Abstract
Pigs are natural host to various zoonotic pathogens including viruses. In this study, we analyzed the viral communities in the feces of 89 piglets with diarrhea under one month old which were collected from six farms in Jiangsu Province of the Eastern China, using the unbiased virus metagenomic method. A total of 89 libraries were constructed, and 46937894 unique sequence reads were generated by Illumina sequencing. Overall, the family Picornaviridae accounted for the majority of the total reads of putative mammalian viruses. Ten novel virus genomes from different family members were discovered, including Parvoviridae (n = 2), Picobirnaviridae (n = 4) and CRESS DNA viruses (n = 4). A large number of phages were identified, which mainly belonged to the order Caudovirales and the family Microviridae. Moreover, some identified viruses were closely related to viruses found in non-porcine hosts, highlighting the potential for cross-species virus dissemination. This study increased our understanding of the fecal virus communities of diarrhea piglets and provided valuable information for virus monitoring and preventing.
Keywords: Diarrhea piglets, Feces, Metagenomic, Viral communities, Virus genome
Highlights
-
•
High diversity of the viral communities existed in the feces of piglets with diarrhea.
-
•
612 different complete or nearly complete CDS of virus genomes were identified.
-
•
Ten newly discovered viral genomes enrich the knowledge about the known viral species diversity.
-
•
Viral infections may endanger the health of piglets.
1. Introduction
Pigs are omnivorous mammals that are frequently kept as livestock and are a significant component of animal protein for consumption by humans all over the world [1]. China is one of the biggest pig farming countries in the world. According to official statistics from the Ministry of Agriculture and Rural Development of the People's Republic of China, the number of pigs slaughtered in 2021 was 1.672 billion [2]. Large-scale high-density pig farming operations are quite vulnerable to virus outbreaks, which may promote the emergence and horizontal transmission of new pathogens, as well as increase the risk of zoonotic infectious diseases spreading to other species, including humans [3]. With the globalization of the pig industry, porcine pathogens are emerging more frequently and spreading over the world [4].
Diarrhea is one of the common diseases of pigs. Piglets' related dehydration symptoms are a common cause of piglet mortality, particularly in intensive pig farms, resulting in major economic losses in the pig industry worldwide [5]. The Porcine Epidemic Diarrhea Virus (PEDV) [6], Swine Fever Virus (SFV), porcine rotavirus [7] and porcine circovirus have been known to cause the diarrhea in piglets [8]. In 2013, the PEDV affected nearly half of America's breeding stock [8]. In China, African swine fever caused $141 billion in direct economic losses [9]. Some of these viruses are zoonotic pathogens that recognize pigs as hosts and transmitted to humans such as Nipah, influenza A, hepatitis E and Japanese encephalitis viruses [2]. Zoonoses can pose a serious threat to public health and the global economy [10]. According to previous studies, the direct costs of zoonotic diseases surpassed $20 billion, while the indirect costs exceed $200 billion [11]. It was reported that there have been outbreaks of new infectious diseases related to diarrhea in piglet farms [12]. These diseases are related to bocaviruses, astroviruses, caliciviruses, kobuviruses, sapeloviruses, etc.
Pigs frequently live in close proximity to humans, and this might be associated with an increased risk of transmission of pathogens including zoonotic agents [13]. Most viruses are spread to humans or other animals through direct contact with living animals, or to the public by contaminated meat [14,15]. Emerging and re-emerging infectious viral diseases have become a constant threat and disease burden to animal and human health. Therefore, it is crucial to investigate the presence of viruses in key host animals such as pigs to find the original hosts or carriers of viruses that may cause disease outbreaks in farmed animals and humans [16]. Some previous studies have investigated the viral composition of feces from diarrheic piglets; however, the fecal viral composition of piglets is affected by a variety of factors such as feeding styles, sanitary conditions, geographic locations, and research methods. Recently, the metagenomic approach has provided us with a new unbiased way to describe the viral community of different sample [17]. In this study, viral metagenomic technique was carried out to analyze the fecal virus communities of diarrheic piglets in Jiangsu province of China, and the potential novel viruses were bioinformatically identified.
2. Materials and methods
2.1. Sample Collection and preparation
To investigate fecal viruses in diarrheic piglets, from April to May in 2018, 89 fecal samples of diarrhea piglets were collected from pig farms in Jiangsu Province of the Eastern China, including Nanjing, Yangzhou, Nantong, Zhenjiang, Changzhou and Suzhou cities (Fig. S1 and Table 1). All piglets were under one month of age (14–30 days of age). Using disposable gloves, feces samples from a single piglet were collected from the floor after the piglets were segregated in a designated area and allowed to fecate on their own. Samples were collected using disposable materials and transported to the laboratory on dry ice. Fecal samples were resuspended with 10 vol of phosphate buffered saline (PBS) and vigorously rotated for 5 min. After centrifugation (10min, 15000 g), the supernatant was placed in a 1.5 ml centrifuge tube and stored at −80 °C for nucleic acid extraction.
Table 1.
Sample information.
| Farm No. | Collection location | Sample size | Sample No. | Age in days |
|---|---|---|---|---|
| A | Nanjing city | 5 | 1–5 | 15 |
| B | Yangzhou city | 3 | 23–25 | 17 |
| C | Nantong city | 29 | 73、74、76-102 | 17∼30 |
| D | Zhenjiang city | 13 | 160–172 | 14 |
| E | Changzhou city | 20 | 177–196 | 21∼28 |
| F | Suzhou city | 19 | 197–215 | 20∼27 |
2.2. Viral particle purification and nucleic acid extraction
A total volume of 500 μl of the mixed supernatant was filtered through 0.45 μm microwell filter to remove eukaryotic cells, and 0.22 μm microwell filter to remove bacterial-sized particles [18]. After centrifugation (13,000 g, 5 min), the filtered filtrate was collected and treated at 37 °C with a mixture of DNases (Ambion Turbo DNase, Epicentre Baseline-ZERO), benzonase (Novagen), and RNase (Fermentas) to digest unprotected nucleic acid. Fecal samples were treated for 90 min [19]. Nucleic acid (including RNA and DNA) was extracted with QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer's instruction [20,21]. Viral nucleic acid was reverse transcribed to cDNA using the six random base primers with reverse transcription Kit (SuperScript IV, Invitrogen), and then the products were denatured at 95 °C for 2 min and rapidly placed on ice for at least 2 min. The second strand was synthesized by using Klenow fragment DNA polymerase (New England Biolabs) and the primer K–8N (GAC CAT CTA GCG ACC TCC ACN NNN NNN NNNN N) [22].
2.3. Library construction for metagenomic sequencing
The PCR procedure was performed in a total reaction volume of 50 μl containing 1 U of Taq DNA polymerase, 0.2 mM dNTP, 2.5 mM MgCl2, 1 × PCR Buffer, 0.8 μM primer K-randoms (GAC CAT CTA GCG ACC TCC AC), and 5 μl of template. Temperature cycling was performed as follows: one cycle at 95 °C for 5 min, followed by 35 amplification cycles (95 °C for 1 min, 59 °C for 1 min, and 72 °C for 1 min), and final extension at 72 °C for 1 min [23]. Products were identified as a smear by agarose gel electrophoresis [24]. Then, Nextera XT DNA sample preparation Kit (Illumina) with dual barcoding was used to construct libraries and the quality was evaluated using agarosegel electrophoresis. Libraries were sequenced on the NovaSeq 6000 Illumina platform, which features 250 base-paired ends with double barcodes for each library [25].
