Abstract
The succession of sulfur-oxidizing bacterial (SOB) community structure and the complex internal sulfur cycle occurring in wastewater biofilms growing under microaerophilic conditions was analyzed by using a polyphasic approach that employed 16S rRNA gene-cloning analysis combined with fluorescence in situ hybridization, microelectrode measurements, and standard batch and reactor experiments. A complete sulfur cycle was established via S0 accumulation within 80 days in the biofilms in replicate. This development was generally split into two phases, (i) a sulfur-accumulating phase and (ii) a sulfate-producing phase. In the first phase (until about 40 days), since the sulfide production rate (sulfate-reducing activity) exceeded the maximum sulfide-oxidizing capacity of SOB in the biofilms, H2S was only partially oxidized to S0 by mainly Thiomicrospira denitirificans with NO3− as an electron acceptor, leading to significant accumulation of S0 in the biofilms. In the second phase, the SOB populations developed further and diversified with time. In particular, S0 accumulation promoted the growth of a novel strain, strain SO07, which predominantly carried out the oxidation of S0 to SO42− under oxic conditions, and Thiothrix sp. strain CT3. In situ hybridization analysis revealed that the dense populations of Thiothrix (ca. 109 cells cm−3) and strain SO07 (ca. 108 cells cm−3) were found at the sulfur-rich surface (100 μm), while the population of Thiomicrospira denitirificans was distributed throughout the biofilms with a density of ca. 107 to 108 cells cm−3. Microelectrode measurements revealed that active sulfide-oxidizing zones overlapped the spatial distributions of different phylogenetic SOB groups in the biofilms. As a consequence, the sulfide-oxidizing capacities of the biofilms became high enough to completely oxidize all H2S produced by SRB to SO42− in the second phase, indicating establishment of the complete sulfur cycle in the biofilms.
In wastewater biofilms, due to the relatively high organic input and low dissolved oxygen (DO) concentration, an internal sulfur cycle consisting of sulfate reduction and subsequent sulfide oxidation is an important process for carrying electrons from the deeper anoxic zone to the oxic surface zone. Consequently, the sulfur cycle could be responsible for mineralization of a substantial part of the organic matter and consumption of dissolved oxygen (19, 25, 34), which prevent the emission of odorous and toxic hydrogen sulfide gas from the biofilms.
The reductive side of the sulfur cycle (i.e., sulfate reduction) occurs only biologically. Thus, the population dynamics, biodiversity, and in situ ecophysiology of sulfate-reducing bacteria (SRB) in wastewater biofilms have been extensively investigated by combining a 16S rRNA gene approach and microelectrode measurements (11, 12, 25, 26, 27, 34). In contrast, the oxidative side of the sulfur cycle (i.e., sulfide oxidation) occurs both biologically and chemically. The chemical oxidation reaction is, however, rather slow at natural pHs and temperatures (e.g., in the range of minutes to several hours) (5, 9, 14) and proceeds via S2O32− as a major intermediate, whereas the biological oxidation reaction occurs via S0 (4, 14, 15). S0 accumulation has often been observed in wastewater biofilms (26), indicating that biological oxidation is a dominant oxidation reaction of H2S. However, only a few studies of the community structure and diversity of sulfur-oxidizing bacteria (SOB) responsible for H2S oxidation and S0 production have been done. Little is known about development of the sulfur cycle that occurs in wastewater biofilms.
One of the reasons for the limited information is probably the phylogenetic diversity of SOB. SOB are phylogenetically distributed in a wide range of Proteobacteria (30, 31). Therefore, complete detection and quantification of SOB with a 16S rRNA-targeting probe is almost impossible, and hence, a genus-specific probe for each SOB group is required. Another reason is the versatile metabolic ability of SOB to utilize various electron donors, such as H2S, S0, S2O32−, and SO3−, and various electron acceptors, such as O2 and NO3− (30, 31), indicating that the sulfide oxidation reactions can take place under various conditions.
It is therefore necessary to multilaterally investigate sulfur oxidation by using several analytical techniques. In the present study, the succession of the SOB community structure and sulfur-oxidizing activity in developing wastewater biofilms was analyzed by combining molecular techniques based on the 16S rRNA gene and microelectrode measurements. In addition, the mass balance for SO42−, S0, and sulfide in biofilm reactors and S0 accumulation in the biofilms were examined to evaluate the development of the internal sulfur cycle in the biofilms. After a complete sulfur cycle was developed, the spatial distributions of numerically important SOB, such as Thiothrix, novel strain SO07, and Thiomicrospira denitrificans, were determined by fluorescence in situ hybridization (FISH) and related to the vertical profiles of H2S, O2, pH, NO3−, and S0 in the biofilms. The data sets resulting from these different approaches were integrated to obtain a clear overall picture of the development of the SOB community structure and complex sulfur cycle occurring in the biofilms.
MATERIALS AND METHODS
Biofilm samples.
Microaerophilic mixed-population biofilms were grown in fully submerged rotating disk reactors (RDRs) on a primary settling tank effluent from a domestic wastewater treatment plant (Sapporo, Japan). Two RDRs (RDR1 and RDR2) were independently operated in the same manner but at different times (for RDR1, May to July 2002; for RDR2, July to September, 2002). The RDRs consisted of nine polymethylmethacrylate disks with a diameter of 18 cm. The reactor volume was 5.6 liters, and the dilution rate in the reactors was kept at 0.2 h−1. Four removal slides (1 by 6 cm) were installed in each disk for sampling biofilms. The disk rotational speed was fixed at 14 rpm for both runs. The influent was supplemented with a KNO3 solution to give a final NO3− concentration of ca. 186 μM. More details concerning the RDR operation have been described elsewhere (25). The average effluent NO3− concentrations were 121 ± 43 μM and 94 ± 69 μM (means ± standard deviations) for RDR1 and RDR2, corresponding to 35% and 49% reductions, respectively. The average DO concentrations in the bulk liquid were 30 ± 19 μM and 40 ± 30 μM for RDR1 and RDR2, respectively. These conditions indicated that the biofilms were grown under microaerophilic conditions in both runs.
SORs.
