broth containing 0.5% glucose than in LB (Fig. 1). The parental strain produced high levels of AI-2 in LB with 0.5% glucose at 28, 37 and 42°C and in M9 minimal medium with 0.5% glucose at 28 and 37°C. The culture supernatant of the LuxS− mutant strain failed to induce luminescence in the Vibrio AI-2 reporter strain, but AI-2 production in the mutant was recovered by complementation with the full luxS clone and its flanking regions harbored on plasmid pBBRLuxST2 (Fig. 1).
FIG. 1.
Induction of bioluminescence in V. harveyi AI-2 reporter strain BB170 by culture supernatants of S. enterica serovar Thompson RM1987 parental, LuxS− mutant, and complemented LuxS− mutant strains. Culture supernatants were obtained from late-log-phase cultures in LB broth (solid bars) and LB broth with 0.5% glucose (hatched bars) at 37°C. Parental strain, RM1987N pBBR1MCS-5; mutant strain, RM1987NLUX pBBR1MCS-5; complemented mutant strain, RM1987NLUX pBBRLuxST2. Values represent the means and standard errors of the means for two replicate cultures.
Growth and survival in chicken.
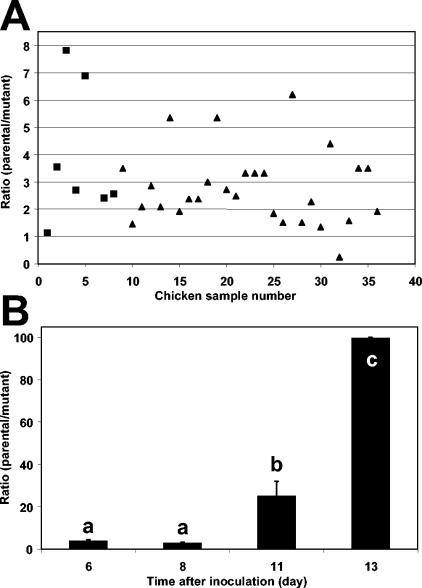
The role of AI-2 production in the competitive fitness of S. enterica serovar Thompson in the chicken intestine was tested by oral coinoculation of the parental strain and the LuxS− mutant in a ratio of 1:1. After 12 days of incubation in chickens, the total S. enterica serovar Thompson population size (parental and mutant strains) in the chicken intestine reached on average 1 × 107.6 CFU g−1 tissue (data not shown), corresponding to an approximate cell density increase of at least 100-fold after inoculation. The estimated ratio of the parental strain population size to that of the LuxS− strain in the intestine of 36 chickens in total from two independent replicate experiments is presented in Fig. 2A. The value of the initial ratios of the concentration of the parental strain to that of the mutant strain in the inoculum were 1.30 and 1.09 for replicate experiments 1 and 2, respectively. In 72.2% of the chickens, the value of the estimated ratio was equal to or higher than 2.0, with one chicken showing a ratio as high as 50.0 (data not shown); the value of the estimated ratio fell below 1.0 in one chicken only (Fig. 2A). Statistical analysis of the log-transformed value of the ratios revealed that the proportion of the parental strain cells to that of the LuxS− mutant cells was significantly higher in the chicken intestines than in the inoculum mixture in both replicate experiments (paired t test; t = 3.08, P < 0.05, and t = 6.85, P < 0.0001, experiments 1 and 2, respectively). Regression analysis of the log-transformed ratio over the log-transformed S. enterica serovar Thompson parental strain population size and over the log-transformed population size of the natural aerobic culturable bacteria, in all chicken intestine samples, yielded r2 values of 0.019 and 0.0023, respectively. Also, the slopes for these regressions were not significantly different from zero (F = 0.49, P = 0.49; F = 0.06, P = 0.81, respectively). Thus, there was a lack of correlation between the value of the population ratio and (i) the population size that the AI-2-producing strain reached in the intestine and (ii) the population size of the natural intestinal culturable bacteria.
FIG. 2.
