Abstract
After the introduction of the rotavirus vaccine into the Universal Immunization Program in India in 2016, relatively few studies have assessed the prevalence and epidemiological patterns of acute gastroenteritis (AGE) among hospitalized children ≤5 years of age. We used a uniform protocol to recruit children with AGE as well as standardized testing and typing protocols. Stool specimens from children with AGE younger than 5 years of age admitted to six hospitals in three cities in India were collected from January 2017 through December 2019. Norovirus was detected by real-time reverse transcription-polymerase chain reaction (RT-qPCR) followed by typing positive specimens by conventional RT-PCR and Sanger sequencing. Norovirus was detected in 322 (14.8%) of 2182 specimens with the highest rate in 2018 (17.6%, 146/829), followed by 2019 (14.4%, 122/849) and 2017 (10.7%, 54/504). Rotavirus vaccine status was known for 91.6% of the children of which 70.4% were vaccinated and 29.6% not. Norovirus positivity in rotavirus-vaccinated children was 16.3% and 12% in unvaccinated children. GII.4 Sydney[P16] (39.3%), GII.4 Sydney[P31] (18.7%), GII.2[P16] (10%), GI.3[P13] (6.8%), GII.3[P16] (5.9%), and GII.13[P16] (5%) accounted for 85.8% (188/219) of the typed strains. Our data highlight the importance of norovirus in Indian children hospitalized with AGE.
Keywords: acute gastroenteritis, hospitalized children, India, norovirus
1 |. INTRODUCTION
Acute gastroenteritis (AGE) is a major cause of morbidity and mortality in infants and young children causing up to 525 000 estimated deaths annually worldwide of which viruses account for approximately 70% of the cases.1 After the successful implementation of rotavirus vaccines, noroviruses have become the most important cause of viral gastroenteritis in many developed countries.2 Globally, the estimated societal economic burden associated with norovirus disease is approximately $60 billion, of which disease in children <5 years of age is responsible for nearly $40 billion per year.3
Noroviruses belong to the family Caliciviridae and are small non-enveloped positive-sense, single-stranded RNA viruses.4,5 The genome is approximately 7.3–8.3 kb in length and is organized into three open reading frames (ORF1–ORF3).4,5 ORF1 encodes nonstructural proteins including RNA-dependent RNA polymerase (RdRp), whereas ORF2 and ORF3 encode the major and minor structural proteins VP1 and VP2, respectively.5,6 Based on genetic differences in the major capsid protein (VP1), noroviruses have been classified into 10 genogroups (GI–GX) with genogroup GI, GII, GIV, GVIII, and GIX infecting humans.7 Of these viruses, GI and GII are associated with the majority of human disease and are further subdivided into at least nine GI and 26 GII (capsid) genotypes and 14 GI and 37 GII P-types.7
Globally, GII.4 noroviruses are the most frequently detected genotype.4 There are few reports on the disease burden of noroviruses among children less than 5 years of age in India; these range from 1.2% to 15.1%, depending on the setting which includes hospital-based, community-based, or in a birth cohort setting.8–13 There are no studies describing norovirus disease burden in rotavirus vaccinated children after the introduction of rotavirus vaccine into the Universal Immunization Program (UIP) in India in 2016.14
In this study, we determined the prevalence and genetic diversity of norovirus among children ≤5 years of age hospitalized with AGE in six hospitals in Southern and Western India after introduction of rotavirus vaccine.
2 |. MATERIALS AND METHODS
2.1 |. Study sites
The study was conducted during 2017–2019 in six hospitals across three cities in India, with two sites in Chennai (Sri Ramachandra Institute of Higher Education & Research [SRIHER] and Indian Council for Medical Research-National Institute of Epidemiology [ICMR-NIE]), one site in Vellore (Christian Medical College [CMC]), and one site in Pune (Interactive Research School for Health Affairs [IRSHA]). Of these, two sites (SRIHER and CMC) have a tertiary care hospital set up. IRSHA in Pune received samples from Bharati Hospital, Mankar Hospital, and Chinmay Hospital in Pune. ICMR-NIE received samples for norovirus testing from the Institute of Child Health and Hospital for Children in Chennai. This was a cross sectional study involving sentinel hospital-based surveillance sites having in-patient facilities for management of AGE in pediatric populations. The study was approved by the institutional review boards/ethics committees of all four sites.
