Abstract
Pathogenic strains of Escherichia coli, such as E. coli O157:H7, have a low infectious dose and an ability to survive in acidic foods. These bacteria have evolved at least three distinct mechanisms of acid resistance (AR), including two amino acid decarboxylase-dependent systems (arginine and glutamate) and a glucose catabolite-repressed system. We quantified the survival rates for each AR mechanism separately in clinical isolates representing three groups of Shiga toxin-producing E. coli (STEC) clones (O157:H7, O26:H11/O111:H8, and O121:H19) and six commensal strains from ECOR group A. Members of the STEC clones were not significantly more acid resistant than the commensal strains when analyzed using any individual AR mechanism. The glutamate system provided the best protection in a highly acidic environment for all groups of isolates (<0.1 log reduction in CFU/ml per hour at pH 2.0). Under these conditions, there was notable variation in survival rates among the 30 O157:H7 strains, which depended in part on Mg2+ concentration. The arginine system provided better protection at pH 2.5, with a range of 0.03 to 0.41 log reduction per hour, compared to the oxidative system, with a range of 0.13 to 0.64 log reduction per hour. The average survival rate for the O157:H7 clonal group was significantly less than that of the other STEC clones in the glutamate and arginine systems and significantly less than that of the O26/O111 clone in the oxidative system, indicating that this clonal group is not exceptionally acid resistant with these specific mechanisms.
Escherichia coli is an ecologically versatile bacterium that has adapted to a variety of ecological conditions encountered in both animal hosts and the external environment. This organism has evolved multiple mechanisms to survive under low-pH conditions. E. coli O157:H7, a food- and waterborne pathogen, has been considered highly acid resistant in nature because of its low infectious dose (17) and ability to survive in acidic foods (32, 36, 45, 49, 50). In the laboratory, E. coli O157:H7 was shown to be exceptionally acid resistant compared to other enteric bacteria, such as Salmonella enterica (16), especially when exposed to mild acid (pH > 4) prior to exposure to strong acid (pH < 3) (4, 26). Outbreaks of human disease caused by E. coli O157:H7 in acidic foods, such as apple juice (13) and salami (12), have stimulated interest in determining the mechanisms behind the acid resistance (AR) of this organism.
Lin et al. developed assays to separate three different AR mechanisms by which E. coli can survive in low-pH environments for extended periods of time: two amino acid decarboxylase-dependent systems (arginine and glutamate) and a glucose catabolite-repressed system (27, 28). Their findings suggest that these mechanisms promote survival in low-pH environments, such as those encountered in the stomach and in acidic food products. The AR systems are governed, in part, by the alternate sigma factor RpoS (10, 34), which plays a central role in the regulation and expression of many proteins involved in stationary phase and the stress response (19, 25, 39). Mutant RpoS alleles exist in natural populations of enterohemorrhagic E. coli (EHEC) and can lead to acid sensitivity (44).
Studies of AR have focused on E. coli O157:H7 because it is one of the most common E. coli types associated with outbreaks and sporadic cases of food- and waterborne disease. Diarrheal disease caused by Shiga toxin-producing E. coli (STEC) of serotypes other than O157:H7 (non-O157 STEC) has increased worldwide, with recent outbreaks attributable to serotypes O26:H11, O111:H8, and O121:H19 (11, 29, 46). These serotypes mark three genetically distinct clonal groups of STEC: EHEC clonal group 1 consists of O157:H7 and its nonmotile relatives, EHEC clonal group 2 consists of serotypes O26:H11 and O111:H8, and strains of serotype O121:H19 represent a distinct clonal group of STEC (35, 41). Non-O157 STEC are common in animal reservoirs, capable of causing the same severe disease as E. coli O157:H7, and are thought to also have a low infectious dose (17); however, these pathogens are associated with disease outbreaks much less frequently than E. coli O157:H7. One reason for this disparity in prevalence may result from differences in the inherent acid resistance of the STEC clones and concomitant survival in acidic foods and low infectious dose.
Here we address the question of whether non-O157 STEC strains possess the same ability to survive using the defined AR mechanisms as E. coli O157:H7. To address this question, we assembled a collection of clinical isolates representing three clonal groups of STEC (O121:H19, O26/O111, and O157:H7) that have been screened for RpoS expression, and we assessed strain-to strain variation in survival at low pH using the three different AR mechanisms.
MATERIALS AND METHODS
Bacterial strains.
A total of 66 pathogenic strains, including 30 E. coli O157:H7 strains, 18 O26:H11 strains, 4 O111:H8 strains, and 14 O121:H19 strains, were used in this study (Table 1). The strains were originally isolated by different investigators from sporadic cases of diarrheal illness in the course of surveillance studies of STEC in the United States (1, 14, 15, 21, 23). We also obtained several additional STEC isolates from James Rudrik of the Michigan Department of Community Health, Lothar Beutin in Germany, and Roger Johnson in Canada. The presence of the Shiga toxin 1 and 2 genes (stx1 and stx2) is given in Table 1 as reported in the original publication or to us by the sender. In addition, six nontoxigenic, commensal strains from the ECOR (33) group A (ECOR strains 1, 2, 3, 4, 7, and 10) were used for comparative purposes.
TABLE 1.