Contribution of AI-2 production to the fitness of S. enterica serovar Thompson in chicken. (A) Plot of the ratio of the population size of the S. enterica serovar Thompson parental strain RM1987N to that of its LuxS− mutant strain in the intestine of 36 chickens 12 days after oral inoculation with a ratio of 1:1 of each strain. The total data from two replicate experiments are presented (experiment 1, ▪; experiment 2, ▴). (B) Change in the ratio of the population size of the S. enterica serovar Thompson parental strain to that of the LuxS− mutant over time in the droppings of chickens inoculated with a ratio of 1:1 of each strain. Values marked with the same letter were not significantly different, as determined with the Tukey-Kramer multiple-comparisons test (P < 0.05).
Invasion of the spleen by S. enterica serovar Thompson was detected in 28% of the inoculated chickens. The ratio of the parental strain to LuxS- strain population sizes in the spleen diverged significantly from the inoculum ratio (paired t test on log-transformed ratio values; t = 2.30, P < 0.05). The population size of the parental strain was also significantly higher than that of the LuxS− mutant in the chicken droppings (Fig. 2B). The ratio of the parental population size to that of the mutant increased over time after inoculation to reach an average value of 100, 12 days after inoculation. The change in population ratio in the chicken feces was considerably larger than in the chicken intestine (Fig. 2A and B). The average total S. enterica serovar Thompson cell density in the feces was 107 CFU g−1 (wet weight).
Growth and survival on plants.
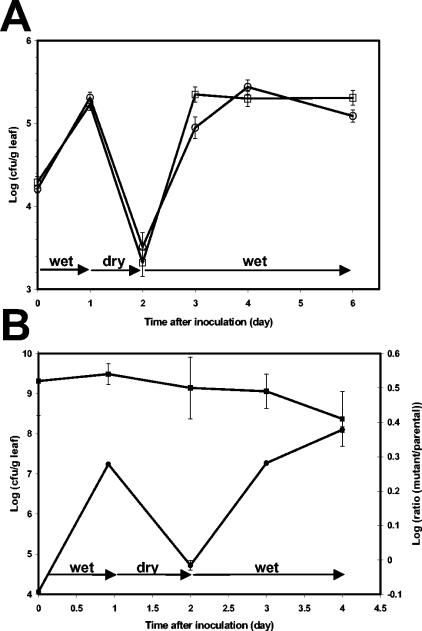
The role of AI-2 production in the ability of S. enterica serovar Thompson to colonize plant surfaces was investigated by inoculation of the parental and mutant strains either singly or in mixtures onto cilantro plants. The plants were incubated under successively wet, dry, and wet conditions to simulate the alternate conditions of free water and dryness on the plant surface under natural conditions. When inoculated singly onto separate cilantro plants, the parental strain and the LuxS− mutant achieved and maintained similar population sizes on wet leaves, survived dry conditions on leaves at comparable rates, and recovered similarly from drought stress under subsequent wet conditions (Fig. 3A). The small differences between the two strains during the first growth phase after inoculation and during growth after drought stress were not consistent across multiple replicate experiments.
FIG. 3.
Population dynamics of the S. enterica serovar Thompson parental strain and its LuxS− mutant over time after inoculation onto the leaves of cilantro plants. The plants were incubated under a regimen of wet-dry-wet conditions. (A) Population sizes of the parental strain (□) and the LuxS− mutant (○) on cilantro leaves when inoculated singly onto separate plants. (B) Dynamics of the total S. enterica serovar Thompson population size (parental and mutant strains) (•) and change in ratio (▪) of the LuxS− mutant strain population size to that of the parental strain on cilantro leaves after their coinoculation in a proportion of 3:1, respectively. Values represent the means and standard errors of the means of the log-transformed population size or ratio for 18 leaves.
In order to examine the competitive ability of the parental and mutant strains in the same microhabitat, the strains were also coinoculated onto cilantro plants. Because of plant-to-plant, and even leaf-to-leaf variations, coinoculation generates more accurate comparative fitness data than when the strains are inoculated singly onto different plants. This approach has been used previously to reveal a fitness trait in Erwinia herbicola on plant surfaces (5). The use of a ratio of 3:1 of the mutant strain to the parental strain in the inoculum mixture aimed to minimize cross-feeding of the AI-2-deficient mutant cells via production of AI-2 by the parental strain cells in their vicinity on the leaf surface. Figure 3B presents the dynamics of the total S. enterica serovar Thompson population size, comprised of the parental and the mutant strains, as well as the change in ratio of the LuxS− mutant population size to that of the parental strain after inoculation onto cilantro plants that were incubated under a wet-dry-wet regimen. The change in ratio of the two strains was analyzed by linear regression analysis of the log-transformed ratio of the mutant strain to the parental strain population sizes for all leaf samples over time after the start of water stress conditions on the leaves (1 day after inoculation). The slope of the linear regression was not significantly different from zero (F = 1.94, P = 0.17), indicating that there was no significant difference between the population dynamics of the parental and mutant strains when they were competing against each other in the cilantro phyllosphere. Similar results were obtained in experiments with an inoculum ratio of 1:1 of each strain after inoculation onto cilantro plants.