2.2 |. Enrollment criteria
Children ≤5 years of age were enrolled if (i) they presented to one of the participating hospitals for treatment of AGE, or (ii) were hospitalized for at least 6h and were given supervised oral, or intravenous (IV) rehydration. Enrollment of all eligible children was done after obtaining written informed consent from the parents/guardian. AGE was defined as having ≥3 episodes of diarrhea (stools of a less formed character than usual) and/or ≥1 episodes of vomiting within a 24 h period, less than 7 days before hospital visit, which was not explained by an underlying medical condition. Exclusion criteria included: children >60 months of age, blood in stool (dysentery), or <3 episodes in a 24 h period. Detailed clinical information on onset and duration of diarrheal and/or vomiting episodes, duration of fever, degree of dehydration, and treatment provided was collected for all enrolled children. Rotavirus vaccination data and other sociodemographic information was also collected during enrollment. One diarrheal stool sample was collected from each child after enrollment. Stool specimens were transported on ice packs to the testing laboratory within 2 h of collection. Specimens were aliquoted and stored at −70°C until testing was performed. All the staff and faculties in this project were trained on collection of clinical data and norovirus testing. The four sites involved in this surveillance used a uniform protocol for recruitment of cases, norovirus testing using real-time reverse transcription-polymerase chain reaction (RT-qPCR), and sequence-based genotyping.
2.3 |. RNA extraction and molecular characterization
Viral RNA was extracted from 140 μL of a 10% stool suspension with a QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer’s instructions. RNA samples were tested for GI/GII norovirus by a multiplex RT-qPCR targeting the ORF1/ORF2 overlap region using an Ag-Path One-Step RT-PCR Kit.15,16 For GI and GII norovirus real-time results, cycle threshold (Ct) cutoff values of 35 and 37 were used as the limits of detection, respectively.16 Positive samples were dual typed by conventional RT-PCR targeting a 579 bp product for GI viruses or a 570 bp product for GII viruses followed by PCR product purification and Sanger sequencing as described previously.17 Genotypes were assigned using an online human calicivirus typing tool (https://calicivirustypingtool.cdc.gov/).7,18
2.4 |. Statistical analysis
Data were analyzed to determine the prevalence, seasonal variation, and genetic diversity of norovirus. Demographic and clinical characteristics between children testing norovirus positive versus negative was compared. STATA 16 (Stata Corp.) was used to calculate p-value using Fisher’s exact test. A p-value of <0.05 was considered statistically significant.
3 |. RESULTS
During the three surveillance years (January 2017 to December 2019), 2182 children were enrolled (Table 1). Of these, 53.1% (1158/2182) were 0–12 months old, 28% (610/2182) were 13–24 months old, while 18.9% (414/2182) children were 25–60 months old.
TABLE 1.
Norovirus positivity among hospitalized children <5 years of age with acute gastroenteritis in six hospitals in three cities in India.
| Site, city | No samples tested | Norovirus positive samples % (N) | GI positive samples % (N) | GII positive samples % (N) | GI/GII positive samples % (N) | GI dual-typed samples | GII dual-typed samples |
|---|---|---|---|---|---|---|---|
| SRIHER, Chennai | 504 | 13.3% (67/504) | 23.9% (16/67) | 76.1% (51/67) | 0 | 16 | 45 |
| CMC, Vellore | 541 | 16.5% (89/541) | 9% (8/89) | 87.6% (78/89) | 3.4% (3/89) | 5 | 61 |
| ICMR- NIE, Chennai | 851 | 14.5% (123/851) | 4.1% (5/123) | 93.5% (115/123) | 2.4% (3/123) | 2 | 72 |
| IRSHA, Pune | 286 | 15% (43/286) | 9.3% (4/43) | 88.4% (38/43) | 2.3% (1/43) | 0 | 18 |
| Total | 2182 | 14.8% (322/2182) | 10.2% (33/322) | 87.6% (282/322) | 2.2% (7/322) | 23 | 196 |
The overall norovirus positivity across the four sites was 14.8% (322/2182) ranging from 13.3% in SRIHER, Chennai to 16.5% in CMC, Vellore (Table 1). There was variation in norovirus positivity across the years, with the highest positivity occurring in 2018 (17.6%, 146/829), followed by 2019 (14.4%, 122/849) and 2017 (10.7%, 54/504). Of the 322 children with norovirus gastroenteritis, 57.8% (186/322) were male and 42.2% (136/322) female (Table 2). 55.9% (180/322) of the children with norovirus gastroenteritis were 0–12 months old, 32% (103/322) were 13–24 months old, while the remaining 12.1% (39/322) were 25–60 months. Norovirus gastroenteritis was observed throughout the year at all sites with a higher prevalence during December to May (Figure 1). Of the 322 specimens, 282 (87.6%) tested positive for GII norovirus, 33 (10.2%) for GI norovirus, and 7 (2.2%) for both GI and GII noroviruses (Table 1).