The potential sulfur (S0) and sulfide (H2S) oxidation rates (SORs) were determined in standard batch experiments by measuring the initial production of sulfate. Two 18-day-old and 65-day-old biofilms were taken from RDR1 and RDR2, homogenized, and inoculated into four serum vials (130 ml) containing 100 ml of synthetic media. A slightly modified medium for neutrophilic Thiobacillus (18) was used, which contained (per liter) 2.0 g of NaHCO3, 1.0 g of KH2PO4, 1.0 g of K2HPO4, 1.0 g of NH4Cl, 0.1 g of CaCO3, 0.2 g of MgSO4, and 1 ml of a trace element solution (18). The media also contained either S0 (1.3 mM) or Na2S (1.3 mM) as the sole electron donor, and the pH was adjusted to 7.0. The vials were incubated for 5 days at 20°C on a shaking table (at 100 rpm) in both oxic conditions and anoxic conditions with nitrate present. Therefore, a total of four different culture conditions were tested for each biofilm sample. At regular time intervals, subsamples were withdrawn, and the sulfate concentrations were measured by using an ion chromatograph equipped with an AS-9 column (Dionex, Japan). The sulfate concentrations during the initial 48-h incubation were used to calculate the SORs. Abiotic controls with no biomass added were incubated in parallel to determine the spontaneous chemical S0 and H2S oxidation rates, which were subsequently subtracted from the overall oxidation rates.
Finally, the duplicate cultures of the 18- and 65-day-old RDR1 biofilms were combined and harvested by centrifugation at 10,000 × g and then subjected to 16S rRNA gene-cloning analysis.
Measurement of reduced sulfur compounds.
Elemental sulfur (S0), acid-volatile sulfide (AVS) (H2S, HS−, S2−, and FeS), and chromium-reducible sulfide (CRS) (FeS2) in the reactor effluents and in the biofilms from RDR1 and RDR2 were determined by the method described by Fossing and Jørgensen (10) and modified by Nielsen et al. (23). Three biofilm samples were taken from each RDR at time intervals and were immediately subjected to measurement. The measurement procedure was described in detail by Okabe et al. (26). Four biofilm samples were taken from each RDR at day 60, and the vertical distributions of S0 in each biofilm were determined by slicing the biofilms into 200-μm sections with a Microslicer (model DTK-1000; Dosaka EM Co., Ltd., Kyoto, Japan) without any pretreatment, as described previously (26). The biofilm sections (200 μm thick) were then subjected to the measurement procedure.
Microelectrode measurements.
A Clark-type O2 microelectrode (28), an H2S microelectrode (29), and liquid ion-exchanging membrane microelectrodes for pH and NO3− (8) were prepared and calibrated as previously described. A biofilm was taken from RDR1 at day 14 and day 60 and incubated in the synthetic medium for about 3 h prior to measurement, which ensured steady-state concentration profiles. The medium contained the following; NaNO3 (0 and 250 μM), MgSO4 · 7H2O (900 μM), NH4Cl (600 μM), Na2HPO4 (570 μM), MgCl2 · 6H2O (84 μM), CaCl2 (200 μM), EDTA · 2Na (270 μM), and sodium propionate (900 μM) as the sole carbon source. Propionate was used because propionate-utilizing Desulfobulbus spp. were the predominant SRB in this type of wastewater biofilms (12, 26). The medium without NaNO3 was used to determine the potential in situ sulfate reduction rates in the biofilm. All measurements were performed in a water chamber containing 1.8 liters of synthetic medium at 20°C. The average liquid flow velocity (2 to 3 cm s−1) above the biofilm was provided by a Pasteur pipette blowing a mixture of air and N2 gas onto the water surface. Each measurement was performed three times at different positions by advancing the microelectrodes in steps of 50 μm through the biofilm. More details concerning the measurement procedure have been described elsewhere (12). In this study, microelectrode measurements for the RDR2 biofilm could not be performed due to time constraints.
Net specific consumption and production rates (in μmol cm−3 h−1) were estimated from the measured microprofiles by using Fick's second law of diffusion. The details of this method have been described previously by Lorenzen et al. (21). The diffusive fluxes of NO3− and H2S in the sulfide-oxidizing zone were calculated by using Fick's first law. We used molecular diffusion coefficients of 1.23 × 10−5 cm2 s−1 for NO3− (3) and 1.39 × 10−5 cm2 s−1 for H2S (19) at 20°C.
16S rRNA gene-cloning and phylogenetic analysis.
An aliquot of the homogenized RDR1 biofilm sample (0.2 ml) and 200-mg portions of centrifuged pellets of the preincubated 18- and 65-day-old RDR1 biofilm cultures were subjected to DNA extraction. DNA was extracted using a Fast DNA Spin kit (BIO 101) as described in the manufacturer's instructions. The nearly full-length 16S rRNA genes from mixed bacterial DNA were amplified by PCR using the universal primer set for Bacteria, 27f and 1492r (20). To minimize nonspecific annealing of the primers to nontarget DNA, a hot-start PCR program was used for all amplifications.
One microliter of the PCR-amplified bacterial 16S rRNA gene was directly ligated into the pGEM-T vector cloning system (Promega) and transformed into competent cells (high-efficiency Escherichia coli JM109; Promega) as described in the manufacturer's instructions. Plasmids were extracted and purified from clones with the Wizard Plus Minipreps DNA purification system (Promega) used in accordance with the manufacturer's instructions. The 16S rRNA gene inserts were sequenced by using an ABI model 310 genetic analyzer with a BigDye terminator Ready Reaction kit (Applied Biosystems).
All sequences were checked for chimeric artifacts by the CHECK_CHIMERA program in Ribosomal Database Project II (7) and were compared with the sequences available in public databases (GenBank and DDBJ) by the BLAST system (1). Sequence data were aligned with the CLUSTAL W package (36). Phylogenetic trees were constructed by the neighbor-joining method (33). Bootstrap resampling analysis for 100 replicates was performed to estimate the confidence of tree topologies.
FISH.