STEC strains used in this study
| Serotype and strain | Locale (yr) | stx gene(s) | % RpoS+ | Sequence type | Stx EIAa | Sourceb |
|---|---|---|---|---|---|---|
| E. coli O157:H7 | ||||||
| TW07591 | Mich. (1997) | stx2 | 27 | 70 | + | 1 |
| TW07695 | Fla. (1997) | stx1, stx2 | 74 | 60 | + | 1 |
| TW07700 | Calif. (1997) | stx1, stx2 | 91 | 66 | + | 1 |
| TW07702 | Ohio (1997) | stx2 | 95 | 59 | + | 1 |
| TW07704 | Ohio (1997) | stx2 | 88 | 66 | + | 1 |
| TW07706 | Utah (1997) | stx1, stx2 | 88 | 66 | + | 1 |
| TW07928 | D.C. (1998) | stx2 | 99 | 66 | + | 1 |
| TW07937 | Mass. (1998) | stx2 | 76 | 66 | + | 1 |
| TW07938 | Mass. (1998) | stx1, stx2 | 100 | 66 | + | 1 |
| TW07939 | Mass. (1998) | stx1, stx2 | 100 | 66 | + | 1 |
| TW07941 | Mass. (1998) | stx1, stx2 | 45 | 66 | + | 1 |
| TW07943 | Mass. (1998) | stx1, stx2 | 96 | 66 | + | 1 |
| TW07945 | Fla. (1998) | stx2 | 94 | 66 | + | 1 |
| TW07949 | D.C. (1999) | stx1, stx2 | 80 | 66 | + | 1 |
| TW07950 | D.C. (1999) | stx1, stx2 | 51 | 66 | + | 1 |
| TW07952 | D.C. (1999) | stx1, stx2 | 82 | 66 | + | 1 |
| TW07953 | D.C. (1999) | stx1, stx2 | 38 | 66 | + | 1 |
| TW07956 | D.C. (1999) | stx1, stx2 | 94 | 66 | + | 1 |
| TW07957 | D.C. (1999) | stx1, stx2 | 63 | 66 | + | 1 |
| TW07958 | D.C. (1999) | stx1, stx2 | 53 | 66 | + | 1 |
| TW07961 | Ohio (1998) | stx1, stx2 | 96 | 66 | + | 1 |
| TW07962 | Ohio (1998) | stx1, stx2 | 82 | 66 | + | 1 |
| TW08022 | Mont. (2000) | stx1, stx2 | 100 | 66 | + | 7 |
| TW08026 | Mont. (2000) | stx1, stx2 | 100 | 71 | + | 7 |
| TW08027 | Mont. (2000) | stx1, stx2 | 100 | 62 | + | 7 |
| TW08030 | Mont. (2000) | stx2 | 45 | 72 | + | 7 |
| TW08080 | Mont. (2000) | stx2 | 61 | 66 | − | 7 |
| TW08609 | Wash. (2000) | stx2 | 100 | 66 | + | 7 |
| TW08610 | Wash. (2000) | stx2 | 100 | 66 | + | 7 |
| TW08612 | Wash. (2000) | stx1, stx2 | 100 | 66 | + | 7 |
| E. coli O26:H11 | ||||||
| TW07594 | Ariz. (1997) | stx1 | 96 | 110 | + | 1 |
| TW07595 | Neb. (1998) | stx1 | 100 | 106 | + | 3 |
| TW07600 | Neb. (1998) | stx1 | 100 | 106 | + | 3 |
| TW07622 | Mich. (2002) | stx1, stx2 | 4 | 106 | + | 5 |
| TW07814 | Idaho (1997) | stx1, stx2 | 100 | 104 | + | 6 |
| TW07936 | Mass. (1998) | stx1 | 100 | 10 | + | 1 |
| TW07948 | D.C. (1999) | stx1 | 97 | 106 | + | 1 |
| TW08024 | Mont. (2000) | stx1 | 100 | 114 | + | 7 |
| TW08033 | Mont. (2000) | stx1 | 100 | 102 | + | 7 |
| TW08038 | Mont. (2000) | stx1 | 98 | 101 | + | 7 |
| TW08052 | Mont. (2000) | stx1 | 76 | 103 | + | 7 |
| TW08060 | Mont. (2000) | stx1 | 100 | 112 | + | 7 |
| TW08084 | Mont. (2000) | stx1 | 99 | 99 | + | 7 |
| TW08569 | Germany (1998) | stx2 | 54 | 115 | − | 2 |
| TW08570 | Germany (1999) | stx2 | 94 | 113 | + | 2 |
| TW08571 | Germany (2000) | stx1, stx2 | 100 | 104 | + | 2 |
| TW08637 | Wash. (2000) | stx1 | 100 | 106 | + | 7 |
| TW09184 | Mich. (2003) | ND | 100 | 106 | + | 5 |
| E. coli O111:H8 | ||||||
| TW07598 | Neb. (1998) | stx1, stx2 | 100 | 106 | + | 3 |
| TW07601 | Neb. (1998) | stx1, stx2 | 100 | 106 | + | 3 |
| TW08642 | Wash. (1999) | stx1 | 100 | 106 | + | 7 |
| TW08643 | Wash. (2001) | stx1 | 100 | 106 | + | 7 |
| E. coli O121:H19 | ||||||
| TW07615 | Mich. (2002) | stx2 | 82 | 182 | + | 5 |
| TW07927 | D.C. (1998) | stx1, stx2 | 100 | 182 | + | 1 |
| TW07931 | Mass. (1998) | stx2 | 100 | 182 | + | 1 |
| TW08036 | Mont. (2000) | stx2 | 100 | 182 | + | 7 |
| TW08040 | Mont. (2000) | stx2 | 100 | 183 | + | 7 |
| TW08042 | Mont. (2000) | stx2 | 100 | 180 | + | 7 |
| TW08043 | Mont. (2000) | stx2 | 90 | 182 | + | 7 |
| TW08055 | Mont. (2000) | stx2 | 88 | 187 | + | 7 |
| TW08063 | Mont. (2000) | stx2 | 99 | 184 | + | 7 |
| TW08091 | Mont. (2000) | stx2 | 100 | 179 | + | 7 |
| TW08646 | Wash. (2000) | stx2 | 100 | 133 | − | 7 |
| TW08649 | Switzerland (2002) | stx2 | 100 | 185 | + | 4 |
| TW08650 | Switzerland (2002) | stx2 | 100 | 181 | + | 4 |
| TW08653 | Canada (2002) | stx2 | 100 | 182 | + | 4 |
Shiga toxin production was tested by enzyme-linked immunoassay on a single RpoS+ colony.
Sequence types and Shiga toxin production.