TABLE 2.
Characteristics of children <5 years of age hospitalized with acute gastroenteritis in six hospitals in three cities in India, 2017–2019.
| Variable | Norovirus positive | Norovirus negative | p Value |
|---|---|---|---|
| (N = 322) | (N = 1860) | ||
| Male | 186 (57.8%) | 1105 (59.4%) | 0.58 |
| Female | 136 (42.2%) | 755 (40.6%) | 0.59 |
| Age, mean months ± SD | 14.9 ± 10.6 | 16.2 ± 12.9 | 0.09 |
| Age group distribution | |||
| ≤1 years (0–12 months) | 180 (55.9%) | 978 (52.6%) | 0.27 |
| >1 years to ≤2 years (13–24 months) | 103 (32%) | 507 (27.3%) | 0.08 |
| >2 years to ≤5 years (25–60 months) | 39 (12.1%) | 375 (20.2%) | <0.05 |
| Duration of illness, mean ± SD | 4.1 ± 3.7 | 4.3 ± 3.6 | 0.36 |
| Fever | 151 (46.9%) | 846 (45.5%) | 0.64 |
| Vomiting | 243 (75.5%) | 1407 (75.6%) | 0.94 |
| Dehydration | |||
| No dehydration | 167 (51.9%) | 860 (46.2%) | 0.06 |
| Some dehydration | 134 (41.6%) | 822 (44.2%) | 0.39 |
| Severe dehydration | 21 (6.5%) | 178 (9.6%) | 0.08 |
| Treatment | |||
| Oral | 95 (29.5%) | 454 (24.4%) | 0.05 |
| IV fluids | 155 (48.1%) | 1036 (55.7%) | <0.05 |
| Oral and IV fluids | 72 (22.4%) | 370 (19.9%) | 0.31 |
FIGURE 1.
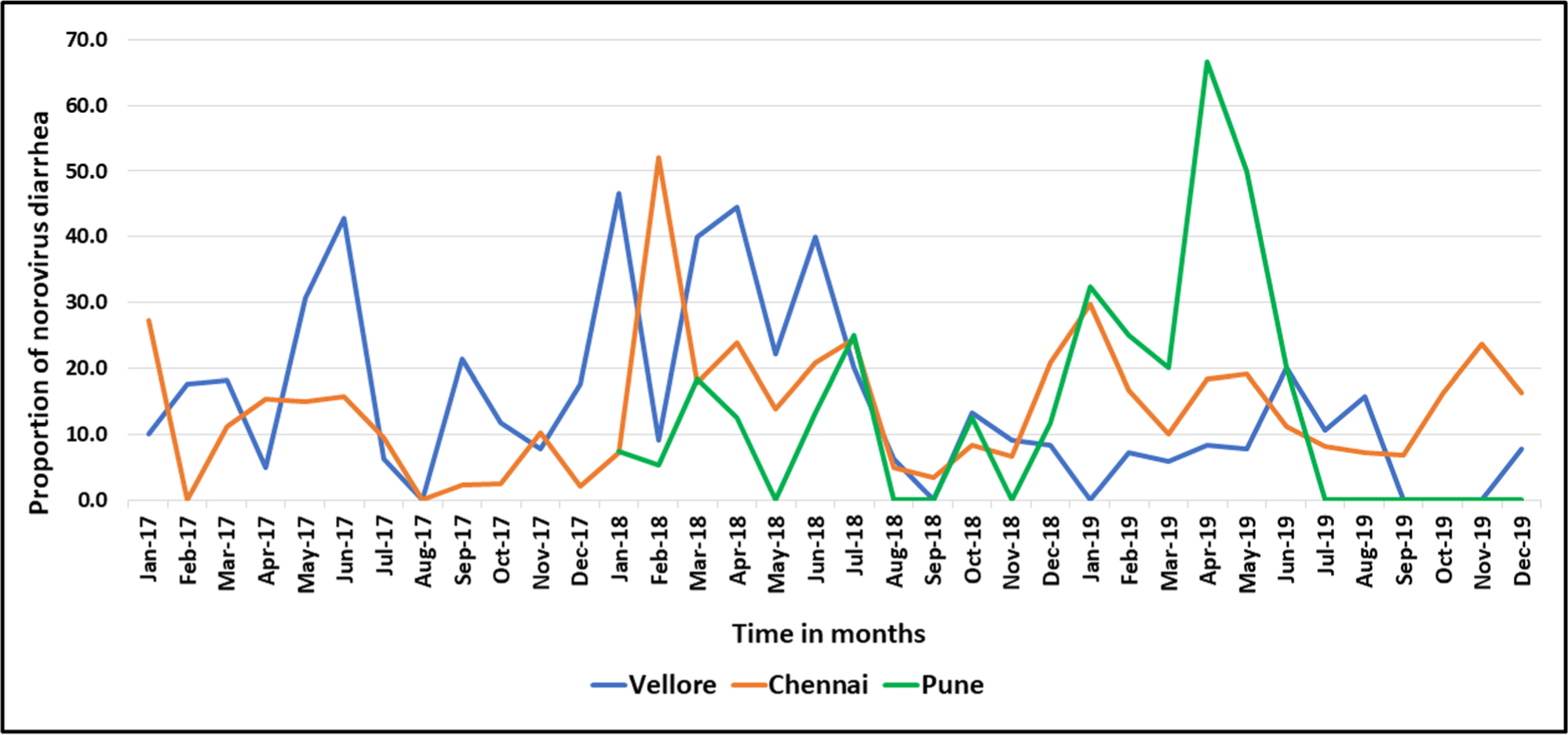
Temporal distribution of norovirus positive cases in children <5 years of age hospitalized with acute gastroenteritis in six hospitals in three cities in India, 2017–2019.