2.4. Bioinformatics analysis
The 250bp paired end reads generated by NovaSeq 6000 sequencing were decoded using Illumina's vendor software, and the data was processed using the internal analysis pipeline running on the 32 node Linux cluster. Clone reads were removed and low-sequencing quality tails were trimmed using Phred (mass fraction threshold set to 10). Adaptors were trimmed using the default parameters of VecScreen which NCBI Blastn with default parameters is designed for adapter removal. The clean readings from Illumina sequencing were De Novo assembled within each barcode group using the integrated assembler Geneious Prime (version 2019.2.3) to combine them into longer contigs. The assembled contigs and singlets were compared with the internal viral proteome database using Blastx, with E-value cut-off <10−526.. Candidate viral hits were then compared with an internal non-virus non-redundant (NVNR) protein database to remove false-positive viral hits [26]. The NVNR database was compiled using non-viral protein sequences extracted from NCBI nr fasta file that based on annotation taxonomy excluding Virus Kingdom. Based on the number of virus reads derived from MEGAN software (MEtaGenome Analyzer, version 6.21.7), the pheatmap package (v1.0.12) was used at the family level of each barcode to establish a heatmap [27].
2.5. Viral diversity, distribution and community analysis
To study the diversity and distribution of viruses in fecal samples from diarrheic piglets, we performed two types of analyses. First, all the reads with sequence length >50 bp in the clean data were compared to the viral proteome database using Blastx as mentioned above and the Blastx results were then loaded into the MEGAN program [28,29]; rarefaction curves were then conducted to visualize differences in the composition of the viral communities. Subsequently, using the “Map to Reference” function in Geneious Prime (version 2019.2.3), the clean data of NGS from the 89 libraries were respectively aligned to each of the 127 viral genomes which were used as reference genomes, the libraries with >10 different reads that consistently matched the reference genome were considered to be positive. Numbers of each library's sequence reads mapped to the 127 genomes are shown in Supplementary Table 1 that is further used to create a heatmap of the viral distribution.
The virus communities were compared using principal coordinate analysis (PcoA), ANOSIM, the alpha diversity index and the Upset plot. The PcoA plot was generated using R v4.1.1 package ggplot2, which implements principal coordinate analysis (PcoA) to visualize beta-diversity patterns. The PcoA plot was based on the Bray-Curtis dissimilarity matrix calculated from the compositional data of operational taxonomic units (OTUs) or amplicon sequence variants (ASVs). The P-value was calculated by ANOSIM. Simpson and Shannon indexes were used to compare the magnitude of differences in the Alpha diversity index between farm groups in box plot. These indices provide measures of species richness and evenness within each sample group. Abundance calculations were performed by summing the relative abundances of all OTUs or ASVs in each sample group. Upset plot is a tool for visualizing intersecting sets of data. The plots were used to represent the distribution of taxonomic groups across samples and to identify patterns of co-occurrence or exclusion between different taxonomic groups. It effectively represented shared and unique viruses among farms. They were all visualized in TUTU cloud analysis platform (https://www.cloudtutu.com/#/demo/page1).
2.6. Viral sequences acquisition and phylogenetic analysis
Viral genomes or fragments were De Novo assembled to obtain full-length genomes in Genious Prime, using the low sensitivity/fastest parameters [30]. The MUSCLE multiple sequence alignment programs in MEGA (version 10.1.8) were run with default parameters to generate amino acid sequence alignments [31]. Based on the predicted amino acid sequence of the viral protein, the closest Blastx match in GenBank (E-value cut-off <10−5 and the selected reference strain with the highest identity with our sequence) and the representative members of the relevant viral species or genera were selected for phylogenetic analysis. Phylogenetic trees were constructed using Bayesian inference (BI) in MrBayes3.2 by mixed models and Markov chain Monte Carlo (MCMC) methods based on amino acid sequences [33]. The run was stopped when the ASDSF dropped below 0.01, and the first 25% of MCMC samples were discarded as burn-in. Figtree v 1.4.4 software, Adobe Illustrator 2020 v24.0.1, iTOL (https://itol.embl.de/) and ChiPlot (https://www.chiplot.online/) were used to visualize the Bayesian inference tree. International Committee on Taxonomy of Viruses (ICTV) criteria (https://ictv.global/report) used to genetically and phylogenetically characterize each of the putative new genus or species of viruses described in this study are listed in Supplementary Table 2.
3. Results
3.1. Viral metagenomic overview
As shown in Supplementary Fig. 1, the sampling sites located in Jiangsu Province, including Nanjing City (Farm A), Yangzhou City (Farm B), Nantong City (Farm C), Zhenjiang City (Farm D), Changzhou City (Farm E), and Suzhou City (Farm F), respectively. A total of 89 libraries were constructed. Each library contained one sample. All Illumina sequencing generated a total of 46,937,894 unique sequence reads, with an average GC content of 44.2% (Supplementary Table 3). Of these reads, 5668086 (12.1%) matched viral proteins through Blastx search based on protein sequence identity. The conduction of taxonomic classification of viral reads was based at the family level. Contigs and singlet readings that resembled viral sequences were divided into 26 viral families (Fig. 1A), and the results showed that the family Picornaviridae and the family Astroviridae were abundant in all libraries. According to Fig. 1B, it was evident that phage viral groups dominated the farm C and were major in Siphoviridae, Microviridae, Podoviridae and Myoviridae in all libraries (Fig. S2). The distribution of mammalian viral reads detected in each library was analyzed and sorted by the abundance of sequence reads (Fig. 1C). The results showed that 2548330 and 2462437 reads have sequence similarity with the Bacteriophages and the family Picornaviridae, respectively, accounting for the 45.0% and 42.5% of the total number of putative viral reads. The next following families were Astroviridae (355280 reads), Caliciviridae (155157 reads), Parvoviridae (116374 reads), Smacoviridae (32027 reads), Circoviridae (23320 reads), Picobirnaviridae (5795 reads), Coronaviridae (3143 reads), genomoviridae (1820 reads), etc. By using sequence assembly combined with Illumina sequencing of gaps between contigs, we identified 612 different complete or nearly complete CDS of virus genomes (Supplementary Table 4).
Fig. 1.
Viral taxonomy analyses at the family level. (A) Heatmap representing the read number of each viral family in each library on log2 scale. The sample groups are shown with corresponding colors (see color legend). (B) Heatmap representing the read number of each phage virus family in each farm on log2 scale. The sample groups are shown with corresponding colors (see color legend). (C) The pie chart shows the composition of fecal virus groups of diarrhea piglets and displays the percentage of virus sequences in different virus families.
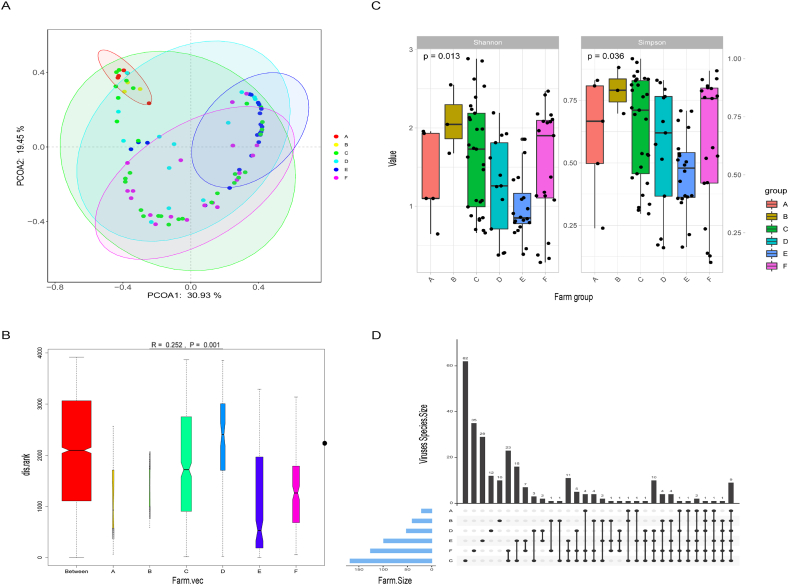
Here, the species rarefaction curve was used to visualize virus species composition in the diarrhea piglets (Fig. S3A). The curve changed to a clear plateau as the number of reads sampled rises, demonstrating that sequencing was sufficiently thorough to detect nearly all viral species in the sample for subsequent analysis. The species accumulation curve was represented the species richness of the sample database, had achieved a steady state, indicating that the sample volume in this study was adequate and accurately reflected the diversity variations among the various communities (Fig. S3). The mapping analysis using the 127 genomes against the 89 NGS data revealed the virus distribution in the 89 sample pools (Fig. S4 and Supplementary Table 1). The PcoA based on Bray-Curtis revealed that the viral community composition of these six farms differed in the group at the family level (Fig. 2A). p-values of 0.05 indicated significant differences in the composition of virus communities in fecal samples from diarrheal piglets on each farm (Fig. 2B and C). The Upset plot is an ideal way to demonstrate both common and specific viruses among the six farms, which revealed the distinctive viral community characteristics and the results of which indicated that all six farms showed the presence of nine common viruses (Fig. 2D). The highest number of viruses at the species level was found in farm C, where 62 viruses were identified; the lowest number was found in Farm B, where only 10 viruses were identified.