Biofilm samples taken on days 18 and 70 from RDR1 and RDR2 were fixed in a 4% paraformaldehyde solution and embedded in Tissue-Tek OCT compound. After freezing at −20°C, vertical 20-μm sections (cross sections) of the fixed biofilms were prepared (26). Dehydration and in situ hybridization were performed according to the procedure described by Amann (2). The following oligonucleotide probes were used: G123T (S-G-Thioth-697-a-A-18), specific for Thiothrix spp. (16); S-*-SO07-0655-a-A-19, specific for the SO07 bacterium belonging to the γ-Proteobacteria (13); and S-S-Tmsde-0257-a-A-18 (5′-AACCCGCTACCCGTCATT-3′), specific for Thiomicrospira denitrificans, designed by using the ARB program (http://www.arb-home.de) in this study. The previously published optimal hybridization conditions were used for each probe. The specificity of probe S-S-Tmsde-0257-a-A-18 was empirically evaluated using the following strains as reference strains: Thiomicrospira denitrificans DSM1251, Thiobacillus denitrificans JCM3870, Atopobium parvulum JCM10300, and Sulfurimonas autotrophica JCM11897. In addition, the specificity of this probe was also tested with the cells in the cultures used for the SOR determination as mentioned above. The hybridization stringency was adjusted by adding formamide to the hybridization buffer (40% for S-S-Tmsde-0257-a-A-18). All probes were synthesized and labeled with tetramethylrhodamine-5-isothiocyanate (TRITC) or with fluorescein isothiocyanate at the 5′ end by TaKaRa Shuzo Co., Ltd. (Shiga, Japan). After hybridization and a washing step, the slides were allowed to air dry and mounted in antifading solution (Slow Fade Light; Molecular Probe, Eugene, Oreg.). Fluorescent and phase-contrast images were recorded with an LSM 510 confocal scanning laser microscope (Carl Zeiss) equipped with an argon laser (488 nm) and an HeNe laser (543 nm). Image combining was performed with the standard software (LSM 510 software, version 2.01) provided by Zeiss.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of the clones used for the phylogenetic analysis are AB181494 to AB181501.
RESULTS
Reactor performance.
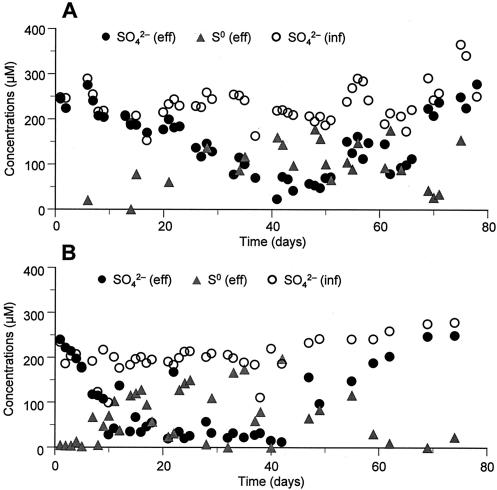
Time courses for different sulfur compounds (sulfate [SO42−] and elemental sulfur [S0]) in RDR1 and RDR2 during the 80 days of operation are presented in Fig. 1. The SO42− concentrations in the effluents started to decrease after the first few days and reached the lowest level (about 10 to 25 μM) at days 20 to 40 in both reactors. Although sulfate reduction occurred slightly earlier in RDR2 than in RDR1 due to a higher water temperature, the concentrations of SO42− and S0 in the effluent were well balanced, indicating that no other intermediate played an important role in the sulfur cycle. Finally, the effluent SO42− concentration returned to the level in the influent (ca. 250 μM SO42−) at day 70. On the other hand, the concentrations of S0 in the effluents were almost equivalent to the amounts of SO42− reduced. The change in the S0 concentration was the inverse of the change in the SO42− concentration. The concentrations of other reduced sulfur compounds, including H2S, HS−, and S2−, and CRS (FeS2) in the effluents of RDR1 and RDR2 were very low (less than 1.0 μM for AVS and CRS, respectively) during the experiments. After day 70, sulfur transformation in the reactors could not be seen from the mass balance of SO42− and S0 in both reactors because the internal sulfur cycle was completely developed. Although sulfate reduction and the subsequent sulfur oxidation occurred slightly earlier in RDR2 than in RDR1, the successional patterns of the sulfur cycle were essentially identical.
FIG. 1.
Time courses for different sulfur compounds in the reactor influent (inf) and effluent (eff) during the experiments. (A) RDR1. (B) RDR2.
Reduced sulfur compounds in the biofilm.
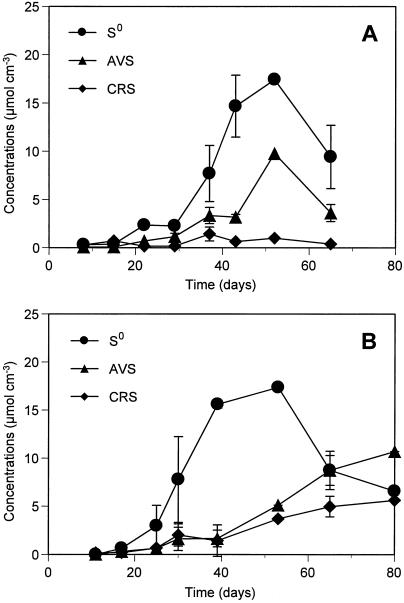
Time-dependent accumulations of reduced sulfur compounds, such as S0, AVS (FeS, H2S, HS−, and S2−), and CRS (FeS2) in the biofilms are shown in Fig. 2. The biofilm thickness gradually increased from ca. 300 to 400 μm at days 8 to 15 to ca. 1,000 to 1,200 μm at days 35 to 42 and thereafter remained more or less constant in both reactors. The biofilm in RDR2 grew faster than that in RDR1, but the thicknesses at steady state were comparable. In addition, the biofilms obviously became denser and darker with time. The reduced sulfur compounds gradually accumulated in the biofilms from the beginning of development. Among the reduced sulfur compounds, S0 was the most abundant sulfur pool in both biofilms. Significant accumulation of S0 up to a concentration of 18 μmol cm−3 was observed during days 43 to 52, when the SO42− concentration in the RDR1 effluent was lowest (Fig. 1). Then the concentration of S0 decreased after day 52. This decrease in the S0 concentration was probably due to an increase in biological S0-oxidizing activity (Fig. 3A). The accumulation pattern for AVS, the second abundant sulfur pool, was similar to that for S0. The CRS concentration was low throughout the experiment. Higher AVS and CRS concentrations were detected in the late biofilm developmental stage (at day 80) in RDR2 (Fig. 3B), which was probably due to the higher sulfate-reducing activity caused by the higher water temperature. The trend toward accumulation of reduced sulfur compounds in the RDR2 biofilm was, however, essentially the same as that in the RDR1 biofilm, suggesting that the two reactors had the same sulfur cycle succession.
FIG. 2.
Accumulation of S0, AVS (H2S, HS−, S2−, and FeS), and CRS (FeS2) in the developing biofilms. (A) RDR1. (B) RDR2. The error bars indicate the standard deviations of the measurements for three biofilm specimens.