No difference in fitness was observed when the strains were exposed to periods of dryness on plants longer than 24 h or when dry conditions were applied after a longer period of wet conditions during which S. enterica serovar Thompson may have reached higher cell densities (data not shown).
Comparison of aggregate formation by parental and LuxS− mutant strains.
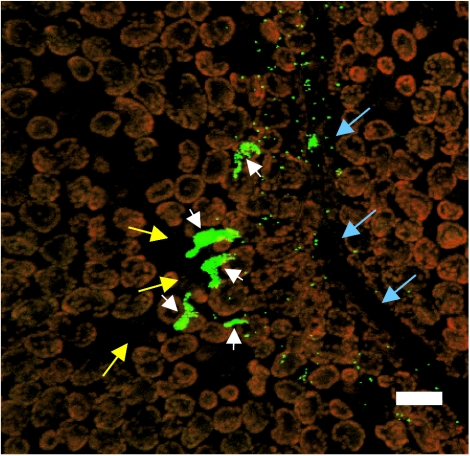
To examine more closely whether there was any difference in growth on plant surfaces between the parental and the LuxS− mutant strain, the distribution of the size of aggregates formed on cilantro leaves by the parental strain was compared to that of the LuxS− mutant using epifluorescence microscopy and image analysis. Figure 4 presents an epifluorescence micrograph of large aggregates of GFP-labeled S. enterica serovar Thompson cells present in the vicinity of stomata and near a vein area of the leaf 3 days after inoculation onto cilantro plants. Such aggregates are composed of at least 100 S. enterica serovar Thompson cells. Due to the low inoculum concentration used in this study, only individual S. enterica serovar Thompson cells scattered at distant locations on the leaf surface were observed at the time of inoculation.
FIG. 4.
Micrograph of GFP-labeled S. enterica serovar Thompson RM1987N cells on the leaf surface of cilantro plants incubated under humid conditions. Large cell aggregates (white arrowheads) are apparent in the vicinity of stomates (yellow arrows) near a leaf vein (blue arrows). Single GFP-S. enterica serovar Thompson cells are also present. The red fluorescent objects are the autofluorescent chloroplasts of the plant epidermal cells. The image is a projected z series obtained with a Leica TCS-NT confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany). Bar, 20 μm.
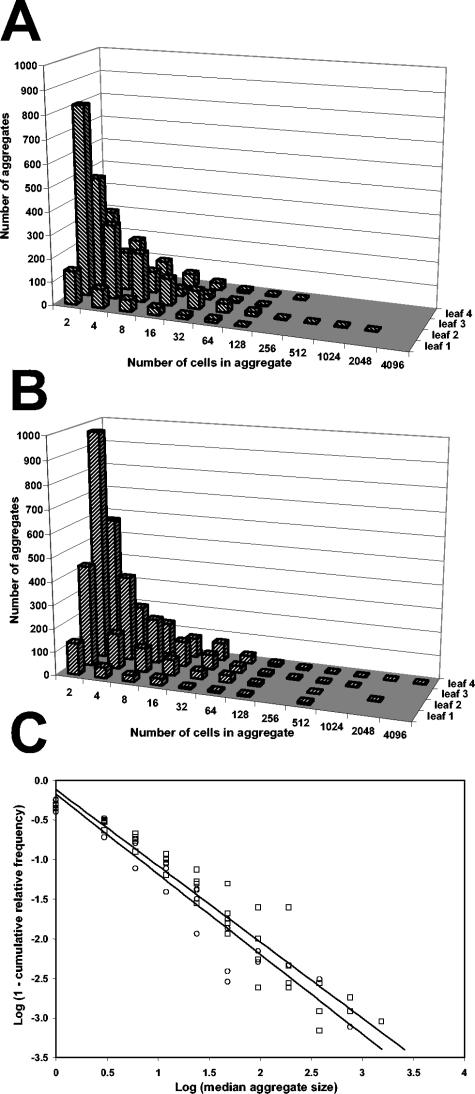
A total of 120 epifluorescence images of S. enterica serovar Thompson cells on the cilantro leaf surface were analyzed for each the parental strain and the LuxS− strain. Both the parental and the mutant strains formed large aggregates on cilantro leaves that varied widely in size (Fig. 5A and B). These large aggregates were observed independently of the average population size achieved by S. enterica serovar Thompson on the leaf surface, since replicate leaves with apparent low population sizes harbored aggregates comparable in size to those on replicate leaves with higher population sizes (Fig. 5A and B). Cumulative frequency data obtained from pooled aggregate sizes for all four replicate leaves revealed that ca. 54% of the parental or mutant cells were located in aggregates composed of more than 128 cells. The mean of the log-transformed population size for all four replicate leaves, when calculated from the frequency of aggregate size data, was not significantly different between the parental and the mutant strains 3 days after inoculation and incubation under wet conditions (t = 1.15, P = 0.294).