Fig. 2.
Diversity analysis of virus difference in six farms. (A) Principal coordinate analysis (PcoA) based on Bray-Curtis of six farms at the family level. (B)The P-value is calculated by ANOSIM. (C) Box plots were used to compare the magnitude of differences in the Alpha diversity index between farm groups using the Simpson and Shannon indexes. Each color box represented a subgroup, and the p-value above the box indicated whether or not that diversity index differed significantly across all subgroups. (D) The Upset diagram effectively represented the farms' shared and unique viruses. The horizontal bar on the left side of the figure depicted the number of virus species in each farm, and the lines connecting the dots represented the intersection of different farms, with individual dots indicating unique to that farm and nodes connected by lines indicating common. The vertical bars represented the number of corresponding intersection elements.
3.2. Parvoviruses
Parvoviruses are some of the common DNA viruses that can infect a variety of animals by causing disease with a single virus or together with other viruses [32]. Parvoviruses have two ORFs, which encode non-structural protein 1 (NS1) and structural protein 1 (VP1) [33]. In this study, a total of six parvoviruses with complete genome sequences were determined from fecal samples of diarrhea piglets, including a member of Chaphamaparvovirus (PFParV2) belonging to the Hamaparvovirinae. The other five sequences were all from Bocaparvovirus (PFParV1, PFParV3, PFParV4, PFParV5, PFParV6). The genome lengths of them were 5287bp, 5095bp, 5649bp, 5446bp, 5137bp and 5245bp, respectively (Fig. 3A). The phylogenetic trees were constructed based on the protein sequences and reference sequences of NS1. In the group of genomes showing similarity to parvovirus, five genomes from five different sample pools were phylogenetically grouped into or close to the genus Bocaparvovirus (Fig. 3B). PFParV1 was closely related to Squirrel Bocaparvovirus (Genbank No. QKV09257), which was another member of the genus Bocaparvovirus detected in Kunming city of China, and formed a cluster in the phylogenetic tree, sharing the highest amino acid sequence identity (56.78%) with the reference sequence. PFParV3-PFParV6 clustered with three different porcine bocaviruses and shared NS1 amino acid identity of 94.01%–99.69%. Only one genome was phylogenetically grouped into the genus Chaphamaparvovirus (Fig. 3C). PFParV2 was closely clustered with rat parvovirus 2 (Genbank No. NC_055465). The amino acid sequence identity of NS1 was 64.09%, indicating that PFParV2 belonged to a new chaphamaparvovirus species.
Fig. 3.
Phylogenies of Parvoviruses identified in the diarrhea piglets. (A) The genomic organization of the six Parvoviruses identified in diarrhea piglets. (B) Bayesian inference tree established based on amino acid sequences of NS protein of the genus Bocaparvovirus. (C) Bayesian inference tree established based on amino acid sequences of NS protein of the genus Chaphamaparvovirus. The viruses identified in this study are labeled with red branches and leave names. Different taxonomic clusters were represented by rectangles filled with different colors, and taxon names are indicated on the right. Tree scales indicate the amino acid substitutions per site.
3.3. Mamstroviruses
Astroviruses, the primary cause of human gastroenteritis, were discovered to be the cause of infant diarrhea in a UK obstetric hospital in 1975 [34]. Astroviruses are composed of three open reading frames [35]: ORF1a encodes the nonstructural polyprotein 1A; ORF1b encodes the polyprotein 1AB, including RNA-dependent RNA polymerase (RdRp) [36] and ORF2, which encodes the viral capsid protein. Twenty-six astrovirus genomes were characterized from 21 different sample pools and were called as PFAstV1-26. The phylogenetic tree based on capsid protein sequences indicated that these astroviruses are grouped into 3 different clades in the genus Mamastrovirus (Fig. 4A). The results showed that most strains detected had the highest identity with Mamastrovirus 3, with amino acid identity ranging from 69.32% to 98.52%. PFAstV3 and Mamastrovirus 22 (Genbank No. MW653750) shared a branch, and the structural and nonstructural proteins of Mamastrovirus 22 shared 95.62% and 96.61% of their amino acid sequences, respectively. PFAstV10 and PFAstV16 formed an independent clade with Porcine astrovirus 5 (GenBank No. YP 009010970), indicating that they belonged to the Unclassified Astroviridae and shared 82.89% and 92.76% of their polyprotein amino acid sequences.
Fig. 4.
Phylogenies of the genus Mamastrovirus and the family Caliciviriade identified in diarrhea piglets. (A) Bayesian inference tree established based on amino acid sequences of Cap protein of the genus Mamastrovirus. (B) Bayesian inference tree established based on amino acid sequences of ORF1 protein of the family Caliciviriade. The viruses identified in this study are labeled with red branches or red dots and leave names. Different taxonomic clusters were represented by rectangles filled with different colors, and taxon names are indicated on the right. Tree scales indicate the amino acid substitutions per site.
3.4. Caliciviruses
Caliciviruses can infect humans and various animals [37]. The Sapovirus genome contains two or three open reading frames (ORFs). ORF1 encodes seven nonstructural proteins, followed by the major capsid protein VP1 [38]; ORF2 encodes the minor structural protein VP2. All five calicivirus sequences obtained from fecal samples of diarrheal pigs were identified as Sapovirus members by alignment in GenBank Blastx. The phylogenetic tree was constructed based on the protein sequence and reference sequence of ORF1 (Fig. 4B). The majority of strains in diarrheal piglets were identified to be clustered inside the Porcine Sapovirus GIII group, which was acknowledged from the tree, however the tree configuration divided the GIII into two branches, which was supported at 100% bootstraps. PFCalV2, PFCalV5 and PFCalV1 were distantly related to each other. Phylogenetic analysis showed that PFCalV1 and PFCalV3, PFCalV 2 and PFCalV5 clustered with Porcine Sapovirus (GenBank No. QHC33959, QHC33963) and Sapovirus GIII (GenBank No. BBA54631, AXO78746) with amino acid sequence identity of 94.4% and 97.9%, 99.1% and 98.8%, respectively. The ORF1 predicted by PFCalV4 encoded a putative polyprotein, which had at least 97.82% amino acid matching with the previously published Sapovirus GVII (GenBank No. AXO78744), which clustered to form GVII with 100% bootstrap support.