Multilocus sequence analysis of seven housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) was used to characterize the multilocus genotype of each strain. Alleles were identified based on sequence comparisons for each gene, and distinct allele combinations were designated as sequence types (STs). The sequencing methods and ST database are available at the STEC website (http://www.shigatox.net/mlst/index.html). The production of Shiga toxins was determined by enzyme immunoassay using the Premier EHEC test (Meridian Diagnostics Inc., Cincinnati, Ohio) according to the manufacturer's instructions.
RpoS status.
Stock cultures of each strain were screened indirectly for the allelic state of rpoS with the hydrogen peroxidase II (HPII) assay (7, 48). For each strain, we picked 93 colonies from a Luria-Bertani (LB) agar plate which were then individually inoculated into wells of microtiter plates containing 100 μl of LB broth. Three wells were used for controls. After 3 h of incubation at 37°C, cultures in the 96 wells were transferred to replicate LB agar plates using a 96-pin replicator tool. Plates were incubated for 24 h at 37°C and were then tested for the expression of catalase by dropping 30% (wt/vol) hydrogen peroxide on each colony. Immediate vigorous bubbling indicated positive HPII activity, which is strongly correlated to expression of RpoS (37). Colonies were thus classified as RpoS+ if they exhibited vigorous bubbling in the HPII assay. To confirm RpoS status, HPII+ colonies were subsequently tested for glycogen production, which is also controlled by RpoS (20, 25). HPII+ colonies were streaked onto LB agar and grown overnight at 37°C. Iodine was dropped onto individual colonies, and glycogen accumulation was detected by the formation of a brown color on the colonies (20). For each strain, a single-pick, RpoS+ colony was grown in LB broth, frozen in 10% glycerol at −70°C, and used as stock for all subsequent AR assays.
AR mechanism assays.
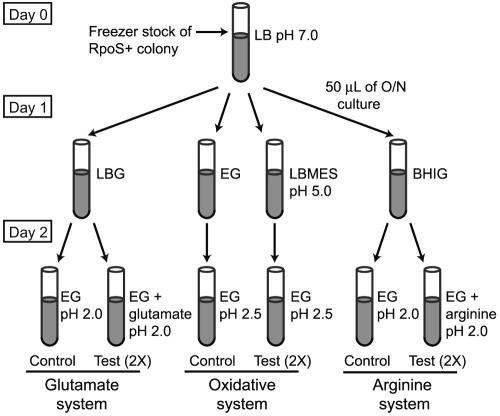
Methods for measuring AR were modified from those of Lin et al. (27, 28) as diagrammed in Fig. 1. On day zero, an RpoS+ isolate of each STEC strain was inoculated from freezer stocks and grown overnight in LB medium (pH 7.0) at 37°C (Fig. 1). On day 1, 50 μl of overnight culture was used to inoculate 10 ml of LB broth with 0.4% glucose (LBG), LB broth buffered to pH 5.5 with 0.1 M morpholineethanesulfonic acid (LBMES), minimal E medium containing 73 mM K2HPO4, 17 mM NaNH4HPO4, 0.8 mM or 12 mM MgSO4, 10 mM sodium citrate, and 0.4% glucose (EG), and brain heart infusion broth with a final concentration of 0.6% glucose (BHIG). All media were prepared by filter sterilization. Strains were grown in these media for 22 h at 37°C, with shaking at 120 rpm. Optical density at 600 nm was measured for each culture after 22 h of growth, and this measurement was used to determine the inoculum volume into the test environment that would have an initial density of ∼106 CFU/ml.
FIG. 1.
Diagram of assay conditions for discriminating the three AR mechanisms.
The ability of the RpoS+ isolates to survive acidic conditions was measured for three AR systems (27, 28): the glutamate-dependent (GLU) system, the glucose-repressed oxidative (OXI) system, and the arginine-dependent (ARG) system (Fig. 1). Cultures grown in LBG were tested in the GLU system (EG supplemented with 5.7 mM sodium glutamate at pH 2.0), cultures grown in LBMES were tested in the OXI system (EG at pH 2.5), and cultures grown in BHIG were tested in the ARG system (EG supplemented with 0.6 mM arginine at pH 2.5). Cultures inoculated in EG at pH 2.0 and pH 2.5 without supplements served as controls for the GLU and ARG systems. The control for the OXI system was cells grown overnight in EG pH 7.0 and tested in EG at pH 2.5. A 4 N HCl solution was used to adjust the pH of the test medium, which was then warmed to 37°C before use. All AR tests were conducted at 37°C. Two replicates of each strain were tested per AR system. Samples were taken at 0, 2, and 6 h and plated in duplicate on LB agar using the Autoplate 4000 (Spiral Biotech, Bethesda, MD). Plates were incubated at 37°C for 24 h and then enumerated using the Q Count software (Spiral Biotech).
To determine if Mg2+ limitation was contributing to cell inactivation (for example, through changing the permeability of the outer membrane [OM]), strains were also tested in the EG test environments described above, with the addition of 12 mM MgSO4 (high Mg2+) instead of the originally described 0.8 mM MgSO4 (low Mg2+).
Statistical analysis.
Plate counts were converted to log CFU/ml values, and log decrease per hour for each assay was determined over the period of the assay. We defined the survival rate, ΔV, as the change in viable cell counts (in log10 CFU/ml) per hour and report the mean and standard deviation for each experiment. Analysis of variance was conducted on the log CFU/ml for each time point and the ΔV values for each assay using SAS (SAS Institute, Cary, NC). Pairwise comparisons were made for each of the clonal groups; significant differences were determined by the Tukey method, adjusted for multiple comparisons. Phylogenetic analysis of the multilocus sequences was conducted with MEGA 2 software (24).
RESULTS
Clonal groups.