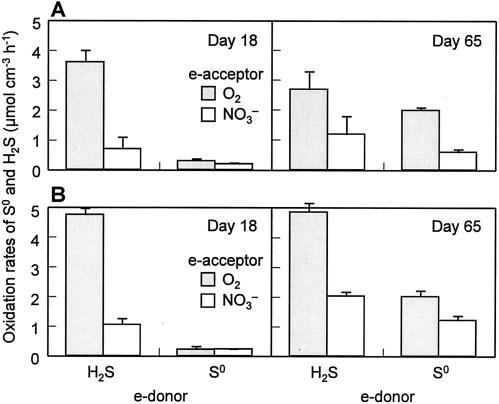
FIG. 3.
Potential sulfur and sulfide oxidation rates of 18-day-old and 65-day-old biofilms taken from RDR1 (A) and RDR2 (B). The rates were determined in batch experiments with O2 or NO3− as the sole electron acceptor (e-acceptor). The error bars indicate the standard deviations for duplicate experiments.
Potential SORs of the biofilm.
The potential S0 and H2S oxidation rates of the biofilms were determined at the early and late developmental stages of the sulfur cycle (Fig. 3). For the 18-day-old biofilm of RDR1, the aerobic and anaerobic H2S oxidation rates were 3.7 ± 0.4 μmol cm−3 h−1 and 0.8 ± 0.4 μmol cm−3 h−1, respectively, whereas the aerobic and anaerobic S0 oxidation rates were less than 0.3 μmol cm−3 h−1. As the biofilm grew, the aerobic and anaerobic S0 oxidation rates increased by seven- and threefold, respectively, at day 65, whereas the aerobic and anaerobic H2S oxidation rates remained relatively constant. The rates of S0 and H2S oxidation obtained for RDR1 at days 18 and 65 did not differ significantly from those determined for RDR2 (t test; α = 0.025). Similar trends (i.e., increase in S0 oxidation rates and no significant change in H2S oxidation rates from day 18 to day 65) were also found in both biofilms (Fig. 3B). These results indicate that SOB populations responsible for aerobic and anaerobic H2S oxidation were already developed to some extent in the early phase and that after this SOB that were capable of oxidizing S0 to SO42− developed in the late-phase biofilm.
Vertical profiles of O2, H2S, NO3−, pH, and S0.
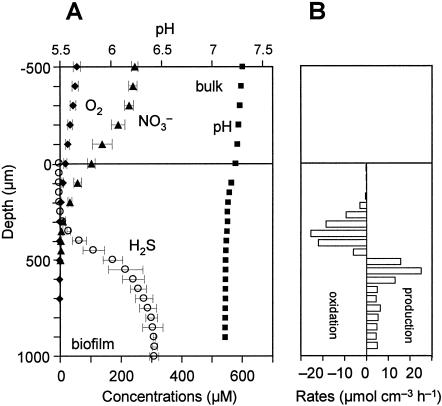
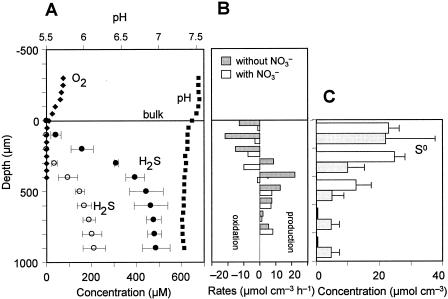
Steady-state microprofiles of O2, H2S (total sulfide), pH, and NO3− in the 14-day-old biofilm taken from RDR1 were measured with microelectrodes (Fig. 4A). Oxygen and nitrate penetrated approximately 200 μm and 500 μm from the surface, respectively. An H2S concentration of 310 μM was detected in the bottom of the biofilm, indicating that high sulfide-producing activity developed within 2 weeks. The H2S profile barely overlapped the O2 profile, but it overlapped the NO3− profile, indicating that H2S was mainly oxidized anaerobically in the 200- to 500-μm zone with a maximum specific H2S oxidation rate of 26 μmol cm−3 h−1 (Fig. 4B). A similar trend was also found in the 60-day-old biofilm taken from RDR1 (Fig. 5A). The H2S concentration was slightly lower than the day 14 concentration, but H2S oxidation occurred in the same zone (200 to 500 μm) when NO3− was present. When NO3− was removed, the H2S concentration rapidly increased and reached 430 μM at a depth of 500 μm. The H2S produced was completely oxidized at the biofilm surface with a maximum specific rate of 23 μmol cm−3 h−1, showing that there was high potential sulfide-oxidizing activity (Fig. 5B).
FIG. 4.
(A) Steady-state concentration profiles for O2, H2S, NO3−, and pH in a 14-day-old wastewater biofilm taken from RDR1. The biofilm was incubated in DO-controlled (DO concentration, approximately 60 μM) synthetic medium with about 250 μM NO3−. The error bars indicate the standard deviations for three measurements at different positions. (B) Spatial distributions of the specific H2S production and oxidation rates. The rates were calculated based on the corresponding microprofiles shown in panel A. The biofilm surface was at a depth of 0 μm.
FIG. 5.
(A) Steady-state concentration profiles for O2, H2S, NO3−, and pH in a 60-day-old wastewater biofilm taken from RDR1. The biofilm was incubated in DO-controlled (DO concentration, approximately 80 μM) synthetic medium with no addition of NO3− (solid circles) and with 90 μM NO3− (open circles). The error bars indicate the standard deviations for three measurements at different positions. (B) Spatial distributions of the specific H2S production and oxidation rates when NO3− was absent and present. The rates were calculated based on the corresponding microprofiles shown in panel A. (C) Vertical distributions of S0 in the biofilms obtained from RDR1 (gray bars) and RDR2 (open bars). The error bars indicate the standard deviations of the measurements for four biofilm specimens. The biofilm surface was at a depth of 0 μm.
The vertical distributions of elemental sulfur (S0) that accumulated in the 60-day-old biofilms taken from RDR1 and RDR2 are shown in Fig. 5C. No significant accumulation of S0 was observed in both biofilms at day 14 (Fig. 2). The concentration of S0 was highest (ca. 22 μmol cm−3) in the surface 200 μm and decreased with depth in the RDR1 biofilm. The large standard deviation at the surface (0 to 200 μm) was probably due to the heterogeneous structure of the biofilm surface. In the RDR2 biofilm, a high concentration of S0 (ca. 23 to 25 μmol cm−3) was also detected at the surface (0 to 400 μm), and S0 became undetectable at depths below 600 μm.
Phylogenetic analysis.