FIG. 5.
Frequency distribution of the size of aggregates formed by the GFP-labeled S. enterica serovar Thompson parental strain (A) and AI-2-deficient strain (B) 3 days after inoculation onto the leaves of cilantro plants incubated under humid conditions. The frequency data represent the number of aggregates of a given size that were observed on a 16-mm2 surface of each of four replicate leaves. Aggregate size is presented as the number of S. enterica serovar Thompson cells per aggregate. (C) Regression analysis of the log-transformed value (1 − cumulative relative frequency of aggregate size) over the log-transformed median aggregate size showing that the frequency distribution of aggregate size did not differ significantly between the S. enterica serovar Thompson parental strain (○) and the LuxS− strain (□) (F = 0.39, P = 0.53).
In order to compare the size of aggregates formed on leaves by the LuxS− mutant strain to those formed by its parental strain, regression analysis was performed on the frequency distribution of aggregate sizes observed for each strain. The frequency distributions for all replicate leaves and for each strain were similarly characterized by heavy tails typical of Pareto distributions (11) (Fig. 5A and B). These heavy tails were generated by the low occurrence of very large aggregates of a given size. In order to perform regression analysis, the median of each aggregate size range was log transformed and the frequency of aggregate size was transformed into the log value (1 − cumulative relative frequency) (11). Figure 5C shows that regression analysis of the transformed frequency over the transformed aggregate size yielded a linear relationship with a slope significantly different from zero for both the parental strain and the mutant strain (F = 310.2, P < 0.0001, and F = 406.1, P < 0.0001, respectively). The distribution function of aggregate size was described as follows for the parental and the LuxS− strains, respectively: y = −0.18 − 1.00x and y = −0.12 − 0.96x (r2 = 0.91 and 0.92). The differences between the slopes obtained for each strain were not significant (F = 0.39, P = 0.53), indicating that the frequency distributions of the size of aggregates formed by the AI-2-producing parental strain and its AI-2-deficient mutant on leaves were not significantly different.
Carbon substrate utilization.
The carbon utilization profile of the parental, LuxS− mutant, and complemented mutant strains was determined by testing carbon compounds present on GN2 Biolog plates. While many carbon sources were utilized in all three strains, differences between the strains in their response to several substrates were observed. Table 2 provides a list of carbon compounds for which utilization in the LuxS− mutant strain was less than 60% of that in the parental strain and was restored at least partially in the complemented mutant. Plasmid pBBR1MCS-5 was introduced into the mutant strain and the parental strain to normalize for any difference in carbon source utilization between these strains and the complemented strain due to the presence of the plasmid alone. The compounds for which the largest difference in utilization was observed were amino acids or their derivatives (l-alanine, l-asparagine, l-glutamic acid, l-serine, and glycyl-l-aspartic acid), organic acids (d-glucuronic acid, succinic acid, and p-hydroxyphenylacetic acid), and M-inositol (Table 2). Other carbon sources, such as l-alanyl-glycine, methyl pyruvic acid, acetic acid, l-gluconic acid, d,l-lactic acid, l-rhamnose, glycerol, and d,l-α-glycerolphosphate, were also utilized consistently less efficiently in the LuxS− mutant and complemented for in the mutant harboring pBBRLuxST2, but their utilization rate was higher than 60% of that in the parental strain (data not shown). All compounds listed above are those for which we observed a difference in utilization between the mutant and parental strains, as well as complementation of the mutant, in at least two out of three replicate experiments.