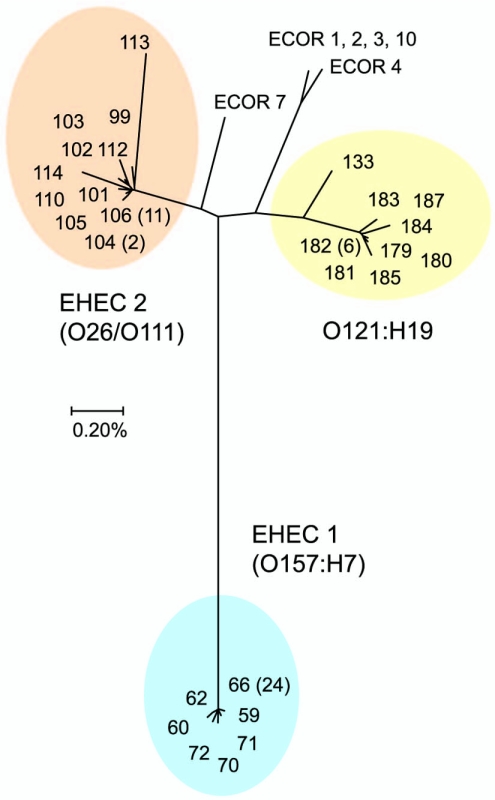
The STEC strains were sequenced at seven loci (3,753 bp; 1,251 codons) and classified into 27 STs based on the alleles resolved (Table 1). Most (24/30) of the O157:H7 strains belong to the most common EHEC 1 sequence type (ST-66). The close genetic relationship between the O26:H11 and O111:H8 strains is reflected by the fact that both serotypes are associated with the common EHEC 2 clone marked by ST-106 (Table 1). A neighbor-joining tree placed the 27 STEC STs into three distinct clusters (Fig. 2). The genetic diversity between strains within a cluster comprised only a fraction of the nucleotide divergence between clusters (Table 2). The nucleotide diversity within clusters ranged from 0.07% for EHEC 1 to 0.23% for the EHEC 2 group, whereas the divergence between clusters ranged from 1.1% to 2.6% (Table 2).
FIG. 2.
Neighbor-joining tree showing the clonal groups of STEC based on multilocus STs. Distances were estimated by the Kimura two-parameter model. The number of isolates of each ST is given in parentheses.
TABLE 2.
Nucleotide diversity within and between three clonal groups of STEC
| Clonal group | No. of isolates | No. of STs | Diversity within group (%) | Distance (%) between groups
|
|
|---|---|---|---|---|---|
| EHEC 1 | EHEC 2 | ||||
| EHEC 1 | 30 | 7 | 0.07 ± 0.03 | ||
| EHEC 2 | 22 | 11 | 0.23 ± 0.04 | 2.47 ± 0.28 | |
| O121:H19 | 14 | 9 | 0.19 ± 0.04 | 2.60 ± 0.26 | 1.10 ± 0.17 |
RpoS status and Stx production.
To determine the variability in RpoS expression, we used the HPII catalase assay to examine 93 colony picks from each of 66 STEC strains (Table 1). For 32 of the strains, 100% of the 93 colonies tested were RpoS+. Of the 30 E. coli O157:H7 strains tested, 15/30 (50%) had more than 90% RpoS+ colonies. In comparison, 19/22 (83%) and 12/14 (86%) strains of E. coli O26/O111 and O121:H19 had more than 90% RpoS+ colonies, indicating significant variation between clonal groups in the proportion of RpoS+ colonies (Gadj = 9.95; P < 0.01, with degrees of freedom = 2). A single RpoS+ colony was selected for each strain and used to create a stock for subsequent experiments.
The RpoS+ derivatives of the 66 STEC strains were all tested for Shiga toxin production using the Stx enzyme immunoassay; 63 of the 66 RpoS+ isolates were Stx positive (Table 1). We further investigated the three Stx-negative isolates by PCR and found that one had lost the stx2 gene in comparison to the original STEC strain and one had stx2 but did not express it. In the third case the original STEC strain was also negative for the stx2 gene, suggesting that the toxin gene had been lost in a previous transfer.
Variation in AR among E. coli O157:H7 strains.
Strains of E. coli O157:H7 exhibited variation in survival when using each of the AR mechanisms. Final cell densities after 6 h in the GLU system approximated a normal distribution (Anderson-Darling normality test, P = 0.121) with a mean log10 CFU/ml of 5.43 ± 0.32. A nested analysis of variance indicated that the among-strain component of the variance was highly significant (F0.05 [29, 50] = 75.6; P < 0.001) and accounted for 92% of the total variation in cell densities at 6 h. Similar distributions were observed after 6 h for the OXI system (3.09 ± 0.71) and for the ARG system (4.79 ± 0.46) (data not shown).
Effect of magnesium on AR of E. coli O157:H7.
The permeability of the bacterial OM can be disturbed by chelators that disrupt the stabilizing interactions of Mg2+, Ca2+, and lipopolysaccharide molecules (42). Decreased OM stability resulting from chelators could amplify cell inactivation that is perceived to be a consequence of acid stress alone. For example, the citrate present in the test environments could chelate Mg2+, leading to a more permeable OM.
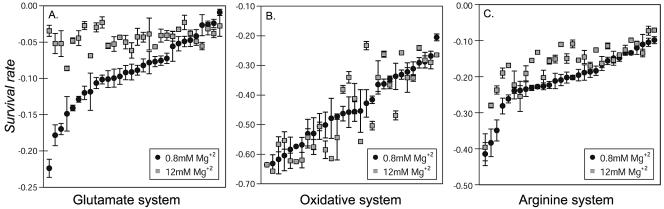
To test for the effects of Mg2+ limitation, we challenged strains in both low (0.8 mM MgSO4) and high (12 mM MgSO4) Mg2+ concentrations. Under conditions of low Mg2+, ΔVGLU values approximated a normal distribution, with a mean ΔVGLU of −0.09 ± 0.05 (Fig. 3A). ΔVARG values also approximated a normal distribution, with a mean ΔVARG of −0.22 ± 0.08 (Fig. 3C). The O157:H7 strains had significantly greater survival rates in the GLU and ARG systems when the MgSO4 levels were increased, and the variation in survival rates decreased. The mean ΔVGLU with high Mg2+ was −0.05 ± 0.02. The mean ΔVARG when MgSO4 was increased was −0.17 ± 0.06. In the GLU system, 22/30 (74%) strains had a significantly greater survival rate with high Mg2+ than low Mg2+. In the ARG system, 14/30 (46%) strains had a significantly higher survival rate with high Mg2+. In contrast, the survival rates were not significantly different between high and low Mg2+ for the OXI system (Fig. 3B).