3.5. Picobirnaviruses
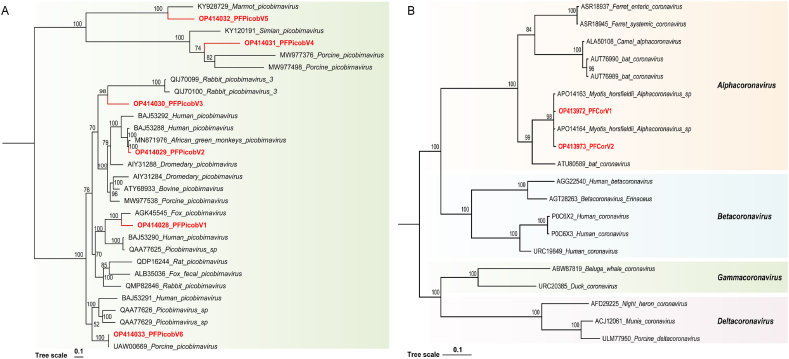
Picobirnaviruses are small, non-enveloped virus with a segmented genome of approximately 4.2 kb double-stranded RNA [39]. The genome of picobirnaviruses is composed of two parts, a large fragment encoding capsid protein and a small fragment encoding RdRp [25]. Six Picobirnavirus fragments in total were discovered in this study. These RdRp proteins shared 65.65%–99.79% amino acid identity with other known picobirnavirus strains. The conserved RdRp amino acid sequence was used to construct phylogenetic tree (Fig. 5A). The tree showed that PFPicobV1 clustered with the Picobirnavirus (GenBank No. AGK45545) detected in fox feces with 65.65% amino acid sequence identity. PFPicobV2 and the closely related Picobirnavirus of African Green monkeys (GenBank No. MN871976) sharing 96.6% of their sequence identities. PFPicobV3 and its distant relative Rabbit picobirnavirus 3 (GenBank No. QIJ70100) and Human Picobirnavirus (GenBank No. BAJ53292) shared only 45.1% and 61.9% sequence identity, respectively. PFPicobV4 and PFPicobV5 were clustered in a large branch, and the support value of the tree was 100. The PFPicobV4 clustered with Simian Picobirnavirus (GenBank No. KY120191), and the amino acid identity was 43.9%. PFPicobV5 and Marmot Picobirnavirus (GenBank No. KY928729) were grouped into one branch with an amino acid identity of 58.4%. PFPicobV6 shared the highest amino acid sequence identity (99.8% identity) with Porcine Picobirnavirus (GenBank No. UAW00669), according to Blastx analysis.
Fig. 5.
Phylogenies of the family Picobirnaviridae and the family Coronaviridae identified in diarrhea piglets. (A) Bayesian inference tree established based on amino acid sequences of RdRp protein of picobirnaviruses. (B) Bayesian inference tree established based on amino acid sequences of RdRp protein of coronaviruses. The viruses identified in this study are labeled with red branches or red dots and leave names. Different taxonomic clusters were represented by rectangles filled with different colors, and taxon names of coronaviruses are indicated on the right. Tree scales indicate the amino acid substitutions per site.
3.6. Coronavirinae
Coronaviruses are envelope-positive single-stranded RNA viruses of the Coronavirinae, including a 5 'cap and a 3′ poly (A) tail [40]. In the current study, fifteen Coronavirus fragments were found in the fecal libraries. These fragments shared more than 99% similarities with porcine epidemic diarrhea virus, which belonged to the genus Alphacoronavirus. Two sequences of the RNA-dependent RNA polymerase (RdRp) were selected for phylogenetic analysis (Fig. 5B). The tree revealed that PFCorV1 and PFCorV2 clustered with Alphacoronavirus sequences (GenBank No. APO14163 and APO14164) detected from Myotis Horsfieldii and shared 99.8% amino acid sequence identity.
3.7. Picornaviruses
Picornaviruses are small, non-enveloped viruses with single-stranded polyadenylated genomic RNA that affect a range of vertebrate hosts and cause a variety of diseases [41]. Typically, viral genomes have a single ORF flanked by the 5 'and 3′ untranslated sequences (UTR) [41]. In this study, a total of 82 complete Picornaviridae genome sequences were assembled in the feces of diarrhea piglets, more than 6000 nucleotides in length. Sequences with more than 99.2% identity were excluded and 34 sequences were chosen to build the phylogenetic tree, and they were temporarily given the name PFPicV1-34. A phylogeny over the amino acid sequence of ORF1 protein showed the relationship of the viruses identified here and their relatives. According to the sequence analysis and phylogeny, six of these picornaviruses clustered with genus Sapelovirus (Fig. 6A), two of them were closely related to viruses in genus Teschovirus (Fig. 6B), thirteen of them were closely related to viruses in genus Enterovirus (Fig. 6C), and twelve can be a member of the genus Kobuvirus (Fig. 6D). These gene sequences shared high amino acid identities, ranging from 90.10%–99.47%.
Fig. 6.
Phylogenies of the Picornaviruses identified in diarrhea piglets. (A) Bayesian inference tree established based on amino acid sequences of ORF1 protein of the genus Sapelovirus. (B) Bayesian inference tree established based on amino acid sequences of ORF1 protein of the genus Teschovirus. (C) Bayesian inference tree established based on amino acid sequences of ORF1 protein of the genus Enterovirus. (D) Bayesian inference tree established based on amino acid sequences of ORF1 protein of the genus Kobuvirus. The viruses identified in this study are labeled with red branches and leave names. Different viral groups are marked with color coding shown by the key in the upper right. Tree scales indicate the amino acid substitutions per site.
3.8. CRESS-DNA viruses
Viruses with small circular rep-encoding ssDNA (CRESS-DNA) genomes encode a replication associated protein (Rep), mainly includes the family Circoviridae [42], Genomoviridae [43], Smacoviridae [44], and others.
3.8.1. Circoviruses
In this study, the full-length genomes of 13 CRESS-DNA viruses were detected from 10 different sample pools. Phylogenetic tree was constructed based on Rep proteins (Fig. 7A). PFCirV1 and PFCirV3 were most closely related to the Rep gene integrated in Entamoeba-associated CRESS DNA virus 4 (GenBank No. QNJ47536), and the amino acid sequence identity was 99.67%. PFCirV2 and PFCirV5 shared 45.57% and 42.2% sequence identity with the closely related Chicken virus (GenBank No. QIR82201), respectively. PFCirV4 and PFCirV10 were members of Po-circo-like virus 51 (GenBank No. QZA75073), sharing 89.7% identity, while PFCirV11 and PFCirV12 belong to Po-circo-like virus 22 (GenBank No. AER30023), with 94.79% and 95.16% amino acid sequence identity, respectively. PFCirV8, PFCirV9 and PFCirV13 were clustered with Circoviruses identified from pigs (GenBank No. QBA83782 and QBA83740), and the amino acid sequence identity was 83.81%, 75.3% and 100.00%, respectively. It was noteworthy that PFCirV7 was found to be clustered with the Circular DNA virus that was detected from Trachemys Scripta elegans (GenBank No. QOW18373), with 49.81% amino acid sequence identity. PFCirV6 was clustered with CRESS virus (GenBank No. QRI44123) detected from water sample along the river ports, and the amino acid identity was 50.73%.
Fig. 7.
Phylogenies of CRESS DNA viruses identified in diarrhea piglets. (A) Bayesian inference tree established based on amino acid sequences of Rep protein of circoviruses. (B) The genomic organization of the four Genomoviruses identified in diarrhea piglets. (C) Bayesian inference tree established based on amino acid sequences of Rep protein of the family Genomoviridae (D) Bayesian inference tree established based on amino acid sequences of Rep protein of the genus Porprismacovirus. The viruses identified in this study are labeled with red dots or red branches and leave names. Different taxonomic clusters of circoviriuses and genomoviriuses were represented by rectangles filled with different colors, and taxon names are indicated on the right. Tree scales indicate the amino acid substitutions per site.
3.8.2. Genomoviruses
In this study, a total of four complete Genomoviridae genome sequences were assembled from three libraries and named PFGenV1-4. The genomic organization of these viruses was shown in Fig. 7B, where the predicted Rep and other proteins were pointing in the opposite directions. The genome length of PFGenV2 is 2206bp and it belongs to Gemycircularvirus. The other three sequences are divided into the unclassified Genomoviridae, whose genome lengths are 2155 bp, 2143bp and 2144 bp, respectively. Phylogenetic analysis based on amino acid sequences of Rep protein (Fig. 7C) showed that PFGenVs were located on two independent clades. PFGenV1 and Crane CRESS DNA virus (GenBank No. UBQ66246) clustered into one branch and shared 96.57% identity. PFGenV2 clustered with Gallus Feces associated Gemycircularvirus 17 (GenBank No. ANC51577) and shared 18.2% identity. It has formed its own deep branch of the Genomoviridae tree. The PFGenV3 shared a 98.53% identity with the chicken stool-associated Gemycircularvirus (GenBank No. YP 009337824). PFGenV4 shared the highest amino acid sequence identity (99.51%) with Genomoviridae (GenBank No. QXN75495) detected from airborne Particulate Matter.
