Abstract
In this study we correlate the presence of genes leading to the synthesis of trehalose and mannosylglycerate (MG) in 17 strains of the genus Thermus with the ability of the strains to grow and accumulate these compatible solutes in a defined medium containing NaCl. The two sets of genes, namely, otsA/otsB for the synthesis of trehalose and mpgS/mpgP for the synthesis of MG, were necessary for the growth of Thermus thermophilus in a defined medium containing up to 6% NaCl. Strains lacking a complete otsA gene did not grow in defined medium containing >2% NaCl. One strain of T. thermophilus lacking the genes for the synthesis of MG did not grow in a medium with >1% NaCl. We did not identify any of these genes in the type strains of the other seven species of Thermus, and none of those strains grew in defined medium with 1% NaCl. The results strongly indicate that the combined accumulation of trehalose and MG is required for optimal osmotic adjustment.
The species of the genus Thermus have optimum growth temperatures of 70 to 75°C, and the majority of them, being isolated from continental hydrothermal springs venting fresh water, do not grow in media containing 1% NaCl. Strains of the species Thermus thermophilus, many of which were isolated from marine hot springs, generally have higher growth rates in media without added salt, but in contrast to the strains of all other species of this genus, they grow in media containing 3 to 6% NaCl (7).
Trehalose is the major compatible solute of T. thermophilus strains HB8, B, RQ-1, and PRQ-14 during salt-induced osmotic stress, but these strains also accumulate smaller amounts of mannosylglycerate (MG) in media containing yeast extract (21). Trehalose is a very common compatible solute of prokaryotes and eukaryotes (11), but MG, which was initially identified in marine red algae of the family Ceramiales (3), has otherwise only been detected in hyperthermophilic archaea and thermophilic bacteria, in which it may be the major or sole compatible solute during osmotic stress (18, 21, 25).
The most common pathway for the synthesis of trehalose in bacteria involves trehalose-phosphate synthase (Tps), encoded by otsA, and trehalose-6-phosphate phosphatase (Tpp), encoded by otsB (1, 29). Trehalose may also be synthesized directly from maltose via trehalose synthase, which is encoded by treS (31). A third pathway found in several bacteria (9, 20) and in the archaeon Sulfolobus acidocaldarius (19) converts the terminal unit of a glucose polymer to trehalose via maltooligosyltrehalose synthase, encoded by treY, and maltooligosyltrehalose trehalohydrolase, encoded by treZ. Some organisms have one of these pathways, others have two, and a few even possess all three pathways (9). Thermus thermophilus RQ-1 possesses otsA, otsB, and treS arranged sequentially in the genome (26).
The synthesis of MG in Thermus thermophilus HB27 proceeds via a two-step pathway by which mannosyl-3-phosphoglycerate synthase (MpgS, encoded by mpgS) catalyzes the conversion of GDP-mannose and 3-phosphoglycerate to mannosyl-3-phosphoglycerate. The phosphorylated intermediate is subsequently converted to MG by mannosyl-3-phosphoglycerate phosphatase (MpgP, encoded by mpgP) (13). This pathway has also been found in the hyperthermophilic archaea Aeropyrum pernix and Pyrococcus horikoshii and the thermophilic bacterium Rhodothermus marinus (2, 12). However, in R. marinus the synthesis of MG can also proceed via a one-step pathway in which GDP-mannose is condensed with d-glycerate to yield MG by mannosylglycerate synthase (Mgs, encoded by mgs) (17).
The role of trehalose during osmotic stress in T. thermophilus strain RQ-1 was recently established by the production of a mutant with a disruption in otsA and otsB that was deficient in the synthesis of this disaccharide. The mutant did not grow in a defined medium (TD) with NaCl above 3% unless trehalose was supplied to the medium. MG was the only compatible solute detected during the growth of this mutant in the defined medium, and it appeared to be essential for low-level osmotic adaptation (26).