With the exception of M-inositol, all carbohydrates present on the Biolog plates were utilized at similar rates in the parental strain and the mutant strain. Similar observations were made when the Biolog plates were incubated for 48 h at 28°C, which reflects the temperature at which the plant experiments were conducted.
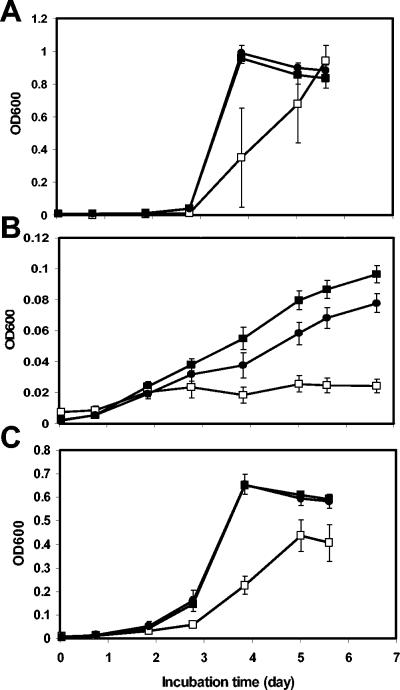
The carbon sources listed in Table 2 were tested for their ability to support growth of the parental, LuxS− mutant, and complemented S. enterica serovar Thompson strains in M9 minimal medium. All three strains had identical growth rates at 37°C when 5, 10, or 25 mM d-glucose was the sole carbon source in the culture; identical growth rates were also observed in M9 medium with 25 mM d-glucose at 28°C. In contrast, the AI-2-deficient mutant had a significantly lower growth rate than the parental strain and was complemented for this deficiency by transformation with pBBRLuxST2 in M9 minimal medium containing 10 mM succinic acid, l-serine, or l-alanine as the sole carbon source (Fig. 6). The remaining carbon sources listed in Table 2 also supported lower growth rates in the LuxS− mutant than in the parental strain in multiple replicate experiments.
FIG. 6.
Growth rate of the S. enterica serovar Thompson parental (▪), LuxS− (□), and complemented mutant (•) strains in M9 minimal medium containing succinic acid (A), l-serine (B), or l-alanine (C) as the sole carbon source. Each value represents the mean OD600 and standard error of the mean for two replicate cultures.
DISCUSSION
Although production of AI-2 has been well characterized in S. enterica serovar Typhimurium (28), the function of the AI-2 synthetic pathway in this strain has remained unclear. Since S. enterica is highly adaptable and capable of growth and survival in a wide variety of habitats (6, 32), we have investigated the role of AI-2 production in S. enterica in a host and a nonhost environment. S. enterica is highly prevalent in chicken, where it can grow to high densities in the gastrointestinal tract. Our comparative population studies revealed clearly that the AI-2-producing parental strain of S. enterica serovar Thompson overall outcompeted the AI-2-deficient mutant strain in the chicken intestine, indicating that AI-2 production conferred on S. enterica serovar Thompson an advantage in the enteric colonization of this host. This was further supported by our observation that S. enterica serovar Thompson invaded the chicken spleen to a significantly higher extent than its AI-2-deficient mutant. The results of our coinoculation experiments in chicken are similar to those reported by Winzer et al. (34), who demonstrated the higher competitiveness of a Neisseria meningitidis wild-type strain compared to its LuxS− mutant in the bloodstream of infant rats following infection with the strains at a 1:1 ratio.
The parental strain also survived at increasingly higher rates than the LuxS− mutant over time in the chicken feces. It is noteworthy that the change in their population size ratio was greater in the feces than in the intestine. This difference may be due to stronger selective pressure in the feces, such as low-moisture conditions. The excretion of high concentrations of S. enterica serovar Thompson observed over a period of 12 days after inoculation is typical of Salmonella-infected young chickens (2). The reinoculation of the chickens due to ingestion of contaminated feces that contained even higher ratios of the parental strain to the mutant strain than the intestine may have contributed to the chicken-to-chicken variability in intestinal population ratios observed in our experiments. The infection of chicks through contaminated feces or inert surfaces of poultry houses, on which Salmonella can survive for prolonged periods of time (8), is a major factor in the spread of Salmonella among young chicken populations. Thus, the high survival of the S. enterica serovar Thompson parental strain compared to its AI-2-deficient mutant in chicken feces indicates that production of AI-2 may be important in the passage of Salmonella from host to host.