3.8.3. Smacoviruses
Recently, smacoviruses was recognized by ICTV as a group of CRESS with genomes ranging from 2.3 to 2.9 kb [45]. A total of 31 complete genome sequences were identified in 27 sample pools. The phylogenetic tree was constructed based on the Rep protein (Fig. 7D) and showed that most of the sequences shared the highest sequence similarity with the different types of Porcine associated Porprismacoviruses, with 79.68%–99.61% in Rep region. Species and genus criteria demarcation of Smacoviridae are based on genome-wide and rep amino acid sequences with cut-off of 77.0% and 40.0%, respectively [44]. Based on these criteria, none of the 31 smacoviruses can be designated as new genera.
3.9. Of virome of the feces
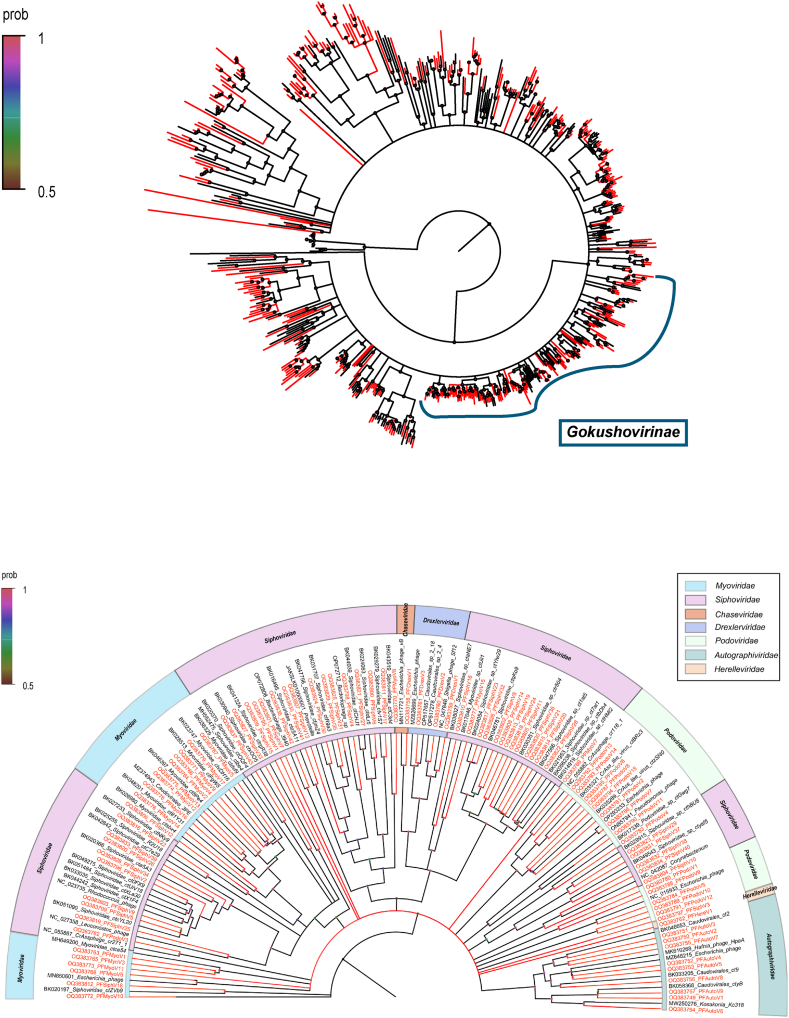
Bacteriophage virus also accounted for a large proportion of the feces of diarrheic piglets, which mainly consisted of the Microvirade and Caudovirales. As one of the most common ssDNA phage viruses, the family Microviridae are small, circular, single-stranded DNA viruses [46]. A phylogenetic analysis was performed using the amino acid sequence of the major capsid protein (MCP) of the 396 Microviridae strains (Fig. 8A). The results revealed that the majority of Microviridae were highly varied and tricky to categorize into recognized subfamilies. A total of 110 Microviridae genomes (from clades 5,6,9) were not clustered within any known families, however formed several potentially new branches. One hundred and nine genomes of Microviridae were found to be clustered within the branch associated with the Gokushovirinae and the other sequences were grouped together in the branch linked to the Unclassified Microvirinae.
Fig. 8.
Phylogenies of the family Microviridae and the order Caudovirales identified in diarrhea piglets. (A) Bayesian inference tree established based on amino acid sequences of MCP protein of the family Microvirdae. The Unclassified Microvirinae were tentatively divided into ten clades artificially. (B) Bayesian inference tree established based on amino acid sequences of TERL protein of the order caudovirales. Representative strains of different subfamilies were included and marked with the color coding in the key on the top right. The viruses identified in this study are labeled with red branches and leave names. The size of the dots on nodes is positively correlated with the corresponding bootstrap score. Tree scales indicate the amino acid substitutions per site.
The order Caudovirales are the most prevalent viruses with double-stranded genomes [47]. A phylogenetic analysis was performed based on the 89 TerL protein sequences (Fig. 8B). The Caudovirales in this study were highly diverse and clustered into multiple groups, according to the phylogenetic tree structure. The majority of the sequences grouped together with known reference sequences of the Siphoviridae, suggesting that they may share similar genetic characteristics and biological functions. Only PFChasV1 and PFHereV1 grouped within the Chaseviridae and Herelleviridae, respectively.
4. Discussion
Researchers found that pigs with diarrhea shed an average of 5.4 different mammalian viruses compared to an average of 4.2 different mammalian viruses shed by healthy piglets [48], which suggested that virus was more easily detected in sick pigs. With the advancement of metagenomics technology in recent years, an increasing number of research have begun to use this technology to examine the pathogenic communities in the feces of diarrheal piglets [49]. Viral metagenomic sequencing help us identify emerging viruses in animals and humans, and was used to characterize pathogens in mammals [17]. Here, we provide a preliminary description of the fecal virome of diarrhea piglets from six farms in Jiangsu, China, using viral metagenomic sequencing. In this study, we identified major viruses such as microviruses, parvoviruses, mamastroviruses, caliciviruses, picobirnaviruses, coronaviruses, picornaviruseses, circoviruses, gemycircularviruses and smacoviruses in the fecal samples of diarrhea piglets. Based on the heatmap and upset plot, we found that more viruses were detected in the fecal samples of diarrhea piglets from Farm C, which may be due to the poor environmental hygiene and management quality. The dirty and crowded conditions of pig farms and the frequent movement of pigs allow viruses to spread and evolve [50]. According to research, co-infection with a large number of viruses may reduce the innate and immune defenses of piglets, leading to diarrhea and other diseases in piglets [48]. Furthermore, the use of animal facilities such as feed delivery and the disposal of animal feces in large lakes may lead to the virus to spread from one farm to another, increasing public health risks [51].
Bocaviruses usually infect the respiratory tract and intestine [25]. The majority of the parvovirus sequences found in this study were clustered with Porcine Bocavirus. Whether it cause disease in pigs remains to be studied [52]. In addition, feces also contained sequences that were very similar to those of Rat Parvovirus and Squirrel Bocavirus. NS1 amino acid sequence identity is used as demarcation criteria for genus and species in Parvoviridae family, according to the ICTV classification criteria, with 35.0% identity as threshold to novel genus and 85.0% to species [53]. It means parvoviruses with NS1 amino acid sequence identity greater than 85% belong to the same species [54]. Therefore, PFParV1 and PFParV2 from this study could potentially be the first members of two novel species. Astroviruses are the main pathogen of human gastroenteritis and are considered to be the main cause of infantile diarrhea [34]. They can also infect different mammals and poultry, including pigs. The majority of the 26 almost complete Astrovirus genome sequences found in diarrheal piglet feces exhibited the highest similarity to Mamastrovirus 3. Currently, the ICTV-recognized Mamastrovirus species should have capsid proteins with amino acid distances greater than 37.8% [55]. The astroviruses found in this research are unlikely to represent a novel species. Caliciviruses are generally transmitted through fecal oral or droplet transmission, which can infect humans and various animals and induce related diseases [37]. Species criteria demarcation of Caliciviridae are based on Cap amino acid sequences with cut-off of 14.7%–26.7% [56]. All calicivirus sequences found in this study were Porcine Sapovirus, and among them, PFCalV4 belonged to Sapovirus GVII.