Overall, there was no significant difference in the mean age (±standard deviation) of children tested positive for norovirus (14.9 ± 10.6 months), compared to those who tested negative for norovirus (16.2 ± 12.9 months) (Table 2). However, norovirus positivity was significantly lower in children >2 years of age compared to the youngest age groups (p < 0.05). Between cases positive for norovirus and those who tested negative, there was no significant difference in magnitude of fever, frequency of vomiting, level of dehydration, and duration of illness (Table 2). A significantly higher proportion of norovirus negative cases received IV fluids compared to norovirus positive cases (p < 0.05).
Rotavirus vaccination status was obtained for 1999 (91.6%) children enrolled in the study. Of these, 1407 (70.4%) were either fully or partially vaccinated, while 592 (29.6%) were not vaccinated. Norovirus positivity in rotavirus-vaccinated children was 16.3% (229/1407) and 12% (71/592) in unvaccinated children. In the 229 rotavirus-vaccinated children with norovirus gastroenteritis, 89.1% (204/229) of the specimens tested positive for GII, 8.7% (20/229) for GI, and 2.2% (5/229) for both GI and GII. Of the 71 unvaccinated children who tested positive for norovirus, 90.1% (64/71) of the specimens tested positive for GII, 7.0% (5/71) positive for GI, and 2.8% (2/71) positive for both GI and GII.