For this study, we assessed the presence of otsA, otsB, treS, mpgS, and mpgP in 10 strains of T. thermophilus and in representative strains of all other species of the genus Thermus. We correlated the presence of these genes with the ability of the organisms to grow and accumulate compatible solutes in a salt-containing defined medium.
MATERIALS AND METHODS
Strains and culture conditions.
Thermus thermophilus strains HB8 (DSM 579T), AT-62 (DSM 674), and HB27 (DSM 7039) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany. T. thermophilus Fiji3 A1 was donated by Hugh Morgan (University of Waikato, New Zealand), and T. thermophilus strains GK24 (30), T-2 (ATCC 27737; American Type Culture Collection, Manassas, Va.), and B (NCIB 11247) were a gift from Richard Sharp, PHLS, Salisbury, United Kingdom. Strains RQ-1 (DSM 9247), CC-16, and PRQ-14, classified as T. thermophilus, are laboratory strains (7). The type strains of Thermus aquaticus YT-1 (DSM 625), Thermus scotoductus (DSM 8553), Thermus antranikianii (DSM 12462), Thermus filiformis (DSM 4687), Thermus igniterrae (DSM 12459), Thermus oshimai (DSM 12092), and Thermus brockianus (NCIB 12676; National Collection of Industrial and Marine Bacteria Ltd., Aberdeen, Scotland) were also used for this study.
The organisms were grown in defined medium (TD) containing basal salts of Degryse medium 162 (8), 1.0 g l−1 tryptone, and a vitamin solution (24) at 70°C; this medium lacked yeast extract, which is a source of trehalose (32). Growth of the organisms was also performed in TD medium to which filter-sterilized trehalose (0.26 mM) or maltose (0.26 mM) was added. These media were used without added NaCl or were supplemented with NaCl to a final concentration of 1 to 7% (wt/vol). Cultures grown in TD medium without NaCl were inoculated into experimental flasks at an initial turbidity (optical density at 610 nm) of 0.05. The growth of the organisms was examined in 1-liter metal-capped Erlenmeyer flasks containing 200 ml of medium in a reciprocal water bath shaker (120 rpm) at 70°C.
DNA manipulations.
The isolation of DNA from Thermus strains was performed as described previously (5). PCR amplifications were performed by use of a GC-rich PCR kit (Roche). The reaction mixtures were preincubated for 5 min at 94°C and then subjected to 30 cycles of denaturation at 94°C for 1 min. Several annealing temperatures between 46 and 60°C were tested for 1 min, and primer extension reactions were done at 72°C for 1 min. The extension step in the last cycle was prolonged for 7 min.
For PCR amplification of the otsA and otsB genes, primers were designed based on the corresponding RQ-1 gene sequences (GenBank accession number AY275558) (Fig. 1A). The primers used for amplification of the treS gene were based on the AT-62 gene sequence (GenBank accession number D86216). Two separate amplifications were necessary to obtain the complete sequence of the treS gene. The first PCR product was a 1.482-kbp fragment and the second PCR product was a 1.683-kbp fragment. Primers for the amplification of mpgS and mpgP were designed based on the HB27 gene sequences (GenBank accession number AY193871). The primers designed for amplification of the mgs gene were based on the Rhodothermus marinus gene sequence (GenBank accession number AF173987).
FIG. 1.