Picobirnavirus has been detected in the feces of many different hosts, including humans, pigs, rabbits, dogs, rats, and birds [25]. In this study, one sequence showed the highest amino acid identity with Picobirnavirus detected in fecal samples from African green monkeys. Several novel fragments clustered with Picobirnavirus detected from fox, Simian and Marmot feces, respectively. We found that PFPicobV1 clustered with the Picobirnavirus detected in fox feces with 65.65% amino acid sequence identity. The PFPicobV4 was clustered with Simian Picobirnavirus and the amino acid identity was 43.9%. PFPicobV5 and Marmot Picobirnavirus were grouped into one branch with an amino acid identity of 58.4%. Phylogenetic distance of RdRp amino acid sequences greater than 78% is one of the species demarcation criteria in these genera [53,57]. Considering those criteria, some of our newly identified viruses can be tentatively considered as new species. The new fragments could simply represent pollutants in the food eaten by piglets. In fact, we cannot decisively comment on the host relationship of any virus found only in feces, as they may also represent food contaminants.
Coronavirus has a wide host range and can infect animals and humans, causing respiratory, intestinal and nervous system diseases [40]. The outbreak of severe acute respiratory syndrome (SARS) in 2003–2004 was caused by an animal-derived Coronavirus [58]. The 90% aa sequence identity threshold now proposed as a species demarcation criterion within each genus has been determined from the analysis of pair-wise aa distances in seven conserved replicase domains (nsp3 ADRP, nsp5 (3CLpro), nsp12 (RdRp), nsp13 (He11), nsp14 (ExoN), nsp15 (NendoU) andnsp16 (O-MT)) of 156 viruses in the Coronaviridae [59]. By phylogenetic analyses, these fragments were grouped with porcine epidemic diarrhea virus, showing more than 99% of amino acid identity, which are unlikely to represent a novel species. Two sequences detected in fecal samples in this study clustered with Alphacoronavirus sequences detected from Myotis Horsfieldii, suggesting the potential for cross-species transmission. The hypothesis that horizontal transmission may occur between domestic animals and bats was based on the PEDV-like viruses detected in Horsfield myositis [60]. This was the first report on the presence of this Coronavirus in Horsfieldii myositis, and at the genetic level, the detected strain was likely to be closely related to PEDV strains infecting pigs and cattles [61]. Furthermore, contamination of food cannot be ruled out. These viruses could be in the diet of the pigs if they are eating something contaminated with bat feces. Myotis bats tend to live in dark places, including farms, houses and barns, and are potential zoonotic host. It is particularly important to investigate whether there are pig farms and bats in the surrounding environment in areas where the coronaviruses were found, which will provide data to explore potential relationships between piglets, bats and other livestock [62]. The fecal virus group of diarrhea piglets includes a large number of vertebrate viruses. The family Picornaviridae reads dominated in all samples. Our study has provided an overview of enteroviruses in diarrhea piglets and has further enriched the current knowledge about picornaviral. Although Porcine Teschovirus and Sapelovirus have been identified as the causes of a variety of diseases including gastrointestinal diseases, poliomyelitis, and respiratory system diseases, the clinical correlation between EV-GS and intestinal or other diseases needs further study [63].
CRESS-DNA viruses with small, circular replication associated protein (Rep)-encoding single stranded (CRESS) DNA genomes, were largely identified based on conserved rolling circle replication proteins [64]. The pathogen causing significant economic losses to the global pig industry is PCV2, which can cause Porcine Circovirus (associated) Disease (PCVD/PCVAD) [65]. Porcine circovirus was not detected in this study, and follow-up studies should expand the sampling area and change the sampling method to detect more viruses. The cut-off criterium for species demarcation in Circoviridade family is 80% complete genome nucleotide sequence identity. The two circoviruses, named PFCirV2 and PFCirV5, respectively, show 30.2% and 31.3% genome-wide identity to known circoviruses. Thus, according to the current species demarcation criteria, they represent two novel circoviruses. PFCirV6 was clustered with CRESS virus detected from water sample along the river ports and the genome-wide nucleotide sequence identity was 39.6%, which may be a new member of this genus. Of course, water from farms may be contaminated. The thirteen CRESS-DNA species described here all belong to unclassified CRESS viruses. The results suggested that pigs may be co-infected by a variety of CRESS DNA viruses. Additionally, the source of the Rep-encoded circular DNA viruses identified in this study was likely to be the intestinal cells of diarrheal piglets, microorganisms or parasites colonising the porcine gut [[66], [67], [68]], or from the porcine diet. The intensive trade of animals and their persistent infection can lead to the spread of various viruses. The microbial communities of the pig intestine is very complex, and to date, little is known about how viruses (including these unclassified porcine associated CRESS DNA viruses) interact with host cells and other microorganisms [69]. Gemycircularviruses have been detected in different organisms and environments, but the host species of these viruses was only known for one fungal virus [70]. Species criteria demarcation of Genomoviridae of ICTV are based on genome-wide amino acid sequences with cut-off of 78% [71]. Gemycircularviruses detected in this study and Gallus feces associated Gemycircularvirus 17 formed their own deep branches in the phylogenetic tree, and the genome-wide nucleotide sequence identity was 51.5%, which may indicate the existence of a new member of the genus Gemycircularvirus. In this study, we identified three new unclassified Genomoviruses strains from fecal samples. Smacoviridae is one of the new families of the group that was recognized by ICTV in 2018 and were thought until recently to have animals as possible hosts [45]. Smacoviruses can be detected in the fecal samples of various vertebrates [72], and most of the sequences belong to Porprismacovirus.
We discovered a very high genetic diversity of bacteriophage viruses in the feces of diarrheal piglets. There are differences in host range, capsid size, serological properties and, insofar as known, DNA sequences between various phage viruses. The majority of the Microviridae phages detected in the feces of diarrheal piglets clustered individually in the evolutionary tree, exhibited a high level of genetic diversity. The symptoms and course of the avian influenza illness can be made worse by phages via facilitating virus replication and transmission [73]. Additionally, phages can attack the intestinal bacteria of some animals, leading to illnesses like diarrhea and intestinal infections.
5. Conclusion
We conducted a comprehensive investigation of the virus communities in the feces of piglets with diarrhea in six different locations in Jiangsu Province, China. We have discovered novel viral genomes including the Parvoviridae, Picobirnaviridae and CRESS DNA viruses. In addition, we also identified a large number of phage viruses in the feces of diarrheic piglets. Our research has increased the knowledge on the diversity of virus communities in diarrheal piglets. We will broaden the scope of sample collecting and enhance the sample size in the future. This can aid in our comprehension of the genetic traits, infection mechanisms, and biological behavior of these viruses, as well as aid in the prediction of potential intra- or interspecies transmission of these viruses. It can also serve as a foundation for future research into the detection, treatment, and prevention of emerging viral infections in mammalian and human hosts.
Ethics statement
This article did not contain any studies with human or animal performed by any of the authors.
Data availability
Genbank has received all genome sequences and stored them there under the accession numbers OP413941-OP414067, OQ383749-OQ383837, OQ387863-OQ388043, OQ388045-OQ388099, OQ388101-OQ388102, OQ388104-OQ388139, OQ388141-OQ388255, OQ388257-OQ388263. The Sequence Read Archive (SRA) has received quality-filtered sequence reads that are listed under the BioProject ID PRJNA874159 and the BioSample ID SAMN30526822. The 89 sets of sequence reads data generated on the Illumina sequencing platform in this study were deposited into the Sequence Read Archive of GenBank database and the accession nos. are shown in Supplementary Table 3. All complete or partial viral genomes acquired in this study were deposited in GenBank and the accession nos. are provided in Supplementary Table 4.