3.1 |. Diversity of norovirus genotypes
Typing information was successfully obtained for 219/322 (68%) norovirus strains (Table 3). GII.4 Sydney was detected in 130 (59.4%) specimens followed by GII.2 (10%, 22/219), GII.3 (5.9%, 13/219), and GII.13 (5.9%, 13/219). GI.3 was the most common GI genotype accounting for 17 (74%, 17/23) of all GI noroviruses, and 7.8% (17/219) of the total dual-typed sequences detected. The remaining 24 (11%) strains included GI.1, GI.4, GI.5, GII.6, GII.7, GII.9, GII.10, GII.12, GII.14, GII.20, and GII.21. The GII.4 Sydney viruses (n = 130) were associated with 3P types: GII.P16 (86/219, 39.3%), GII.P31 (41/219, 18.7%), and GII.P4 (3/219, 1.4%). P16 was the P-type for all (n = 35) GII.2 and GII.3 viruses. GII.13 viruses (n = 13) were found in combination with GII.P16 (84.6% [11/13]) and GII.P21 (15.4% [2/13]). For GI.3 viruses, the P-types were GI.P13 (88.2% [15/17]) and GI.P10 11.8% (2/17) (Table 3).
TABLE 3.
Distribution of norovirus genotypes in six hospitals in three cities of India, 2017–2019.
| Dual genotypes | SRIHER | CMC | ICMR-NIE | IRSHA | Total (%) |
|---|---|---|---|---|---|
| GI.1[P1] | 2 | 0 | 0 | 0 | 2 (0.9) |
| GI.3[P10] | 2 | 0 | 0 | 0 | 2 (0.9) |
| GI.3[P13] | 9 | 4 | 2 | 0 | 15 (6.8) |
| GI.4[P4] | 0 | 1 | 0 | 0 | 1 (0.5) |
| GI.5[P5] | 1 | 0 | 0 | 0 | 1 (0.5) |
| GI.5[P12] | 2 | 0 | 0 | 0 | 2 (0.9) |
| GII.2[P16] | 7 | 5 | 10 | 0 | 22 (10) |
| GII.3[P16] | 5 | 1 | 5 | 2 | 13 (5.9) |
| GII.4 Sydney[P4] | 0 | 0 | 3 | 0 | 3 (1.4) |
| GII.4 Sydney[P16] | 26 | 21 | 29 | 10 | 86 (39.3) |
| GII.4 Sydney[P31] | 5 | 22 | 11 | 3 | 41 (18.7) |
| GII.6[P7] | 0 | 1 | 4 | 1 | 6 (2.7) |
| GII.7[P7] | 0 | 2 | 0 | 0 | 2 (0.9) |
| GII.9[P7] | 0 | 0 | 1 | 0 | 1 (0.5) |
| GII.10[P16] | 0 | 0 | 1 | 0 | 1 (0.5) |
| GII.12[P16] | 0 | 0 | 2 | 0 | 2 (0.9) |
| GII.12[P33] | 1 | 0 | 0 | 0 | 1 (0.5) |
| GII.13[P16] | 0 | 5 | 5 | 1 | 11 (5) |
| GII.13[P21] | 0 | 2 | 0 | 0 | 2 (0.9) |
| GII.14[P7] | 0 | 1 | 0 | 0 | 1 (0.5) |
| GII.20[P20] | 0 | 1 | 0 | 0 | 1 (0.5) |
| GII.21[P16] | 0 | 0 | 1 | 1 | 2 (0.9) |
| GII.21P[21] | 1 | 0 | 0 | 0 | 1 (0.5) |
| Total sample genotyped | 61 | 66 | 74 | 18 | 219 (68) |
The six most detected dual types were GII.4 Sydney[P16], GII.4 Sydney[P31], GII.2[P16], GI.3[P13], GII.3[P16], and GII.13[P16], which accounted for 85.8% (188/219) of all typed strains (Figure 2). The remaining 14.2% (31/219) viruses were composed of 17 other dual types, each accounting for <3% of all sequences (Figure 2). More than 10 different types were detected in Vellore and in the two sites from Chennai (Table 3) while the genotype diversity was less in Pune (six types detected). Rarely reported dual types such as GII.20[P20], GII.21,16 and GII.21[P21] were also detected.
FIGURE 2.