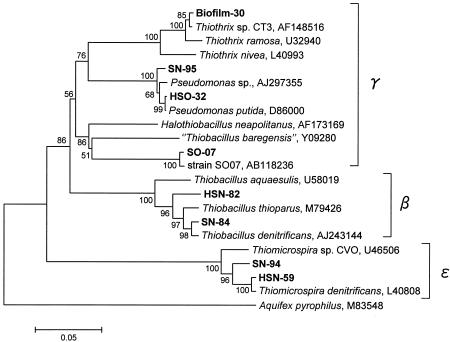
Three 16S rRNA gene clone libraries of bacteria were constructed from the RDR1 biofilm sample and the preincubated biofilm cultures to investigate the diversity of the SOB community structure. A total of 102 clones were analyzed for the biofilm clone library. Only one clone was affiliated with chemolithoautotrophic Thiothrix sp. strain CT3, with 99% sequence similarity (Table 1). None of the other phylotypes related to known SOB was retrieved directly from the biofilm without preincubation. Therefore, the biofilm samples taken from RDR1 at day 18 and day 65 were preincubated for 5 days with different combinations of electron donors (H2S and S0) and acceptors (O2 and NO3−), and then the 16S rRNA gene was analyzed to detect the SOB community. In total, 171 clones were analyzed. Among the clones analyzed, four clone types were closely related to known SOB species: Thiobacillus denitrificans (β-Proteobacteria), Pseudomonas spp. (γ-Proteobacteria), Thiomicrospira denitrificans (ɛ-Proteobacteria), and a novel sulfur-oxidizing bacterium, SO07 (γ-Proteobacteria), distantly related to Halothiobacillus spp. (Table 1 and Fig. 6). In particular, clones related to Thiomicrospira denitrificans (represented by HSN-59) were most frequently detected (detection frequency, 33%) from anaerobic incubation of an 18-day-old biofilm supplemented with NO3− and H2S. In addition, clones closely related to SO07 (represented by SO-07) were also obtained with a high detection frequency (36%) from aerobic incubation of a 65-day-old biofilm with S0 as the electron donor. Clones closely related to Thiobacillus denitrificans and Thiomicrospira denitrificans (represented by SN-84 and SN-94, respectively) were also retrieved from anaerobic incubation of a 65-day-old biofilm with S0 as the electron donor.
TABLE 1.
Phylogenetic analysis of 16S rRNA gene fragments derived from the biofilm taken from RDR1 and cultures incubated with combinations of an electron donor (S0 or H2S) and an electron acceptor (O2 or NO3−)
| Sample origin | Clone name | Closest relative (accession no.) | Taxonomic group | Frequency (%)a | % Similarityb |
|---|---|---|---|---|---|
| 18-day-old biofilm incubated withc: | |||||
| H2S + O2 | HSO-32 | Pseudomonas putida (D86000) | γ-Proteobacteria | 16 (7/44) | 99 (1,470) |
| H2S + NO3− | HSN-59 | Thiomicrospira denitrificans (L40808) | ɛ-Proteobacteria | 33 (11/33) | 99 (1,437) |
| H2S + NO3− | HSN-95 | Pseudomonas sp. (AJ297355) | γ-Proteobacteria | 6 (2/33) | 98 (1,405) |
| H2S + NO3− | HSN-82 | Thiobacillus denitrificans (AJ243144) | β-Proteobacteria | 3 (1/33) | 95 (1,371) |
| 65-day-old biofilm incubated withc: | |||||
| S0 + O2 | SO-07 | Novel sulfur-oxidizing bacterium SO07 (AB118236) | γ-Proteobacteria | 36 (17/47) | 98 (1,412) |
| S0 + NO3− | SN-84 | Thiobacillus denitrificans (AJ243144) | β-Proteobacteria | 6 (3/47) | 98 (1,458) |
| S0 + NO3− | SN-94 | Thiomicrospira denitrificans (L40808) | ɛ-Proteobacteria | 2 (1/47) | 96 (1,447) |
| 65-day-old biofilm | Biofilm-30 | Thiothrix sp. strain CT3 (AF148516) | γ-Proteobacteria | 1 (1/102) | 99 (1,422) |
The numbers in parentheses are number of clones obtained/number of clones analyzed in the library.
The numbers in parentheses are the sequence lengths compared (in base pairs).
The biofilm was taken from RDR1 and then preincubated for 5 days.
FIG. 6.
Phylogenetic distance tree showing the affiliations of 16S rRNA gene clone sequences related to SOB shown in Table 1. The nearly full-length sequences of 16S rRNA genes (>1,380 bp) were retrieved from the RDR1 biofilm (Biofilm-) and RDR1 biofilms precultured with H2S-O2 (HSO-), H2S-NO3− (HSN-), S0-O2 (SO-), and S0-NO3− (SN-) as combinations of sole electron donor and sole electron acceptor. The bar represents 5% estimated divergence, and the numbers at the nodes are bootstrap values (100 replicates) with more than 50% bootstrap support. Aquifex pyrophilius served as an outgroup.
In situ identification of SOB in the biofilm.
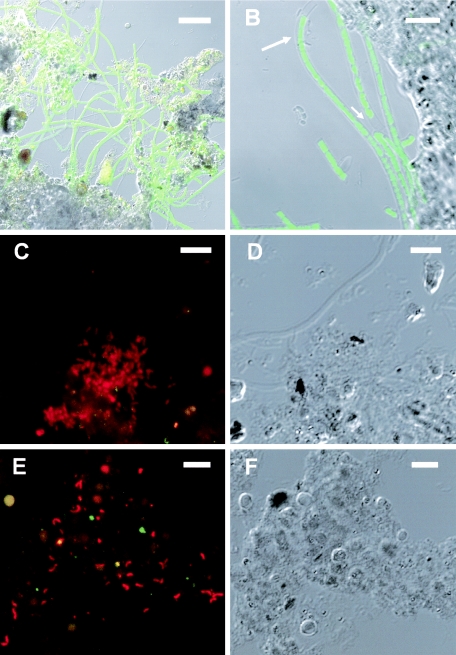
The surfaces of the RDR1 and RDR2 biofilms were whitish and were covered with a number of filamentous bacteria. Based on the 16S rRNA gene clone analyses, FISH targeting for Thiothrix, SO07, and Thiomicrospira denitrificans was conducted to investigate their in situ abundance and distributions in the biofilms obtained at days 18 and 70 from RDR1 and RDR2, respectively. Microscopic observation of a thin vertical section of the 70-day-old RDR1 biofilm revealed that probe G123T-hybridized filamentous Thiothrix was dominant at the surface of the biofilm (Fig. 7A and B). The population density of the probe G123T-hybridized Thiothrix was estimated to be 2 × 109 cells cm−3 (from a conversion of filament length to cell number; cell length, 1.5 μm) at the surface. The cells were sheathed filamentous cells (Fig. 7B), like Thiothrix sp. strain CT3 cells (32), and thus were distinguishable from type 021N Thiothrix species (lacking a sheath). In situ hybridization with the SO07-specific probe revealed that a number of rod-shaped SO07 cells (approximately 108 cells cm−3) were mainly present along with some filamentous bacteria at the surface (100 μm) of the biofilm (Fig. 7C and D). Spiral-shaped Thiomicrospira denitrificans cells that were 0.5 to 1.0 μm thick were distributed throughout the biofilm at a density of approximately 107 to 108 cells cm−3 (Fig. 7E and 7F). It should be noted that the population was larger (ca. 1 × 108 to 5 × 108 cells cm−3) near the oxic-anoxic interface. In the 18-day-old biofilm, only very few cells were detected with the probes targeting Thiothrix and SO07, whereas the population of Thiomicrospira denitrificans was comparable to that in the 70-day-old biofilm. The developmental pattern, spatial distribution, and abundance of these three SOB in the RDR2 biofilm were comparable to those in the RDR1 biofilm.