CRediT authorship contribution statement
Lingling Qian: Writing – original draft, Visualization, Formal analysis, Data curation. Zi Zhuang: Data curation. Juan Lu: Software. Huiying Wang: Resources. Xiaochun Wang: Investigation. Shixing Yang: Software. Likai Ji: Data curation. Quan Shen: Writing – review & editing, Project administration, Methodology, Conceptualization. Wen Zhang: Writing – review & editing, Writing – original draft, Project administration, Conceptualization. Tongling Shan: Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project was financially supported by National Key Research and Development Programs of China for Virome in Important Wildlife (No. 2017YFC1200201), Natural Science Foundation of the Jiangsu Higher Ediucation institutions of China (No. 2020220456), China Agriculture Research System of MOF and MARA (CARS-42-35), and Climbing Plan of Shanīghai Academy of Agricultural Sciences (PG21171).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25616.
Contributor Information
Lingling Qian, Email: qll980096@126.com.
Zi Zhuang, Email: 248439206@qq.com.
Juan Lu, Email: ljuan73@126.com.
Huiying Wang, Email: yjshywang@sina.com.
Xiaochun Wang, Email: 1000003208@ujs.edu.cn.
Shixing Yang, Email: johnsonyang1979@163.com.
Likai Ji, Email: jilikai01@ujs.edu.cn.
Quan Shen, Email: shenquan@ujs.edu.cn.
Wen Zhang, Email: z0216wen@yahoo.com.
Tongling Shan, Email: shantongling@shvri.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Lima D.A., Cibulski S.P., Finkler F., et al. Faecal virome of healthy chickens reveals a large diversity of the eukaryote viral community, including novel circular ssDNA viruses [J] J. Gen. Virol. 2017;98(4):690–703. doi: 10.1099/jgv.0.000711. [DOI] [PubMed] [Google Scholar]
- 2.Yang S., Zhang D., Ji Z., et al. Viral metagenomics reveals diverse viruses in tissue samples of diseased pigs [J] Viruses. 2022;14(9) doi: 10.3390/v14092048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith T.C., Thapaliya D., Bhatta S., et al. Geographic distribution of livestock-associated Staphylococcus aureus in the United States. Microbes Infect. 2018;20(6):323–327. doi: 10.1016/j.micinf.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Urbano A.C., Ferreira F. African swine fever control and prevention: an update on vaccine development [J] Emerg Microbes Infect. 2022;11(1):2021–2033. doi: 10.1080/22221751.2022.2108342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin P., Li H., Wang J.-W., et al. Genetic and pathogenic characterization of a novel reassortant mammalian orthoreovirus 3 (MRV3) from a diarrheic piglet and seroepidemiological survey of MRV3 in diarrheic pigs from east China [J] Vet. Microbiol. 2017;208:126–136. doi: 10.1016/j.vetmic.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine [J] Arch. Virol. 1978;58(3):243–247. doi: 10.1007/bf01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes M.K., Palmer E.L., Obijeski J.F. Rotaviruses: a review [J] Curr. Top. Microbiol. Immunol. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3. [DOI] [PubMed] [Google Scholar]
- 8.Hu J., Ma L., Nie Y., et al. A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets [J] Cell Host Microbe. 2018;24(6) doi: 10.1016/j.chom.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen S.S., Alvarez J., Bicout D.J., et al. Scientific opinion on the assessment of the control measures of the category A diseases of animal health law: african swine fever [J] EFSA J. 2021;19(1) doi: 10.2903/j.efsa.2021.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Altan E., Reyes G., et al. Virome of bat guano from nine northern California roosts [J] J. Virol. 2021;95(3) doi: 10.1128/JVI.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suminda G.G.D., Bhandari S., Won Y., et al. High-throughput sequencing technologies in the detection of livestock pathogens, diagnosis, and zoonotic surveillance [J] Comput. Struct. Biotechnol. J. 2022;20:5378–5392. doi: 10.1016/j.csbj.2022.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bányai K., Martella V., Á Bogdán, et al. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains [J] J. Gen. Virol. 2008;89(Pt 2):534–539. doi: 10.1099/vir.0.83134-0. [DOI] [PubMed] [Google Scholar]
- 13.Amimo J.O., El Zowalaty M.E., Githae D., et al. Metagenomic analysis demonstrates the diversity of the fecal virome in asymptomatic pigs in East Africa [J] Arch. Virol. 2016;161(4):887–897. doi: 10.1007/s00705-016-2819-6. [DOI] [PubMed] [Google Scholar]
- 14.Leung N.H.L. Transmissibility and transmission of respiratory viruses [J] Nat. Rev. Microbiol. 2021;19(8):528–545. doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith T.C., Harper A.L., Nair R., et al. Emerging swine zoonoses [J] Vector Borne Zoonotic Dis. 2011;11(9):1225–1234. doi: 10.1089/vbz.2010.0182. [DOI] [PubMed] [Google Scholar]
- 16.Morse S.S., Mazet J.A., Woolhouse M., et al. Prediction and prevention of the next pandemic zoonosis [J] Lancet. 2012;380(9857):1956–1965. doi: 10.1016/s0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoenfeld T., Liles M., Wommack K.E., et al. Functional viral metagenomics and the next generation of molecular tools [J] Trends Microbiol. 2010;18(1):20–29. doi: 10.1016/j.tim.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X.U., He Y., Li W., et al. Identification and characterization of a novel recombinant porcine astrovirus from pigs in anhui, China [J] Pol. J. Microbiol. 2020;69(4):471–478. doi: 10.33073/pjm-2020-051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Li L., Deng X., et al. Viral nucleic acids in human plasma pools [J] Transfusion. 2016;56(9):2248–2255. doi: 10.1111/trf.13692. [DOI] [PubMed] [Google Scholar]
- 20.Zhao M., Yue C., Yang Z., et al. Viral metagenomics unveiled extensive communications of viruses within giant pandas and their associated organisms in the same ecosystem [J] Sci. Total Environ. 2022;820 doi: 10.1016/j.scitotenv.2022.153317. [DOI] [PubMed] [Google Scholar]
- 21.Allander T., Tammi M.T., Eriksson M., et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples [J] Proc Natl Acad Sci U S A. 2005;102(36):12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stang A., Korn K., Wildner O., et al. Characterization of virus isolates by particle-associated nucleic acid PCR [J] J. Clin. Microbiol. 2005;43(2):716–720. doi: 10.1128/jcm.43.2.716-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluge M., Campos F.S., Tavares M., et al. Metagenomic survey of viral diversity obtained from feces of subantarctic and south American Fur seals [J] PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victoria J.G., Kapoor A., Dupuis K., et al. Rapid identification of known and new RNA viruses from animal tissues [J] PLoS Pathog. 2008;4(9) doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y., Wang H., Feng L., et al. Fecal, oral, blood and skin virome of laboratory rabbits [J] Arch. Virol. 2020;165(12):2847–2856. doi: 10.1007/s00705-020-04808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Z., Wang H., Feng Z., et al. Identification of a novel circovirus in blood sample of giant pandas (Ailuropoda melanoleuca) [J] Infect. Genet. Evol. 2021;95 doi: 10.1016/j.meegid.2021.105077. [DOI] [PubMed] [Google Scholar]
- 27.Huson D.H., Beier S., Flade I., et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data [J] PLoS Comput. Biol. 2016;12(6) doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huson D.H., Beier S., Flade I., et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data [J] PLoS Comput. Biol. 2016;12(6) doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shan T., Yang S., Wang H., et al. Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses [J] Microbiome. 2022;10(1):60. doi: 10.1186/s40168-022-01246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J., Yang S., Wang C., et al. Gut virome of the world's highest-elevation lizard species (and) reveals versatile commensal viruses [J] Microbiol. Spectr. 2022;10(1) doi: 10.1128/spectrum.01872-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhie A., McCarthy S.A., Fedrigo O., et al. Towards complete and error-free genome assemblies of all vertebrate species [J] Nature. 2021;592(7856):737–746. doi: 10.1038/s41586-021-03451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S., Liu Z., Wang Y., et al. A novel rodent Chapparvovirus in feces of wild rats [J] Virol. J. 2016;13:133. doi: 10.1186/s12985-016-0589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotmore S.F., Agbandje-McKenna M., Canuti M., et al. ICTV virus taxonomy profile: Parvoviridae [J] J. Gen. Virol. 2019;100(3):367–368. doi: 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appleton H., Higgins P., Letter G. Viruses and gastroenteritis in infants [J] Lancet. 1975;1(7919):1297. doi: 10.1016/s0140-6736(75)92581-7. [DOI] [PubMed] [Google Scholar]