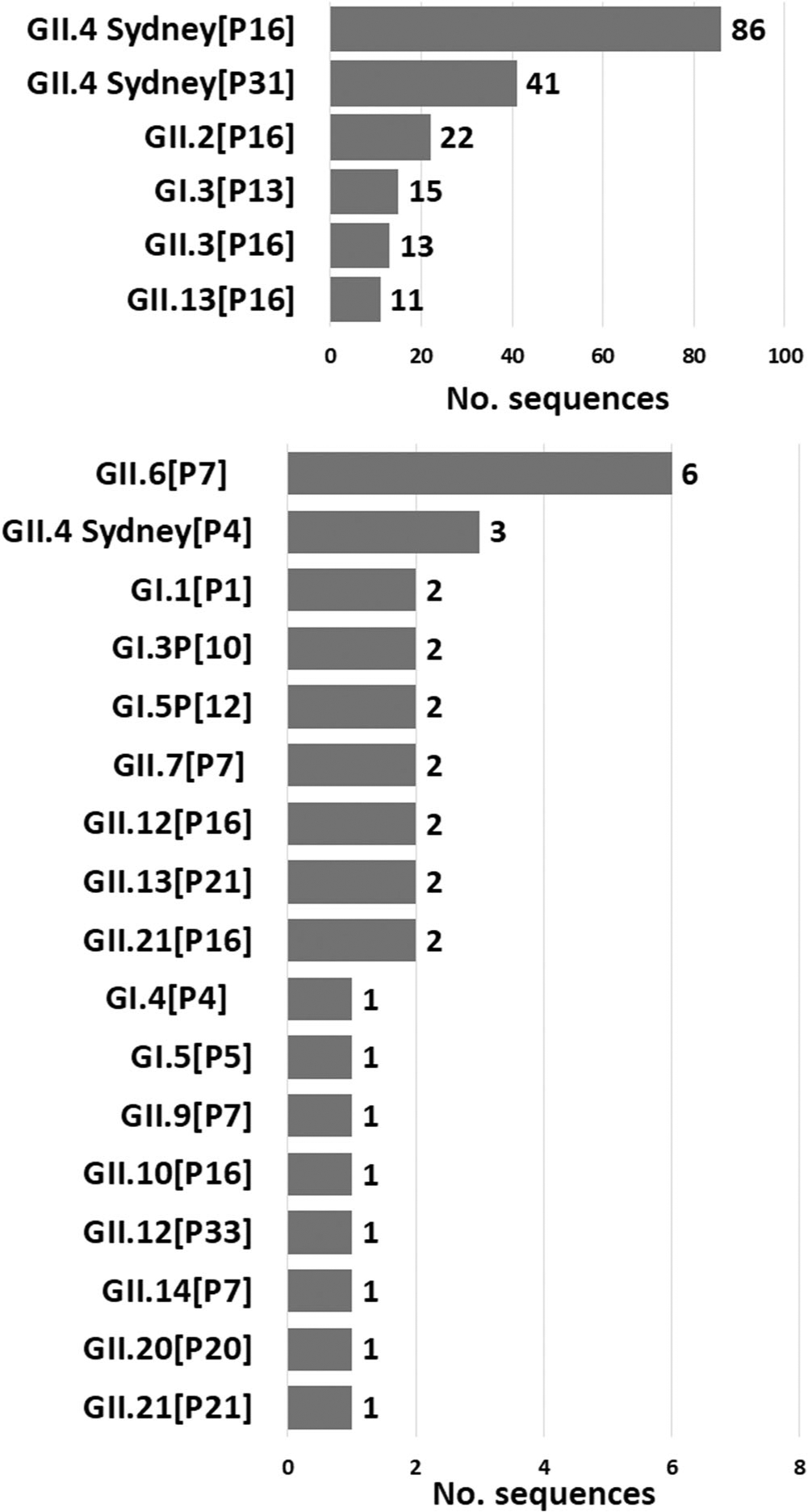
Genotype distribution of pediatric norovirus cases in India, 2017–2019. Number to the right of each bar indicates the number of sequences detected during the surveillance (the six most detected dual types are shown above in the figure, while the less common dual types are shown below).
4 |. DISCUSSION
Between January 2017 and December 2019, norovirus prevalence in children ≤5 years of age hospitalized with AGE ranged from 13.3% to 16.5% across four sites in India. Norovirus positivity was 16.3% in rotavirus-vaccinated children compared to 12% in non-vaccinated children. GII.4 Sydney was the predominant (59.4%) genotype.