FIG. 3.
Variation in survival rates (ΔV = change in the density of viable cells per h) among 30 E. coli O157:H7 strains using each AR system. Strains are ranked by the average ΔV from lowest to highest survival and plotted in order from left to right. Error bars indicate standard deviations of two replicate samples. Closed circles indicate survival under low-Mg2+ (0.8 mM) conditions, and open squares indicate survival under high-Mg2+ (12 mM) conditions. Strains were ranked separately for each AR system. A. ΔV values at pH 2.0 for the glutamate system. B. ΔV values at pH 2.5 for the oxidative system. C. ΔV values at pH 2.5 for the arginine system.
Comparison of STEC clones.
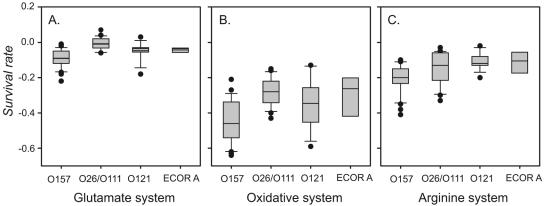
The glutamate system provided the most protection under acidic conditions, with average ΔVGLU values ranging from 0.004 ± 0.04 for the O26/O111 clone to −0.09 ± 0.05 for the O157:H7 group (Fig. 4A). In the first 2 h of the assay, the O157 strains had a ΔVGLU similar to that of the O26/O111 strains. However, in the 2- to 6-h time period (t2-t6), the density of bacteria of the O157:H7 group decreased at a faster rate [ΔVGLU(t2-t6) = −0.10 ± 0.01] than strains of the other clonal groups. As a result, the ability of E. coli O157:H7 strains to survive using the GLU system was significantly less (P < 0.001) than the other clonal groups in the 6-hour assay period. Strains of the non-O157 clonal groups were also tested under high-Mg2+ conditions, but there were no significant differences in survival rates for any of the clonal groups with the exception of O157:H7 (see Table S3 in the supplemental material), and so all ΔV values reported are from low-Mg2+ test environments.
FIG. 4.
Box plots of survival rates (ΔV) for strains in each clonal group. The horizontal bar indicates the mean for each group. A. ΔV values for each serogroup in the glutamate system at pH 2.0. B. ΔV values for each serogroup in the oxidative system at pH 2.5. C. ΔV values for each group in the arginine system at pH 2.0.
The oxidative system provided the least amount of protection against acidic environments, with average ΔVOXI values ranging from −0.28 ± 0.08 for the O26/O111 strains to −0.44 ± 0.12 for the O157:H7 strains (Fig. 4B). In this assay, strains in the O157:H7 clonal group decreased in number at a significantly (P < 0.01) greater rate (ΔVOXI = −0.44 ± 0.12) than the O26/O111 clonal group. The greatest variability in survival rate among strains of each clonal group was seen in the OXI system (Fig. 4B). The E. coli O121:H19 and ECOR strains had a similar ability to survive acidic conditions and were not significantly different from the O26/O111 or O157:H7 groups when using the oxidative system as measured by the average ΔVOXI (Fig. 4B).
The arginine system did not provide as much protection as the glutamate system but was more effective than the oxidative system at pH 2.5. ΔVARG values ranged from −0.11 ± 0.05 for O121 strains to −0.22 ± 0.08 for the O157:H7 strains (Fig. 4C). Pairwise comparisons of the average ΔVARG value for each clonal group indicated that the O157:H7 strains were significantly different from the O26/O111 and O121 strains (P < 0.003) but not the ECOR A group in the arginine system (Fig. 4C). The majority of strains exhibited high survival with the GLU system, whereas the abilities to utilize the OXI and ARG systems were more variable across strains. Strain TW08650 (O121:H19) has high survival rates with all three mechanisms, with ΔVGLU = −0.01, ΔVOXI = −0.13, and ΔVARG = −0.12. Strain TW07962 (O157:H7) exhibits low survival rates in two of the systems, with ΔVGLU = −0.09, ΔVOXI = −0.45, and ΔVARG = −0.35. Overall, the survival rate using one mechanism is not significantly correlated with the ability to survive using another (r2 for GLU versus OXI = 0.42; r2 for OXI versus ARG = 0.19; r2 for GLU versus ARG = 0.15). ΔV values for each strain can be found in Table S4 of the supplemental material.
We partitioned the total phenotypic variance in survival rate into within-clonal and between-clonal group components. For the glutamate system, 42% of the variation was due to differences between strains within a clonal group and 52% was due to differences between clonal groups. For the oxidative system, 62% was due to differences between strains within group while 33% was due to differences between clonal groups. For the arginine system, 67% was due to differences between strains within a clonal group and 30% was due to differences between the clonal groups. These results indicate that a substantial fraction of the total variation in acid resistance for a given mechanism is accounted for by the differences between STEC lineages.
RpoS+ versus RpoS− colonies.