In contrast to our observation of AI-2 activity in chickens, the ability to synthesize AI-2 conferred no apparent advantage on S. enterica serovar Thompson in its colonization of cilantro leaf surfaces. Our comparative population studies show that the AI-2-deficient strain was as fit as the parental strain under a variety of conditions in the cilantro phyllosphere. It is possible that some cross-feeding of the mutant occurred despite a 3:1 ratio of mutant to parental strains when both were coinoculated onto leaves, thereby obscuring any small difference in fitness between the two strains. However, the following observations do not support a significant role for AI-2 production in the growth or survival of S. enterica serovar Thompson in the phyllosphere: (i) the great similarity in the behavior of the LuxS− mutant and the parental strain when they were inoculated alone onto plants, (ii) their similar population sizes on leaves based on microscopy and image analysis at the single-cell level, and (iii) the significantly greater fitness of the parental strain than that of the mutant in the chicken, despite a lower inoculation ratio of mutant strain to parental strain (1:1), with which cross-feeding would be more likely to occur than with the 3:1 ratio used on plants.
Although the natural plant bacterial flora may have influenced the population sizes of S. enterica serovar Thompson on cilantro by releasing AI-2, experimental evidence in our laboratory suggests that cross-feeding of AI-2 by plant epiphytes to the parental strain or the LuxS− mutant strain does not influence the dynamics of these strains in the cilantro phyllosphere. More precisely, coinoculation with AI-2-producing Pantoea agglomerans or Erwinia chrysanthemi, two common colonizers of plants, did not have any effect on the population sizes that the parental or AI-2-deficient S. enterica serovar Thompson strain reached and maintained on cilantro leaves (M. Brandl, unpublished data). Thus, our observations indicate that the synthesis of AI-2 by S. enterica serovar Thompson or by its cohabitants on leaf surfaces does not measurably affect its fitness in that environment.
Quorum sensing via AI-2 is involved in biofilm formation in Streptococcus mutans (17) and Streptococcus gordonii (15). Prouty et al. (22) reported that mutation of luxS in S. enterica serovar Typhi prevented its development of complete biofilms on gallstones. In our study, regression analysis failed to reveal any significant difference in the frequency distribution of aggregate sizes on leaves between the parental strain and the AI-2-deficient mutant. This suggests that under the conditions of our experiments, AI-2 production in S. enterica serovar Thompson did not play a major role in its formation and development of aggregates in the phyllosphere.
On the other hand, our image analysis data revealed that as much as 70% of the S. enterica serovar Thompson cell population on leaves of plants incubated under humid conditions is located in aggregates composed of more than 128 cells and that aggregates as large as 4,096 cells can occur. Previous studies demonstrated that the majority of Pseudomonas syringae cells that colonized bean leaves after plant inoculation in the laboratory was accounted for in large aggregates (20) and that a higher proportion of the leaf viable bacterial cell population after desiccation stress was present in aggregates than as single cells (19). Thus, it is noteworthy that a human pathogenic bacterial species, such as S. enterica, follows similar colonization patterns as a plant-associated bacterial species. The localization of the majority of the S. enterica cell population in large aggregates on plants under natural conditions in the field would have important implications not only for the ability of S. enterica to survive stressful conditions in the plant environment but also for the accurate assessment of S. enterica contamination levels on produce and its efficient sanitization. In addition, this observation raises the possibility that S. enterica cells within aggregates on plant surfaces have a particular physiology that affects their virulence or pathogenicity.