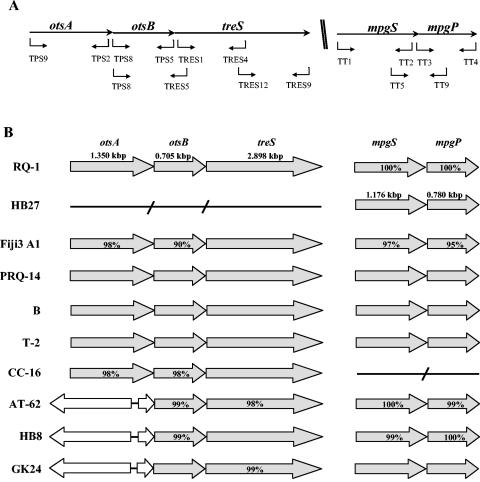
Schematic representation of primers used for PCR amplifications (A) and comparison of the organizations of genes for the synthesis of trehalose and mannosylglycerate (B) in T. thermophilus strains RQ-1, HB27, Fiji3 A1, PRQ-14, AT-62, B, T-2, CC-16, HB8, and GK24. The mpgS and mpgP sequences of strain HB27; the otsA, otsB, and treS sequences of strain RQ-1; the treS sequence of strain AT-62; and the otsB and treS sequences of strain GK24 were previously described (12, 16, 26, 31). The identities (%) of the deduced amino acid sequences to the corresponding proteins from RQ-1 and HB27 are shown inside the arrows. Unshaded arrows represent the α-glucosidase gene (EC 3.2.1.20).
For sequencing purposes, PCR-amplified genes were visualized by agarose gel electrophoresis, purified by use of a DNA purification kit (Promega), and cloned into the pGEM-T Easy vector (Promega). Transformations of Escherichia coli XL1-Blue were carried out as described previously (22). Ampicillin was added to the medium at a final concentration of 100 μg/ml for plasmid selection.
Southern blot analysis was performed essentially as described previously (22). Purified DNAs (5 μg) were digested overnight with BamHI and electrophoresed in agarose gels (1% [wt/vol]). DNAs were capillary transferred for 16 h onto a nylon membrane (positively charged; Roche) in 0.4 N NaOH, 1 M NaCl buffer. Hybridization steps were carried out overnight at temperatures between 55 and 68°C, using a DIG High Prime DNA labeling and detection starter kit II (Roche). The membrane was washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 5 min at room temperature and in 0.5× SSC-0.1% sodium dodecyl sulfate for 15 min at 65°C with shaking. After the immunological detection steps, the membrane was sealed in a hybridization bag, exposed to Lumi-Film (chemiluminescent detection film; Boehringer Mannheim) in a hypercassette, and incubated for 45 min at 37°C to enhance the luminescent reaction. The probes used were PCR-amplified individual otsA (1.350 kbp) and otsB (0.705 kbp) or otsA/otsB-linked genes (2.055 kbp), the 5′ end of the treS (1.482 kbp) gene from strain RQ-1, mpgS (1.194 kbp) and mpgP (0.780 kbp) from strain HB27, and the mgs (1.191 kbp) gene from Rhodothermus marinus.
Inverse PCR to obtain the genes flanking otsB was performed by use of the primers TPP132 (5′-GCGCCGTGACCACGTAGACG) and TPP640 (5′-GCGCGGCTCAAGGACGTGGAGG), which were designed from the sequence of the otsB gene of strain HB8. Genomic DNAs (3.5 μg) from strains HB8, AT-62, and GK24 were digested with 5 U of HindIII for 30 min at 37°C, and the restriction endonuclease was then inactivated by heating at 80°C for 20 min. The digested DNAs were diluted to 2.8 ng of DNA per μl (total, 50 μl) and circularized with 2 U of T4 DNA ligase (Roche) for 16 h at 16°C. Inverse PCRs were performed with a GC-rich PCR kit (Roche) in mixtures containing 56 ng circularized DNA, 0.5 μM primers, and 125 μM of each deoxynucleoside triphosphate. The thermal cycler parameters were as follows: preincubation for 5 min at 94°C; an initial denaturation step of 1 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C; and a final extension step of 7 min at 72°C. Inverse PCR products were visualized by agarose gel electrophoresis, purified by use of a DNA purification kit (Promega), cloned into the pGEM-T Easy vector (Promega), and sequenced by AGOWA GmbH (Berlin, Germany).
Gene sequence analysis.