FIG.7.
In situ detection of sulfur-oxidizing bacteria in a thin vertical section (thickness, 20 μm) of a 70-day-old wastewater biofilm taken from RDR1. (A and B) FISH with fluorescein isothiocyanate-labeled probe G123T (specific for Thiothrix sp.) at the surface of the biofilm (0 to 100 μm): confocal scanning laser microscope projection image (A) and differential interference contrast (DIC) image (B). (C) FISH with TRITC-labeled probe for strain SO07 in the surface 100 μm of the biofilm. (D) DIC image of the field shown in the panel C. (E) FISH with TRITC-labeled probe for Thiomicrospira denitrificans at a depth of approximately 400 μm in the biofilm. (F) DIC image of the field shown in panel E. The yellowish signals were autofluorescence. Bars = 20 μm (A), 10 μm (B), and 5 μm (C to F).
DISCUSSION
Development of a complete sulfur cycle in the biofilm.
The succession of the SOB populations and their in situ activity in the wastewater biofilms was analyzed by a polyphasic approach that employed analyses of the biofilm reactor performance, potential SOR measurement, the 16S rRNA gene clone library, FISH, and microelectrode measurement. Two biofilm systems were independently operated, and consistent results were obtained. This clearly indicated that the succession of the internal sulfur cycle and responsible SOB populations in the complex wastewater biofilms was reproducible. The results obtained from all these experiments demonstrated that a complete internal sulfur cycle was established via S0 accumulation within approximately 80 days in the biofilm system. The formation of S0 in H2S oxidation is mainly dependent on the rate of sulfide production by SRB, the availability of electron acceptors (i.e., O2 and NO3−), and the size and type of the SOB populations in the biofilm. In general, the development of the complete sulfur cycle in this biofilm can be split into two phases, (i) a sulfur-accumulating phase and (ii) a sulfate-producing phase.
Sulfur-accumulating phase.
In the first phase (until about day 40), partial oxidation of H2S to S0 occurred as a result of the rapid development of sulfate-reducing activity (Fig. 4A and B), resulting in the accumulation of S0 in the biofilm (Fig. 2 and 5C). During this period, although sulfate-reducing activity had developed (Fig. 4A), the SOB communities were not sufficiently developed yet, and available electron acceptors, such as O2 and NO3− for SOB, were somewhat limited in the biofilm (Fig. 4A). Thus, the sulfide production rate (SRB activity) exceeded the maximum sulfide-oxidizing capacity of the SOB present in the biofilm. In order to be able to oxidize increasing amounts of H2S produced by SRB, the SOB have to convert H2S to S0 (HS− → S0 + H+ + 2e−) instead of SO42− (HS− + 4H2O→ SO42− + 9H+ + 8e−), thereby keeping the electron flux constant (39). This prevents the accumulation of sulfide in the biofilm.
The 16S rRNA gene-cloning analysis of the 18-day-old biofilm sample revealed that Thiomicrospira denitrificans, Thiobacillus denitrificans, and Pseudomonas spp. (6, 35) first appeared in the biofilm during the first phase (Table 1). In addition, the FISH analysis revealed that a high concentration (ca. 107 to 108 cells cm−3) of Thiomicrospira denitrificans was distributed throughout the biofilm, especially near the oxic-anoxic interface in the 18-day-old biofilm (ca. 1 ×108 to 5 ×108 cells cm−3). This is because Thiobacillus denitrificans and Thiomicrospira denitrificans prefer extremely low oxygen concentrations (optimum growth occurs at ca. 1% of air saturation) (17, 37, 38). Microelectrode measurements showed that the H2S profiles did not really overlap the O2 profile when NO3− was present, indicating that H2S was oxidized mainly anaerobically during the first phase (Fig. 4A). The anaerobic H2S oxidation zone (200 to 500 μm from the surface) shown in Fig. 4B overlapped the main location of Thiomicrospira denitrificans in the biofilm. Based on these results, it could be speculated that Thiomicrospira denitrificans was one of the main contributors to the partial anaerobic oxidation of H2S to S0, which led to the accumulation of S0 in the biofilm during the first phase.
The contribution of these nitrate-reducing SOB to sulfide oxidation in the biofilm systems was roughly estimated. When the oxidation of H2S was carried out with NO3− as the electron acceptor and S0 as the end product, it could be described by the following stoichiometric equation: 5HS− + 2NO3− + 7H+ → 5S0 + N2 + 6H2O (equation 1). Based on this equation, the oxidation of 1 mol of HS− requires 0.4 mol of NO3− and produces 1 mol of S0 (HS−:NO3−:S0, 1:0.4:1; HS−/NO3−, 2.5). The average concentrations of SO42− reduced, NO3− reduced, and S0 produced in RDR1 were 142 μM SO42−, 67 μM NO3−, and 125 μM S0, respectively, during days 40 to 55, which gives a ratio of HS− to NO3− to S0 of 1:0.47:0.88. A similar ratio (HS−:NO3−:S0, 1:0.53:0.86) was found in RDR2 during days 30 to 45. These ratios agree reasonably well with the theoretical ratio. The slightly higher values for NO3− could be explained by the occurrence of heterotrophic denitrification and aerobic oxidation of H2S. Furthermore, the ratio of H2S flux to NO3− flux determined from the concentration gradients of H2S and NO3− (i.e., in situ H2S oxidation and NO3− consumption rates) in the H2S-oxidizing zone (ca. 200 to 400 μm), where both profiles overlapped, was calculated to be 2.7 (Fig. 4A). This ratio was also close to the theoretical value, 2.5, as described in equation 1. It should be noted that the sulfur (S0) production was maximal at this ratio.