The major difference observed in the contribution of AI-2 synthesis to the fitness of S. enterica serovar Thompson in chickens compared to that on plants may be related to differences in the chemical environment in the chicken intestine and in the phyllosphere. Indeed, examination of carbon utilization profiles revealed that the parental strain utilized amino acids and their derivatives, as well as organic acids, more efficiently than the AI-2-deficient strain but that most carbohydrates were utilized at a similar rate in both strains. This observation was supported by our growth rate studies in minimal medium with single carbon sources. Complementation of the mutant with a functional luxS in the carbon utilization studies as well as in the growth rate experiments suggests that full metabolic potential in S. enterica serovar Thompson is dependent on a functional luxS and implicitly on AI-2 production. Although a wide variety of amino acids and organic acids are present on plant surfaces, albeit in small amounts (21), the population size of epiphytic bacteria appears to be limited by the amount of sugars, such as glucose, fructose, and sucrose, present on leaves, rather than by the amounts of amino and organic acids (16, 31). Assuming that S. enterica behaves similarly on plants, it could be predicted that, based on our carbon utilization and growth rate data, AI-2 production would be of little benefit to the growth of S. enterica in the phyllosphere. In contrast, amino and organic acids, which are likely to be present in high amounts in the chicken intestine due to the breakdown of proteins from food, may be an important nutrient source for S. enterica, in addition to simple carbohydrates. Thus, the parental strain of S. enterica serovar Thompson may have benefited from higher utilization rates of these compounds and competitively excluded its isogenic AI-2-deficient strain in the chicken intestine.
The lack of a correlation between the ratio of the parental strain to the mutant strain and the population size of the AI-2-producing parental strain as well as that of the natural intestinal bacterial flora over all chicken intestine samples indicates that the benefit of AI-2 production in S. enterica serovar Thompson may not be related directly to quorum sensing. The above-mentioned observations and the results of our comparative studies on the carbon utilization profiles of the parental and LuxS− mutant strains support the hypotheses that the synthesis of AI-2 in S. enterica serovar Typhimurium is linked to the metabolic state of the cells rather than cell density (4) and that LuxS is involved in bacterial metabolism (33).
Our studies provide evidence that AI-2 production has a niche-specific benefit in S. enterica. Because of the high prevalence of S. enterica in poultry production, and the role of chicken feces in its contamination cycle, the results of our studies open new opportunities for the control of this human pathogen by disruption of the AI-2 biosynthetic pathway.
Acknowledgments
We thank Steven Huyhn, Aileen Haxo, and Feli Bautista for excellent technical assistance. We are grateful to Peter Piet for helpful discussion about statistical analysis. We thank Bonnie Bassler for the gift of V. harveyi strain BB170 and Sharon Abbot for the gift of S. enterica serovar Thompson.
This work was funded by the U.S. Department of Agriculture, Agriculture Research Service CRIS projects 201-5325-210-40 and 201-5325-210-41.
REFERENCES
- 1.Anonymous. 2002. Outbreak alert: closing the gaps in our federal food safety net. Report from the Center for Science in the Public Interest. Center for Science in the Public Interest, Washington, D.C.
- 2.Barrow, P. A., J. M. Simpson, and M. A. Lovell. 1988. Intestinal colonization in the chicken of food-poisoning Salmonella serotypes: microbial characteristics associated with fecal excretion. Avian Pathol. 17:571-588. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 4.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandl, M. T., and S. E. Lindow. 1998. Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Appl. Environ. Microbiol. 64:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandl, M. T., and R. E. Mandrell. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 8.Davies, R. H., and C. Wray. 1995. Observations on disinfection regimens used on Salmonella enteritidis infected poultry units. Poultry Sci. 74:638-647. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heim, R., A. B. Cubitt, and R. Y. Tsien. 1995. Improved green fluorescence. Nature 373:663-664. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, N. L., and S. Kotz. 1970. Distributions in statistics: continuous univariate distributions-1. John Wiley & Sons, New York, N.Y.
- 12.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 13.Lowder, M., A. Unge, N. Maraha, J. K. Jansson, J. Swiggett, and J. D. Oliver. 2000. Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl. Environ. Microbiol. 66:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 15.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercier, J., and S. E. Lindow. 2000. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 19.Monier, J.-M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monier, J.-M., and S. E. Lindow. 2004. Frequency, size and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan, J. V., and H. B. Tukey, Jr. 1964. Characterization of leachate from plant foliage. Plant Physiol. 39:590-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 24.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, J. N., and B. M. Ahmer. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 28.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 30.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, M., and S. E. Lindow. 1994. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 34.Winzer, K., Y. H. Sun, A. Green, M. Delory, D. Blackley, K. R. Hardie, T. J. Baldwin, and C. M. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]