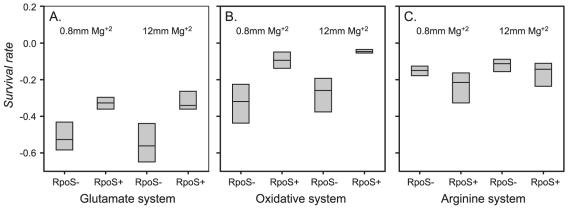
Eight strains of E. coli O157:H7 (TW07591, TW07695, TW07702, TW07704, TW07928, TW07941, TW07962, and TW08030) were selected, and the RpoS+ and RpoS− isolates derived from single colonies for each strain were tested for their abilities to survive using the three AR mechanisms. In the glutamate system, the RpoS− isolates (colonies that exhibited slow bubbling in the HPII assay) all exhibited significantly lower mean survival rates (t[0.05, 60] = 35.97; P < 0.001) than the corresponding RpoS+ isolate at both levels of Mg2+, with the exception of strain TW07962 in low Mg2+ (Fig. 5A). In the oxidative system, only strains TW07962 and TW07928 did not exhibit significantly lower survival rates (t[0.05, 60] = 24.39; P < 0.001) for the RpoS− isolates compared to the corresponding RpoS+ isolate at both levels of Mg2+ (Fig. 5B). In contrast, the RpoS− isolates exhibited similar or greater survival than the RpoS+ isolates in the arginine system (Fig. 5C).
FIG. 5.
Box plots of survival rates (ΔV) for paired RpoS+ and RpoS− isolates from eight strains of E. coli O157:H7 in each AR system. Tests were conducted under low-Mg2+ (0.8 mM) and high-Mg2+ (12 mM) conditions.
DISCUSSION
E. coli O157:H7 was first thought to have enhanced abilities to resist acid and other environmental stresses because of its low infective dose and ability to survive in and be transmitted by acidic foods. Strain-to-strain variation in AR ability has been reported (albeit with a small number of strains), with a continuum of survival abilities classified from highly resistant to sensitive. For example, Buchanan and Edelson (9) observed that the ability to survive in acidic environments varies substantially among isolates within a single serotype; AR of EHEC strains grown to stationary phase was dependent on the type of acidulant and whether the pH-dependent AR system had been induced (9). Benjamin and Datta (5) compared survival of six O157:H7 strains in LB at pH 2.5 and found that two of the strains were considered highly acid tolerant while the other four strains were moderately and slightly acid tolerant. Leyer et al. (26) measured survival of acid-adapted and nonadapted cells of five strains of E. coli O157:H7; although the acid-adapted cells had a better survival rate than the nonadapted, there was variability in survival rates among the strains. Lin et al. (28) assayed 11 strains of E. coli O157:H7 for their ability to use three different AR mechanisms to protect against high acidity. Considerable variation existed in the abilities of strains to use a particular AR mechanism, indicating that not all O157:H7 strains had the same capability to survive in a low-pH environment using a specific AR mechanism (28). Our analysis with 30 O157:H7 isolates collected independently from sporadic cases of disease shows continuous variation in ability to survive at low pH using these AR mechanisms and that survival rates fit a normal distribution. In addition, more than half (∼50% to 70%) of the total phenotypic variation in acid resistance among STEC clones is a result of between-strain variation within a clonal group. This finding suggests that there are additional factors that contribute to variability in acid resistance of STEC strains.
Understanding the extent of variation among pathogens of a group, for example of E. coli O157:H7 strains, is of considerable importance, especially for purposes of predictive modeling. Whiting and Golden have identified the differences among strains of the same species as a major source of variation in growth and survival studies; variations determined for four different parameters (growth, survival, thermal inactivation, and toxin production) of E. coli O157:H7 strains were larger than the error calculated from experimental procedures using a single strain or cocktail (47). We also observed that the strain-to-strain variation within a clonal group was larger than variation between replicates or due to experimental error. Thus, drawing conclusions about differences in the acid resistance abilities of a variable group of bacteria will require examination of large numbers of isolates.
The alternate sigma factor RpoS governs gene expression during stationary phase and in response to stressful environments. Variation in the expression of this global regulator can contribute to overall variability in the stress response. It is clear that expression of the alternate sigma factor rpoS is required for stationary-phase induction of acid resistance in E. coli and Shigella (39), and Arnold and Kaspar found that as the growth of E. coli O157:H7 neared stationary phase, survival of cells in tryptic soy broth at pH 2.0 increased dramatically (4).
Expression of rpoS can vary in natural populations of E. coli, possibly because rpoS null mutants have a competitive advantage in prolonged exposure under starvation conditions (48). Variation in rpoS, ranging from null to full expression, could account for a component of the variability observed in the AR ability of E. coli strains. Waterman and Small found during a survey of AR of 58 STEC strains that mutant rpoS alleles exist in natural populations of STEC. Complementation with rpoS on a plasmid conferred AR to 9 of 13 acid-sensitive strains (44). We also observed variation in the allelic state of RpoS among STEC, with E. coli O157:H7 strains having the most variability. In this study, we attempted to correct for the potential confounding effects of rpoS null mutants in our isolates by assaying RpoS activity indirectly through the HPII assay (7) and selecting RpoS+ colonies to test AR.
Lin et al. determined that functional rpoS is required for a strain to be able to use the oxidative mechanism of acid resistance. A mutant rpoS allele resulted in a reduced ability to utilize the arginine and glutamate mechanisms of AR (28). In a comparison of survival using a specific AR mechanism of eight E. coli O157:H7 strains that had rpoS+ and rpoS− isolates, our results did not agree with those of Lin et al. In the arginine system, mutant rpoS isolates survived better than the wild-type isolates. The allelic state of RpoS was confirmed in these strains by testing colonies for HPII activity with hydrogen peroxide and for glycogen production with iodine. Arginine decarboxylase, encoded by adiA, is positively regulated by CysB (10, 38) and by AdiY, a transcriptional regulator encoded downstream of adiA (40). It is possible that RpoS does not contribute to regulating expression of adiA. However, further studies are needed to determine the role of RpoS and other regulators in governing the arginine AR system.