- 35.Cortez V., Meliopoulos V.A., Karlsson E.A., et al. Astrovirus biology and pathogenesis [J] Annu Rev Virol. 2017;4(1):327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- 36.Du Y., Ji C., Liu T., et al. Identification of a novel protein in porcine astrovirus that is important for virus replication [J] Vet. Microbiol. 2021;255 doi: 10.1016/j.vetmic.2021.108984. [DOI] [PubMed] [Google Scholar]
- 37.Bank-Wolf B.R., König M., Thiel H.-J. Zoonotic aspects of infections with noroviruses and sapoviruses [J] Vet. Microbiol. 2010;140(3–4):204–212. doi: 10.1016/j.vetmic.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Oka T., Yokoyama M., Katayama K., et al. Structural and biological constraints on diversity of regions immediately upstream of cleavage sites in calicivirus precursor proteins [J] Virology. 2009;394(1):119–129. doi: 10.1016/j.virol.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 39.Ganesh B., Masachessi G., Mladenova Z. Animal picobirnavirus [J] Virusdisease. 2014;25(2):223–238. doi: 10.1007/s13337-014-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses [J] Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zell R., Delwart E., Gorbalenya A.E., et al. ICTV virus taxonomy profile: Picornaviridae [J] J. Gen. Virol. 2017;98(10):2421–2422. doi: 10.1099/jgv.0.000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright P.F., Neumann G., Kawaoka Y. 2007. In Fields Virology, 5th Edition [J] [Google Scholar]
- 43.Varsani A., Krupovic M. Family Genomoviridae: 2021 taxonomy update [J] Arch. Virol. 2021;166(10):2911–2926. doi: 10.1007/s00705-021-05183-y. [DOI] [PubMed] [Google Scholar]
- 44.Krupovic M., Varsani A.A. 2021 taxonomy update for the family Smacoviridae [J] Arch. Virol. 2021;166(11):3245–3253. doi: 10.1007/s00705-021-05224-6. [DOI] [PubMed] [Google Scholar]
- 45.Varsani A., Krupovic M. Smacoviridae: a new family of animal-associated single-stranded DNA viruses [J] Arch. Virol. 2018;163(7):2005–2015. doi: 10.1007/s00705-018-3820-z. [DOI] [PubMed] [Google Scholar]
- 46.Kirchberger P.C., Ochman H. Microviruses: a world beyond phiX174 [J] Annu Rev Virol. 2023;10(1):99–118. doi: 10.1146/annurev-virology-100120-011239. [DOI] [PubMed] [Google Scholar]
- 47.Zinke M., Schröder G.F., Lange A. Major tail proteins of bacteriophages of the order Caudovirales [J] J. Biol. Chem. 2022;298(1) doi: 10.1016/j.jbc.2021.101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan T., Li L., Simmonds P., et al. The fecal virome of pigs on a high-density farm [J] J. Virol. 2011;85(22):11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachsenröder J., Twardziok S., Hammerl J.A., et al. Simultaneous identification of DNA and RNA viruses present in pig faeces using process-controlled deep sequencing [J] PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi M., Lam T.T.-Y., Hon C.-C., et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses [J] J. Virol. 2010;84(17):8700–8711. doi: 10.1128/JVI.02551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramesh A., Bailey E.S., Ahyong V., et al. Metagenomic characterization of swine slurry in a North American swine farm operation [J] Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-95804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou F., Sun H., Wang Y. Porcine bocavirus: achievements in the past five years [J] Viruses. 2014;6(12):4946–4960. doi: 10.3390/v6124946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A D M., Jm F.S., R B C., et al. Faecal virome analysis of wild animals from Brazil [J] Viruses. 2019;11(9) doi: 10.3390/v11090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang S., He Y., Chen X., et al. Viral metagenomics reveals diverse viruses in the feces samples of raccoon dogs [J] Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.693564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams S.H., Che X., Garcia J.A., et al. Viral diversity of house mice in New York city [J] mBio. 2018;9(2) doi: 10.1128/mBio.01354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smiley J.R., Chang K.O., Hayes J., et al. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus [J] J. Virol. 2002;76(20):10089–10098. doi: 10.1128/jvi.76.20.10089-10098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Da Costa B., Duquerroy S., Tarus B., et al. Picobirnaviruses encode a protein with repeats of the ExxRxNxxxE motif [J] Virus Res. 2011;158(1–2):251–256. doi: 10.1016/j.virusres.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Lacroix A., Duong V., Hul V., et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia [J] Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorbalenya A.E., Enjuanes L., Ziebuhr J., et al. Nidovirales: evolving the largest RNA virus genome [J] Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simas P.V.M., Barnabé A.C. d S., Durães-Carvalho R., et al. Bat coronavirus in Brazil related to appalachian ridge and porcine epidemic diarrhea viruses [J] Emerg. Infect. Dis. 2015;21(4):729–731. doi: 10.3201/eid2104.141783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines [J] Virus Gene. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou H., Ji J., Chen X., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses [J] Cell. 2021;184(17) doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuchiaka S., Naoi Y., Imai R., et al. Genetic diversity and recombination of enterovirus G strains in Japanese pigs: high prevalence of strains carrying a papain-like cysteine protease sequence in the enterovirus G population [J] PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0190819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Z., He Q., Tang C., et al. Identification and genomic characterization of a novel CRESS DNA virus from a calf with severe hemorrhagic enteritis in China [J] Virus Res. 2018;255:141–146. doi: 10.1016/j.virusres.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palinski R., Piñeyro P., Shang P., et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure [J] J. Virol. 2017;91(1) doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aiemjoy K., Altan E., Aragie S., et al. Viral species richness and composition in young children with loose or watery stool in Ethiopia [J] BMC Infect. Dis. 2019;19(1):53. doi: 10.1186/s12879-019-3674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kinsella C.M., Bart A., Deijs M., et al. Entamoeba and Giardia parasites implicated as hosts of CRESS viruses [J] Nat. Commun. 2020;11(1):4620. doi: 10.1038/s41467-020-18474-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siqueira J.D., Dominguez-Bello M.G., Contreras M., et al. Complex virome in feces from Amerindian children in isolated Amazonian villages [J] Nat. Commun. 2018;9(1):4270. doi: 10.1038/s41467-018-06502-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fehér E., Mihalov-Kovács E., Kaszab E., et al. Genomic diversity of CRESS DNA viruses in the eukaryotic virome of swine feces [J] Microorganisms. 2021;9(7) doi: 10.3390/microorganisms9071426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu X., Li B., Fu Y., et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus [J] Proc Natl Acad Sci U S A. 2010;107(18):8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varsani A., Krupovic M. Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae [J] Virus Evol. 2017;3(1):vew037. doi: 10.1093/ve/vew037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anindita P.D., Sasaki M., Gonzalez G., et al. Discovery and genetic characterization of diverse smacoviruses in Zambian non-human primates [J] Sci. Rep. 2019;9(1):5045. doi: 10.1038/s41598-019-41358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weigel C., Seitz H. Bacteriophage replication modules [J] FEMS Microbiol. Rev. 2006;30(3):321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genbank has received all genome sequences and stored them there under the accession numbers OP413941-OP414067, OQ383749-OQ383837, OQ387863-OQ388043, OQ388045-OQ388099, OQ388101-OQ388102, OQ388104-OQ388139, OQ388141-OQ388255, OQ388257-OQ388263. The Sequence Read Archive (SRA) has received quality-filtered sequence reads that are listed under the BioProject ID PRJNA874159 and the BioSample ID SAMN30526822. The 89 sets of sequence reads data generated on the Illumina sequencing platform in this study were deposited into the Sequence Read Archive of GenBank database and the accession nos. are shown in Supplementary Table 3. All complete or partial viral genomes acquired in this study were deposited in GenBank and the accession nos. are provided in Supplementary Table 4.