The otsA genes from T. thermophilus strains CC-16 and Fiji3 A1, the otsB genes from strains AT-62, HB8, GK24, and Fiji3 A1, and the mpgS and mpgP genes from T. thermophilus strains Fiji3 A1, AT-62, HB8, and RQ-1 were cloned into the pGEM-T Easy vector and sequenced in one direction by the use of vector- and insert-specific oligodeoxynucleotide primers by AGOWA GmbH (Berlin, Germany). Nucleic acid and protein sequence analyses were performed with programs in the Wisconsin Genetics Computer Groups software package (10).
Extraction of organic solutes and their quantification by nuclear magnetic resonance.
Thermus thermophilus cells were harvested by centrifugation (7,000 × g, 10 min, 4°C) during mid-exponential growth (optical density at 610 nm, 0.4) and washed twice with a NaCl solution identical in concentration to that of the growth medium. Cell pellets were extracted twice with boiling 80% ethanol as described previously (26). The protein content of the cells was determined by the Bradford assay (4) after sonication of the cells, using an aliquot of the suspension before the extraction of compatible solutes. Freeze-dried extracts were analyzed by nuclear magnetic resonance as described previously (26).
Nucleotide sequence accession numbers.
Sequences containing partial otsA and complete otsB genes and their flanking regions from Thermus thermophilus strains HB8 and AT-62 have been deposited in GenBank under the accession numbers AY836576 and AY836577.
RESULTS
Identification and organization of genes for synthesis of trehalose and mannosylglycerate.
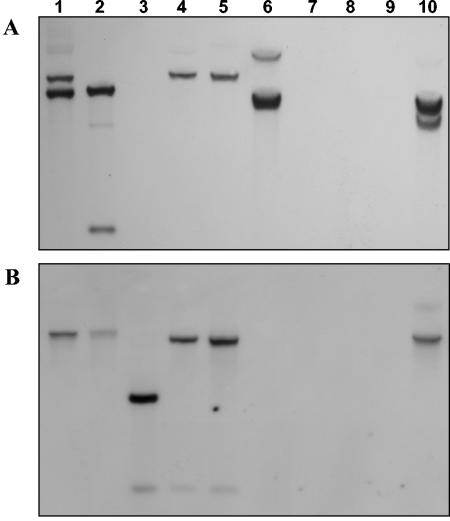
The genes involved in the synthesis of trehalose, namely otsA, otsB, and treS, are structurally linked in T. thermophilus RQ-1 (26), and those leading to the synthesis of MG are structurally linked in T. thermophilus HB27 (13). Different primer combinations ensured that the associations between otsA and otsB, between otsB and treS, and between mpgS and mpgP could be determined in this study (Fig. 1A). All PCR results were confirmed by Southern blot analysis using otsA, otsB, treS, mpgS, mpgP, and mgs sequences as probes (Fig. 2). Thermus thermophilus strains Fiji3 A1, CC-16, T-2, PRQ-14, and B have otsA, otsB, and treS arranged sequentially in their genomes. However, strains GK24, HB8, and AT-62 lack a complete otsA gene, whereas strain HB27 lacks otsA, otsB, and treS (Fig. 1B). Although our PCR results suggested that otsA was absent from strains GK24, HB8, and AT-62, Southern blots using the otsA probe showed one hybridization band at 60°C (Fig. 2A). This band became progressively weaker with higher stringencies (65 and 68°C), unlike the bands showing otsA hybridization in the lanes corresponding to strains RQ-1, Fiji3 A1, CC-16, and B. One recognition site for BamHI was present in all complete otsA sequences, except for that of strain Fiji3 A1, which had two sites. The partial otsA gene of strains GK24, HB8, and AT-62 lacked the BamHI site, explaining the single hybridization signal with the otsA probe (Fig. 2A, lanes 4 and 5). The presence of a partial otsA gene in strains GK24, HB8, and AT-62 was demonstrated by inverse