Sulfate-producing phase.
The biofilm S0 oxidation rates determined by the batch experiments significantly increased from day 18 to day 65 in both biofilms (Fig. 3). This result suggested that the SOB populations particularly responsible for S0 oxidation developed in the second phase. The 16S rRNA gene-cloning and FISH analyses revealed that the SOB populations developed further and diversified with time; in particular, novel strain SO07 and Thiothrix sp. became numerically important members of the SOB in the biofilms (Table 1). High concentrations of novel strain SO07 (ca. 108 cells cm−3) and Thiothrix sp. (ca. 2 × 109 cells cm−3) were present mainly in the oxic surface zone (0 to 200 μm), where S0 mainly accumulated (Fig. 7A to D). This result suggested that the novel SOB strain SO07 could be responsible for oxidation of S0 to SO42− under oxic conditions. Further isolation and characterization of strain SO07 revealed that strain SO07 was an aerobic chemolithoautotrophic sulfur-oxidizing bacterium capable of oxidizing S0 and H2S to SO42− (13). More details concerning the ecophysiological characteristics of strain SO07 have been reported previously (13). Consequently, the overall sulfide-oxidizing capacity of the biofilm was high enough to completely oxidize or efficiently utilize all of the H2S produced by SRB and the accumulated S0 to SO42− among the SOB community members. This is probably a reason for the decrease in the S0 concentrations in the reactor effluents and in the biofilms after around day 40 (Fig. 1 and 2).
In the final stage of development of the sulfur cycle, the probe G123T-hybridized filamentous organism Thiothrix sp. became the most abundant SOB in the 70-day-old biofilms (Fig. 7A and B). The sheathed filaments and 16S rRNA gene sequence analysis suggested that the G123T-hybridized Thiothrix was most likely a member of Thiothrix sp. strain CT3. The versatile physiological ability of Thiothrix sp. strain CT3 to utilize various reduced sulfur compounds with either O2 or NO3− as the electron acceptor and the heterotrophic growth on acetate (32) might provide a competitive advantage in a wastewater biofilm growing under microaerophilic conditions in which acetate was often detected (25).
Recently, specific thiosulfate oxidation rates of pure cultures of Thiothrix isolated from activated sludge (32) and Thiothrix-dominated activated sludge (24) have been determined to be 1.9 × 10−11 and 0.9 × 10−11 mmol S2O32− cell−1 h−1, respectively. By multiplying these oxidation rates by the population density of Thiothrix determined in the biofilms (ca. 2 × 109 cells cm−3), the potential thiosulfate oxidation rate of Thiothrix in the biofilms was calculated to be 18 to 38 μmol S2O32− cm−3 h−1. This oxidation rate is 2 orders of magnitude higher than the average sulfate reduction rate (0.26 μmol cm−3 h−1) of the biofilm reactors. The thiosulfate oxidation rate may be slightly different from the sulfide oxidation rate, and the availability of electron acceptors in the biofilms was uncertain. Nevertheless, this calculation suggests that the population of Thiothrix in the surface biofilms was large enough to completely oxidize all of the H2S produced to SO42−, which was clearly demonstrated by microelectrode measurements when NO3− was absent (Fig. 5A). Therefore, it could be speculated that the probe G123T-hybridized Thiothrix must have contributed significantly to H2S oxidation in the late stage of development. Although an abundant Thiothrix population was detected by FISH analysis, no clone related to Thiothrix was retrieved from the biofilm cultures incubated with S0 and H2S. The incubation in liquid synthetic media with vigorous shaking probably inhibited the growth of filamentous Thiothrix.
As the biofilm grew, the active sulfide production zone moved upward to the oxic-anoxic interface and intensified with time due to the increasing SRB activity (12). Sulfate reduction and sulfide oxidation were therefore found to occur in close proximity at the oxic-anoxic interface in the biofilm (Fig. 5A), as in other wastewater biofilms (12, 19, 22, 26). This probably enabled SOB to efficiently utilize electrons that H2S carried, and thus the turnover rate of H2S was very high. The average turnover rates of H2S in the H2S-oxidizing zone in the biofilm were 36 s and 18 s when NO3− was present and absent, respectively, which were determined from the microprofiles in Fig. 5A. Such high H2S turnover rates could be attributed to biological oxidation because the time scale of the spontaneous chemical reaction of H2S and O2 could be in the range of minutes to several hours (9, 22). This indicates that the chemical oxidation of H2S does not play an important role in wastewater biofilms growing under microaerophilic conditions.
In summary, the data sets resulting from these different approaches were integrated to obtain a clear understanding of how the SOB community structure and the complex internal sulfur cycle are established in a wastewater biofilm. Although the water quality of wastewater fluctuated with time and the wastewater biofilms were generally heterogeneous, the succession of the internal sulfur cycle and the responsible SOB populations in the complex wastewater biofilms was reproducible. However, the contributions of each phylogenetic SOB group to the overall sulfide oxidation in the biofilm were not really clear in this study. Therefore, further molecular ecological studies using, for example, microautoradiography combined with fluorescence in situ hybridization with 35SO42− or H14CO3− are required to analyze which phylogenetic groups of SOB preferentially utilize which types of reduced sulfur compounds (H2S, S0, and so on) under in situ conditions. This obviously would provide more detailed information on a complex sulfur cycle that occurs in microaerophilic biofilms.
Acknowledgments
This work was partially supported by grant-in-aid 13650593 for developmental scientific research from the Ministry of Education, Science and Culture of Japan. This study was also carried out as part of “The Project for Development of Technologies for Analyzing and Controlling the Mechanism of Biodegrading and Processing,” which was entrusted by the New Energy and Industrial Technology Development Organization (NEDO). T.I. is supported by a research fellowship from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole-cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Andrussow, L. 1969. Diffusion, p. 513-727. In Landolt-Börnstein Zahlenw Funkt, 6th ed., vol. II5a. Springer, Berlin, Germany.