To date, few studies from India have reported the prevalence of norovirus gastroenteritis in children ≤5 years hospitalized for AGE with positivity rates ranging from 1.2% to 15.1%.8–12,19,20 Most of these studies were conducted before the introduction of oral rotavirus vaccine into the UIP in India.14 In contrast to our study, these studies used conventional RT-PCR to detect norovirus, which has been shown to be less sensitive than real-time RT-PCR.21 Our data show a slight increase of norovirus prevalence in rotavirus-vaccinated children compared to unvaccinated children which is expected as numerous countries have seen an increase in the proportion of AGE due to norovirus after rotavirus vaccine introduction because norovirus makes up an increased proportion of the AGE that is remaining.2,22 Similarly, studies in several countries reported a higher proportion of norovirus in children that had been vaccinated for rotavirus.23–29
Globally, GII.4 Sydney is the most common genotype causing norovirus disease in hospitalized children with AGE.30 In our 3-year study, GII.4 Sydney was detected in 59.4% of the cases while in a recent study from eastern India, GII.4 Sydney was associated with 83.3% of the norovirus positive cases.19 GII.4 Sydney[P16] was the most common strain in our study followed by GII.4 Sydney[P31]. In a recent study on global genotype trends among children with norovirus gastroenteritis, GII.4 Sydney[P16] and GII.4 Sydney[P31] viruses comprised more than 50% of the strains.30 In our study, GI viruses were detected in 10.2% of the norovirus positive cases, which is higher than reported in other studies from India.8–12,19
Our study had several limitations. With six hospitals from three cities in south (Chennai, Vellore) and west (Pune) India, our data cannot be generalized to the entire country. In addition, we did not test for other gastroenteritis viruses as they have been frequently reported as coinfections in specimens from low and middle income countries.31 Time of enrollment varied across the sites with two sites enrolling cases between January 2017 and December 2019, whereas the remaining two sites (ICMR-NIE, Chennai and IRSHA, Pune) started enrollment 7 month later (August 2017 to December 2019), and 1 year later (January 2018 to December 2019), respectively.
In conclusion, we conducted a post rotavirus vaccine hospital-based surveillance of norovirus gastroenteritis in Indian children that were eligible for rotavirus vaccination using a uniform protocol for recruitment of cases, norovirus detection, and genotyping. Future studies on the molecular norovirus surveillance in India should ideally include multiple different geographic sites across the country to enhance our understanding of the epidemiology and strain distribution of norovirus disease. Such data will help guide introduction of pediatric norovirus vaccines that are currently in clinical trials.
ACKNOWLEDGMENTS
We thank the clinicians and families of the children who participated in the study.
Funding information
Centers for Disease Control and Prevention
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Data will be available on request due to privacy/ethical reasons.
REFERENCES
- 1.World Health Organization. Diarrhoeal disease [Internet] 2017. https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease
- 2.Payne DC, Vinjé J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in US children. N Engl J Med. 2013;368(12): 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One. 2016;11(4): e0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robilotti E, Deresinski S, Pinsky BA. Norovirus. Clin Microbiol Rev. 2015;28(1):134–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinjé J Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53(2):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorne LG, Goodfellow IG. Norovirus gene expression and replication. J Gen Virol. 2014;95(2):278–291. [DOI] [PubMed] [Google Scholar]
- 7.Chhabra P, de Graaf M, Parra GI, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100(10): 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhabra P, Chitambar SD. Norovirus genotype IIb associated acute gastroenteritis in India. J Clin Virol. 2008;42(4):429–432. [DOI] [PubMed] [Google Scholar]
- 9.Gopalkrishna V, Ganorkar N, Patil P, Hedda G, Ranshing S, Kulkarni R. Clinical, epidemiological, and molecular aspects of picornaviruses (entero, parecho) in acute gastroenteritis: a study from Pune (Maharashtra), Western India. J Med Virol. 2021;93(6): 3590–3600. [DOI] [PubMed] [Google Scholar]