PCR. The complete otsA gene of strains RQ-1, Fiji3 A1, CC-16, B, T-2, and PRQ-14 was 1,350 bp long, while the partial otsA sequences of strains GK24, HB8, and AT-62 contained 232, 216, and 216 bp of the 3′ end of the gene, respectively. We detected an intergenic region of 174 bp followed by an α-glucosidase gene (EC 3.2.1.20) upstream of the truncated otsA gene in GK24, HB8, and AT-62. With the exception of strain CC-16, consecutive mpgS and mpgP genes, involved in the synthesis of MG, were detected in all strains of T. thermophilus examined (Fig. 1B and 2B). Homologues of the mgs gene found in R. marinus were not detected in any Thermus strain. Moreover, genes encoding enzymes for the synthesis of trehalose or MG were not detected by PCR and Southern analysis in the type strains of T. aquaticus, T. scotoductus, T. filiformis, T. igniterrae, T. brockianus, T. oshimai, and T. antranikianii (Fig. 2). The otsA genes of strains Fiji3 A1 and CC-16 and the otsB genes of strains Fiji3 A1, CC-16, AT-62, and HB8, as well as the mpgS and mpgP genes of strains RQ-1, Fiji3 A1, AT-62, and HB8, were cloned and sequenced to confirm their authenticity and to evaluate their degrees of identity to those in strains RQ-1 and HB27. The predicted amino acid sequences of the genes for the synthesis of trehalose and MG had extremely high identities that ranged between 90 and 100% (Fig. 1B).
FIG. 2.
Southern blot analysis of BamHI-digested genomic DNA (5 μg) probed with the otsA gene of strain RQ-1 (A) and the mpgS gene of strain HB27 (B) at 60°C. Lane 1, strain RQ-1; lane 2, strain Fiji3 A1; lane 3, strain HB27; lane 4, strain HB8; lane 5, strain AT-62; lane 6, strain CC-16; lane 7, Thermus aquaticus (DSM 625); lane 8, Thermus scotoductus (DSM 8553); lane 9, Thermus filiformis (DSM 4687); lane 10, strain B.
Halotolerance of Thermus strains.
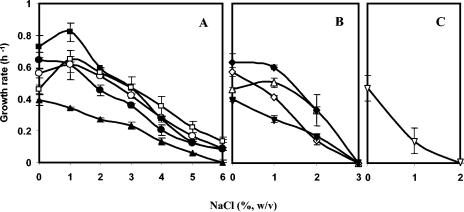
The strains of T. thermophilus examined had higher growth rates in TD medium without added NaCl or containing 1.0% NaCl than in TD medium containing higher levels of NaCl. Moreover, only the strains of T. thermophilus, except for strain CC-16, which lacks mpgS/mpgP, grew in TD medium with NaCl concentrations higher than 1.0%. Strains RQ-1, B, Fiji3 A1, T-2, and PRQ-14, which have the complete otsA/otsB/treS and mpgS/mpgP gene clusters, grew in TD medium containing up to 5 or 6% NaCl (Fig. 3A). Strains HB8, AT-62, and GK24, which completely lack the otsA gene, and strain HB27, which lacks otsA/otsB/treS, grew only in TD medium containing 2% NaCl (Fig. 3B). Strains HB8 and GK24 grew in medium containing 3% NaCl after the addition of exogenous trehalose to the growth medium, but strains HB27 and AT-62 did not (results not shown). The addition of maltose to the medium as a substrate for TreS did not lead to the growth of strains HB8, AT-62, GK24, and HB27 at salinities higher than 2% (results not shown). Strain CC-16, which has the three genes involved in the synthesis of trehalose but lacks mpgS and mpgP, only grew in TD medium with 1% NaCl (Fig. 3C). The addition of trehalose or maltose did not lead to the growth of this organism in medium with 2% NaCl.
FIG. 3.