- 4.Buisman, C., P. Ijspeert, A. Janssen, and G. Lettinga. 1990. Kinetics of chemical and biological sulphide oxidation in aqueous solutions. Water Res. 24:667-671. [Google Scholar]
- 5.Chen, K. Y., and J. C. Morris. 1972. Kinetics of oxidation of aqueous sulfide by O2. Environ. Sci. Technol. 6:529-537. [Google Scholar]
- 6.Chung, Y.-C, C. Huang, and C.-P. Tseng. 1996. Biodegradation of hydrogen sulfide by a laboratory-scale immobilized Pseudomonas putida CH11 biofilter. Biotechnol. Prog. 12:773-778. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Beer, D., A. Schramm, C. M. Santegoeds, and M. Kühl. 1997. A nitrite microsensor for profiling environmental biofilms. Appl. Environ. Microbiol. 63:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eary, L. E., and J. A. Schramke. 1990. Rates of inorganic oxidation reactions involving dissolved oxygen, p. 379-396. In D. C. Melchior and R. L. Basset (ed.), Chemical modeling of aqueous systems II. American Chemical Society, Washington, D.C.
- 10.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 11.Ito, T., J. L. Nielsen, S. Okabe, Y. Watanabe, and P. H. Nielsen. 2002. Phylogenetic identification and substrate uptake patterns of sulfate-reducing bacteria inhabiting an oxic-anoxic sewer biofilm determined by combining microautoradiography and fluorescent in situ hybridization. Appl. Environ. Microbiol. 68:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, T., S. Okabe, H. Satoh, and Y. Watanabe. 2002. Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl. Environ. Microbiol. 68:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., K. Sugita, and S. Okabe. 2004. Isolation, characterization and in situ detection of a novel chemolithoautotrophic sulfur-oxidizing bacterium in wastewater biofilms growing under microaerophilic conditions. Appl. Environ. Microbiol. 70:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, A. J. H., R. Sleyster, C. van der Kaa, A. Jochemsen, J. Bontsema, and G. Lettinga. 1995. Biological sulphide oxidation in a fed-batch reactor. Biotechnol. Bioeng. 47:327-333. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen, B. B. 1982. Ecology of the bacteria of the sulphur cycle with special reference to anoxic-oxic interface environments. Phil. Trans. R. Soc. Lond. 298:543-561. [DOI] [PubMed] [Google Scholar]
- 16.Kanagawa, T., Y. Kamagata, S. Aruga, T. Kohno, M. Horn, and M. Wagner. 2000. Phylogenetic analysis of and oligonucleotide probe development for Eikelboom type 021N filamentous bacteria isolated from bulking activated sludge. Appl. Environ. Microbiol. 66:5043-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly, D. P., and A. H. Harrison. 1989. Genus Thiobacillus, p. 1842-1858. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, 1st ed., vol. 3, Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 18.Kuenen, J. G., L. A. Robertson, and O. H. Tuovinen. 1991. The genera Thiobacillus, Thiomicrospira, and Thiosphaera, p. 2638-2657. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, N.Y. [Google Scholar]
- 19.Kühl, M., and B. B. Jørgensen. 1992. Microsensor measurements of sulfate reduction and sulfide oxidation in compact microbial communities of aerobic biofilms. Appl. Environ. Microbiol. 58:1164-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, England.
- 21.Lorenzen, J., L. H. Larsen, T. Kjar, and N. P. Revsbech. 1998. Biosensor detection of the microscale distribution of nitrate, nitrate assimilation, nitrification, and denitrification in a diatom-inhabited freshwater sediment. Appl. Environ. Microbiol. 64:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, D. C., B. B. Jørgensen, and N. P. Revesbech. 1986. Growth pattern and yield of a chemoautotrophic Beggiatoa sp. in oxygen-sulfide microgradients. Appl. Environ. Microbiol. 52:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, P. H., W. Lee, Z. Lewandowski, M. Morison, and W. G. Characklis. 1993. Corrosion of mild steel in an alternating oxic and anoxic biofilm system. Biofouling 7:267-284. [Google Scholar]
- 24.Nielsen, P. H., M. A. de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 25.Okabe, S., T. Ito, and H. Satoh. 2003. Sulfate-reducing bacterial community structure and their contribution to carbon mineralization in a wastewater biofilm growing under microaerophilic conditions. Appl. Microbiol. Biotechnol. 63:322-334. [DOI] [PubMed] [Google Scholar]
- 26.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsing, N. B., M. Kühl, and B. B. Jørgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 55:1907-1910. [Google Scholar]
- 29.Revsbech, N. P., and B. B. Jorgensen. 1986. Microelectrodes: their use in microbial ecology. Adv. Microb. Ecol. 9:293-352. [Google Scholar]
- 30.Robertson, L., and J. G. Kuenen. 1991. The colorless sulfur bacteria, p. 385-413. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, N.Y. [Google Scholar]
- 31.Robertson, L., and J. G. Kuenen. 1. April 2002, posting date. The genus Thiobacillus. In M. Dworkin et al. (ed.), Prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.9. Springer-Verlag, Berlin, Germany. [Online.] http://link.springer-ny.com/link/service/books/10125/.
- 32.Rossetti, S., L. L. Blackall, C. Levantesi, D. Uccelletti, and V. Tandoi. 2003. Phylogenetic and physiological characterization of a heterotrophic, chemolithoautotrophic Thiothrix strain isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53:1271-1276. [DOI] [PubMed] [Google Scholar]
- 33.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 34.Santegoeds, C. M., T. G. Ferdelman, G. Muyzer, and D. de Beer. 1998. Structure and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl. Environ. Microbiol. 64:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schook, L. B., and R. S. Berk. 1978. Nitritional studies with Pseudomonas aeruginosa grown on inorganic sulfur sources. J. Bacteriol. 133:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmer-ten Hoor, A. 1975. A new type of thiosulfate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth. J. Sea Res. 9:344-350. [Google Scholar]
- 38.Timmer-ten Hoor, A. 1981. Cell yield and bioenergetics of Thiomicrospira denitrificans compared with Thiobacillus denitrificans. Antonie Leeuwenhoek 47:231-243. [DOI] [PubMed] [Google Scholar]
- 39.Visser, J. M., L. A. Robertson, H. W. van Verseveld, and J. G. Kuenen. 1997. Sulfur production by obligately chemolithoautotrophic Thiobacillus species. Appl. Environ. Microbiol. 63:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]