Chelating compounds, as well as organic and inorganic acids, increase the permeability of the OM (2, 18), which has been linked to loss of viability during acid stress in E. coli O157:H7 (22). Bacterial inactivation by acid could be aided by the presence of chelators in the test environment. Citric acid has been shown to be a potent OM permeabilizer in O157:H7, and the permeabilization effect can be abolished with the addition of 5 mM Mg (18). Mg2+ stabilizes cell membranes (8) by interacting with adjacent lipopolysaccharide molecules (42). Our results indicate that high levels of magnesium in the low-pH environment significantly increase the survival rates of most E. coli O157:H7 strains in the glutamate system. The increase in survival is not as dramatic for the arginine system, possibly because this system is tested at a higher pH. Interestingly, bacteria of the other STEC clonal groups, as well as isolates of the ECOR group, were not significantly affected by the increased levels of magnesium in these two AR systems. No differences were observed in survival for the O157 and O26/O111 clones in the OXI system at the two levels of magnesium, but the O121 group showed a significantly lower survival rate at 12 mM Mg in the OXI system. Further work is needed to understand the mechanisms by which Mg2+ concentration influences survival in acidic environments.
In vitro studies on AR of E. coli O157:H7 strains, other pathogenic E. coli strains, and different enteric bacteria have indicated that E. coli O157:H7 can exhibit AR, but whether O157:H7 strains have superior ability to survive in acid has not been substantiated. The consensus seems to be that E. coli O157:H7 is well suited to adverse conditions even though little direct evidence supports the hypothesis that this pathogen is substantially different from other E. coli serotypes in AR abilities (30). Most AR studies are based on only a few non-O157 STEC strains of the same serotype, which reduces the statistical power to detect significant differences between groups. For example, Miller and Kaspar observed survival of two strains of E. coli O157:H7 and a single nonpathogenic strain under acidic conditions (32). While a significant difference in survival was reported between the O157:H7 and nonpathogenic strains, their suggestion that resistance to acidic pH is an additional characteristic that distinguishes O157:H7 from other E. coli serotypes is not warranted, because other pathogenic E. coli serotypes were not tested. Benjamin and Datta compared survival of 14 EHEC strains in LB at pH 2.5 and 3.0 and found that no correlation could be drawn between acid tolerance and serotype (5), but again only a few non-O157 serotypes were evaluated. McKellar and Knight surveyed a set of 19 EHEC strains for ability to survive in tryptic soy broth acidified with HCl (pH 2.0) and with acetic acid (pH 4.0). When strains were grouped by source, they found that survival of outbreak strains in acid was significantly greater than in strains from human or animal sources. When strains were grouped by serotype, however, strains of E. coli O157:H7 were among the more acid sensitive, but clear conclusions could not be drawn since other EHEC serotypes were only represented by a single strain each (31). Berry et al. recently reported a comprehensive comparison of AR between 39 isolates of O157 and 20 isolates of non-O157 biotype I E. coli and found no significant differences in the ability of strains from either group to survive in BHI at pH 2.5; they concluded that high acid tolerance is not unique to strains of pathogenic E. coli (6). Our results indicate that strains of E. coli O157:H7 do not have superior AR abilities compared to strains of E. coli O121:H19 and O26:H11 using the three AR mechanisms. While E. coli O157:H7 clones can survive in low pH using the three described AR mechanisms, there is no evidence that O157:H7 has greater acid resistance in any single system than other clones of STEC in these laboratory test environments.
In a comparison of three clonal groups of STEC and a clonal group of nonpathogenic ECOR strains, we have shown that strains of E. coli O157:H7 do not have superior AR abilities using three specific AR mechanisms. Under natural conditions, however, E. coli O157:H7 may have the ability to use multiple mechanisms in combination, through some form of epistasis, to achieve higher AR than bacteria of other clonal groups. It is also possible that E. coli O157:H7 has evolved alternative mechanisms, yet to be identified, which contribute to acid resistance in nature. This study suggests that there is no evidence for enhanced AR of O157:H7 by a measurable superiority of one of the three major AR mechanisms of E. coli. There is also compelling evidence for a substantial strain-to-strain component of variation in survival rate; more than 60% of the total variance in survival rates for the oxidative and arginine systems was attributed to between-strain differences. Our findings demonstrate that because of the high interstrain variability, a large number of isolates should be examined in order to test and draw conclusions about survival of E. coli O157:H7 and non-O157 STEC. A narrow focus on a few isolates or a single serotype may seriously underestimate the variability that underlies important phenotypic variation in pathogen populations.
Supplementary Material
Acknowledgments
We thank the following people for kindly providing the strains used in this study: David W. Acheson, CFSAN; Lothar Beutin, Robert Koch Institute; Paul D. Fey, University of Nebraska; Alison D. O'Brien, Uniformed Services University of the Health Sciences; Phillip I. Tarr, Washington University at St. Louis; Roger Johnson, University of Guelph; and James Rudrik from the Michigan Department of Community Health. We also thank Lukas Wick and Shannon Manning for helpful discussion.
This research was supported in part by the Michigan Agriculture Experimental Station.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acheson, D. W., K. Frankson, and D. Willis. 1998. Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. C-205.