Effect of NaCl concentration in TD medium on the growth rate of T. thermophilus. (A) Strains B (•), Fiji3 A1 (○), T-2 (▪), PRQ-14 (□), and RQ-1 (▴); (B) strains HB8 (▵), HB27 (♦), AT-62 (⋄), and GK24 (▾); (C) strain CC-16 (▿).
Accumulation of compatible solutes.
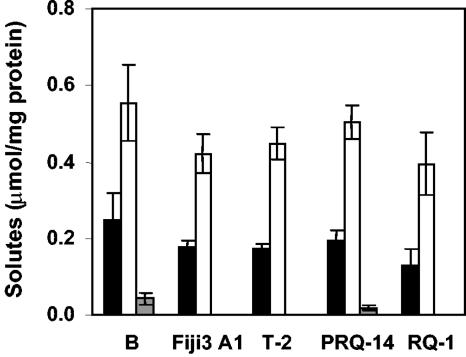
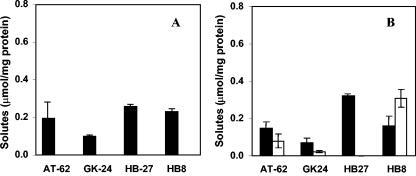
An analysis of the accumulation of compatible solutes by T. thermophilus strains showed that the strains with the full set of genes for the synthesis of trehalose and MG, namely, strains B, RQ-1, Fiji3 A1, T-2, and PRQ-14, accumulated high levels of trehalose and small amounts of MG in response to salt stress in TD medium. Strains B and PRQ-14 also accumulated trace levels of glycine betaine (Fig. 4). Strains HB8, AT-62, GK24, and HB27 only accumulated MG in TD medium containing 2% NaCl without exogenous trehalose (Fig. 5A). Strains HB8, AT-62, and GK24 accumulated trehalose when this disaccharide was added to the salt-containing medium with a concomitant decrease in the accumulation of MG, but with the exception of strain HB8, trehalose was not the major compatible solute of these organisms under salt stress (Fig. 5B). Strain HB27, on the other hand, did not accumulate trehalose in TD medium to which trehalose had been added (Fig. 5B). None of the T. thermophilus strains lacking the complete otsA gene or the otsA/otsB/treS gene cluster accumulated trehalose after the addition of exogenous maltose (results not shown). Strain CC-16 accumulated small amounts of trehalose (0.075 ± 0.015 μmol/mg protein), but not MG, in TD medium containing 1% NaCl.
FIG. 4.
Accumulation of compatible solutes in TD medium by Thermus thermophilus strains B, Fiji3 A1, T-2, PRQ-14, and RQ-1. The concentrations of compatible solutes were determined for cultures grown in medium containing 4% NaCl. Bars represent the intracellular concentrations of mannosylglycerate (black), trehalose (white), and glycine betaine (gray).
FIG. 5.
Accumulation of compatible solutes by Thermus thermophilus strains AT-62, HB27, GK24, and HB8. The concentrations of compatible solutes were determined for cultures grown in TD medium containing 2% NaCl (A) and TD medium containing 2% NaCl and exogenous trehalose (B). Bars represent the intracellular concentrations of mannosylglycerate (black) and trehalose (white).
DISCUSSION
Previous studies had identified complete otsB and treS genes in T. thermophilus strain GK24 (16) and incomplete otsB and treS genes in strain AT-62 (31), but the presence of a complete otsA gene was only recently shown for strain RQ-1, which possesses three genes involved in the synthesis of trehalose (26), leading us to assume that the otsB gene of strain AT-62 had not been completely sequenced. Our PCR results showed a complete sequence for otsB from this strain but failed to confirm the presence of full-length otsA genes in strains GK24, AT-62, and HB8. Southern blot experiments with these strains indicated weak hybridization signals with otsA probes at high stringencies, while strains RQ-1, T-2, PRQ-14, Fiji3 A1, CC-16, and B clearly showed strong signals for the otsA genes at the highest hybridization temperature. Only an inverse PCR strategy unambiguously demonstrated the presence of a partial otsA sequence immediately upstream of otsB, comprising about 20% of the full-length otsA gene from T. thermophilus strains RQ-1, T-2, PRQ-14, Fiji3 A1, CC-16, and B.