- 10.Jain A, Srivastava S, Kumar V, Singh M, Gupta S, Singh K. Aetiology of childhood viral gastroenteritis in Lucknow, north India. Indian J Med Res. 2015;141(4):469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain S, Thakur N, Grover N, Vashistt J, Changotra H. Prevalence of rotavirus, norovirus and enterovirus in diarrheal diseases in Himachal Pradesh, India. Virusdisease. 2016;27(1):77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni R, Patel A, Bhalla S, Chhabra P, Cherian S, Chitambar SD. Characterization of GII.4 noroviruses circulating among children with acute gastroenteritis in Pune, India: 2005–2013. Infect, Genet Evol. 2016;37:163–173. [DOI] [PubMed] [Google Scholar]
- 13.Menon VK, George S, Sarkar R, et al. Norovirus gastroenteritis in a birth cohort in Southern India. PLoS One. 2016;11(6):e0157007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik A, Haldar P, Ray A, et al. Introducing rotavirus vaccine in the Universal Immunization Programme in India: from evidence to policy to implementation. Vaccine. 2019;37(39):5817–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park GW, Chhabra P, Vinjé J. Swab sampling method for the detection of human norovirus on surfaces. J Vis Exp. 2017; 6(120):e55205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon JL, Barclay L, Collins NR, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII. 4 recombinant viruses. J Clin Microbiol. 2017;55(7):2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chhabra P, Browne H, Huynh T, et al. Single-step RT-PCR assay for dual genotyping of GI and GII norovirus strains. J Clin Virol. 2021;134:104689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusov RL, Chhabra P, Diez-Valcarce M, Barclay L, Cannon JL, Vinjé J. Human calicivirus typing tool: a web-based tool for genotyping human norovirus and sapovirus sequences. J Clin Virol. 2021;134:104718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo M, Mitra S, De P, et al. Genetic characterization and evolutionary analysis of norovirus genotypes circulating among children in eastern India during 2018–2019. Arch Virol. 2021;166:2989–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal I, Moses PD, Sarkar R, Kang G, Menon VK, Simon A. Norovirus genogroup II gastroenteritis in hospitalized children in South India. Am J Trop Med Hyg. 2013;89(5):1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreier J, Störmer M, Mäde D, Burkhardt S, Kleesiek K. Enhanced reverse transcription-PCR assay for detection of norovirus genogroup I. J Clin Microbiol. 2006;44(8):2714–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo HL, Neill FH, Estes MK, et al. Noroviruses: the most common pediatric viral enteric pathogen at a large university hospital after introduction of rotavirus vaccination. J Pediatric Infect Dis Soc. 2012;2(1):57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentile Á, Areso MS, Degiuseppe JI, et al. Role of noroviruses in sporadic acute gastroenteritis cases from children attending a large referral children’s hospital in Buenos Aires City, Argentina. Pediatr Infect Dis J. 2023;42(2):94–98. [DOI] [PubMed] [Google Scholar]
- 24.Bucardo F, Reyes Y, Svensson L, Nordgren J. Predominance of norovirus and sapovirus in Nicaragua after implementation of universal rotavirus vaccination. PLoS ONE. 2014;9(5):e98201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challappa R, Saito M, Mejia C, et al. Burden of norovirus and rotavirus in children after rotavirus vaccine introduction, Cochabamba, Bolivia. Am J Trop Med Hyg. 2016;94(1):212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rönnelid Y, Bonkoungou I, Ouedraogo N, Barro N, Svensson L, Nordgren J. Norovirus and rotavirus in children hospitalised with diarrhoea after rotavirus vaccine introduction in Burkina Faso. Epidemiol Infect. 2020;148:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos VS, Gurgel RQ, Cavalcante SMM, et al. Acute norovirus gastroenteritis in children in a highly rotavirus-vaccinated population in Northeast Brazil. J Clin Virol. 2017;88:33–38. [DOI] [PubMed] [Google Scholar]
- 28.Quintero-Ochoa G, Romero-Argüelles R, Aviles-Hernández A, et al. Viral agents of gastroenteritis and their correlation with clinical symptoms in rotavirus-vaccinated children. Infect, Genet Evol. 2019;73:190–196. [DOI] [PubMed] [Google Scholar]
- 29.Lambisia AW, Onchaga S, Murunga N, Lewa CS, Nyanjom SG, Agoti CN. Epidemiological trends of five common diarrhea-associated enteric viruses pre- and post-rotavirus vaccine introduction in Coastal Kenya. Pathogens. 2020;9(8):660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannon JL, Bonifacio J, Bucardo F, et al. Global trends in norovirus genotype distribution among children with acute gastroenteritis. Emerging Infect Dis. 2021;27(5):1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888): 209–222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Data will be available on request due to privacy/ethical reasons.