- 2.Alakomi, H. L., E. Skytta, M. Saarela, T. Mattila-Sandholm, K. Latva-Kala, and I. M. Helander. 2000. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, K. W., and C. W. Kaspar. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin, M. M., and A. R. Datta. 1995. Acid tolerance of enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 61:1669-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, E. D., G. A. Barkocy-Gallagher, and G. R. Siragusa. 2004. Stationary-phase acid resistance and injury of recent bovine Escherichia coli O157 and non-O157 biotype I Escherichia coli isolates. J. Food Prot. 67:583-590. [DOI] [PubMed] [Google Scholar]
- 7.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brock, T. D. 1962. Effects of magnesium ion deficiency on Escherichia coli and possible relation to the mode of action of novobiocin. J. Bacteriol. 84:679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchanan, R. L., and S. G. Edelson. 1999. pH-dependent stationary-phase acid resistance response of enterohemorrhagic Escherichia coli in the presence of various acidulants. J. Food Prot. 62:211-218. [DOI] [PubMed] [Google Scholar]
- 10.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2000. Escherichia coli O111:H8 outbreak among teenage campers—Texas, 1999. Morb. Mortal. Wkly. Rep. 49:321-324. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1995. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. Morb. Mortal. Wkly. Rep. 44:157-160. [PubMed] [Google Scholar]
- 13.Cody, S. H., M. K. Glynn, J. A. Farrar, K. L. Cairns, P. M. Griffin, J. Kobayashi, M. Fyfe, R. Hoffman, A. S. King, J. H. Lewis, B. Swaminathan, R. G. Bryant, and D. J. Vugia. 1999. An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Intern. Med. 130:202-209. [DOI] [PubMed] [Google Scholar]
- 14.Fey, P. D., R. S. Wickert, M. E. Rupp, T. J. Safranek, and S. H. Hinrichs. 2000. Prevalence of non-O157:H7 Shiga toxin-producing Escherichia coli in diarrheal stool samples from Nebraska. Emerg. Infect. Dis. 6:530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gansheroff, L. J., M. R. Wachtel, and A. D. O'Brien. 1999. Decreased adherence of enterohemorrhagic Escherichia coli to HEp-2 cells in the presence of antibodies that recognize the C-terminal region of intimin. Infect. Immun. 67:6409-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorden, J., and P. L. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 18.Helander, I. M., and T. Mattila-Sandholm. 2000. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 88:213-219. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877-1886. [DOI] [PubMed] [Google Scholar]
- 21.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 22.Jordan, K. N., L. Oxford, and C. P. O'Byrne. 1999. Survival of low-pH stress by Escherichia coli O157:H7: correlation between alterations in the cell envelope and increased acid tolerance. Appl. Environ. Microbiol. 65:3048-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein, E. J., J. R. Stapp, C. R. Clausen, D. R. Boster, J. G. Wells, X. Qin, D. L. Swerdlow, and P. I. Tarr. 2002. Shiga toxin-producing Escherichia coli in children with diarrhea: a prospective point-of-care study. J. Pediatr. 141:172-177. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. Jakobsen, and M. Nei. 2000. MEGA 2: molecular evolutionary genetics analysis program, version 2.0. Pennsylvania State University, University Park.
- 25.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 26.Leyer, G. J., L. L. Wang, and E. A. Johnson. 1995. Acid adaptation of Escherichia coli O157:H7 increases survival in acidic foods. Appl. Environ. Microbiol. 61:3752-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy, T. A., N. L. Barrett, J. L. Hadler, B. Salsbury, R. T. Howard, D. W. Dingman, C. D. Brinkman, W. F. Bibb, and M. L. Cartter. 2001. Hemolytic-uremic syndrome and Escherichia coli O121 at a lake in Connecticut, 1999. Pediatrics 108:E59. [DOI] [PubMed] [Google Scholar]
- 30.McClure, P. J., and S. Hall. 2000. Survival of Escherichia coli in foods. Symp. Ser. Soc. Appl. Microbiol. 2000:61S-70S. [DOI] [PubMed] [Google Scholar]
- 31.McKellar, R. C., and K. P. Knight. 1999. Growth and survival of various strains of enterohemorrhagic Escherichia coli in hydrochloric and acetic acid. J. Food Prot. 62:1466-1469. [DOI] [PubMed] [Google Scholar]
- 32.Miller, L. G., and C. W. Kaspar. 1994. Escherichia coli O157:H7 acid tolerance and survival in apple cider. J. Food Prot. 57:460-464. [DOI] [PubMed] [Google Scholar]
- 33.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price, S. B., C. M. Cheng, C. W. Kaspar, J. C. Wright, F. J. DeGraves, T. A. Penfound, M. P. Castanie-Cornet, and J. W. Foster. 2000. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 36.Riordan, D. C., G. Duffy, J. J. Sheridan, R. C. Whiting, I. S. Blair, and D. A. McDowell. 2000. Effects of acid adaptation, product pH, and heating on survival of Escherichia coli O157:H7 in pepperoni. Appl. Environ. Microbiol. 66:1726-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc. Natl. Acad. Sci. USA 86:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi, X., and G. N. Bennett. 1994. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J. Bacteriol. 176:7017-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stim-Herndon, K. P., T. M. Flores, and G. N. Bennett. 1996. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311-1320. [DOI] [PubMed] [Google Scholar]
- 41.Tarr, C. L., T. M. Large, C. L. Moeller, D. W. Lacher, P. I. Tarr, D. W. Acheson, and T. S. Whittam. 2002. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70:6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterman, S. R., and P. L. Small. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weagant, S. D., J. L. Bryant, and D. H. Bark. 1994. Survival of Escherichia coli O157:H7 in mayonnaise and mayonnaise-based sauces at room and refrigerated temperatures. J. Food Prot. 57:629-631. [DOI] [PubMed] [Google Scholar]
- 46.Werber, D., A. Fruth, A. Liesegang, M. Littmann, U. Buchholz, R. Prager, H. Karch, T. Breuer, H. Tschape, and A. Ammon. 2002. A multistate outbreak of Shiga toxin-producing Escherichia coli O26:H11 infections in Germany, detected by molecular subtyping surveillance. J. Infect. Dis. 186:419-422. [DOI] [PubMed] [Google Scholar]
- 47.Whiting, R. C., and M. H. Golden. 2002. Variation among Escherichia coli O157:H7 strains relative to their growth, survival, thermal inactivation, and toxin production in broth. Int. J. Food Microbiol. 75:127-133. [DOI] [PubMed] [Google Scholar]
- 48.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, T., and M. P. Doyle. 1994. Fate of enterohemorrhagic Escherichia coli O157:H7 in commercial mayonnaise. J. Food Prot. 57:780-783. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, T., M. P. Doyle, and R. E. Besser. 1993. Fate of enterohemorrhagic Escherichia coli O157:H7 in apple cider with and without preservatives. Appl. Environ. Microbiol. 59:2526-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.