To our surprise, there was a large amount of diversity with respect to the presence of otsA, otsB, and treS among these organisms. Some strains possessed all three genes, other strains had a truncated otsA gene and complete otsB and treS genes, and strain HB27 had none of the genes. The absence of the three genes from the genome sequence of strain HB27 was, in fact, the first hint that some strains lacked some or all of the genes for trehalose synthesis (15). We also wrongly assumed that all T. thermophilus strains have the genes leading to the synthesis of MG. However, strain CC-16 does not possess these genes, and unlike all other T. thermophilus strains known, is not halotolerant, making it more difficult to identify strains of this species by the ability to grow in media containing 2 to 3% NaCl (7).
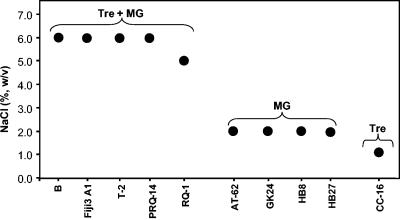
Our results show that T. thermophilus strains can be divided into three groups based on their osmotic relations. Strains RQ-1, Fiji3 A1, PRQ-14, B, and T-2, with the full set of genes leading to the synthesis of trehalose and MG, grew in TD medium containing up to about 6% NaCl (Fig. 6). Strains HB8, HB27, AT-62, and GK24, all of which were isolated from hot springs in Japan and appear to be closely related (7), have an incomplete otsA gene or lack the otsA/otsB/treS gene cluster and did not grow in TD medium containing more than 2% NaCl. Strain CC-16, which lacks mpgS/mpgP but possesses the three genes leading to the synthesis of trehalose, grew only in TD medium containing up to 1% NaCl.
FIG. 6.
Representation of maximum NaCl concentrations for strains of Thermus thermophilus during growth in TD medium at 70°C. The strains clustered into three groups according to their halotolerance and the nature of accumulated compatible solutes. MG, mannosylglycerate; Tre, trehalose.
This study reinforces our earlier observations that trehalose is crucial for the growth of strain RQ-1 in salt concentrations ranging from 3 to 6% (26). Strains RQ-1, B, Fiji3 A1, T-2, and PRQ-14 accumulated primarily trehalose and small amounts of MG, corroborating previous results (21, 26), while strains HB8, AT-62, GK24, and HB27, which only accumulated MG in TD medium, only grew in medium containing up to 2% NaCl. The accumulation of trehalose and small amounts of MG by strain HB8 had been reported before for medium containing yeast extract (a source of trehalose) with 3% NaCl (21). The present study confirms these results, since the addition of trehalose to TD medium also led to the accumulation of the disaccharide and the growth of strain HB8 in medium containing 3% NaCl.
The presence of Tps and Tpp, encoded by otsA and otsB, respectively, appears to be crucial for the synthesis of trehalose in the otsA/otsB-negative mutant RQ-1M6 under osmotic stress, as the maltose-converting TreS (encoded by treS) protein did not lead to the accumulation of trehalose when exogenous maltose was added to salt-containing medium (26). Similar results were obtained in this study with otsA-negative variants (HB8, AT-62, and GK24), which also did not accumulate trehalose when maltose was added to the medium, even though they possess treS. Moreover, trehalose was not formed in cell extracts after the addition of radiolabeled maltose, nor was maltose formed from radiolabeled trehalose (26), although a recombinant TreS protein has been shown to convert maltose to trehalose (16). It is possible, therefore, that the gene is not expressed under the conditions examined.
The absence of a full-length otsA gene, which codes for Tps, from the genomes of some Thermus strains raises the question of the function of Tpp, encoded by otsB, in strains that lack a functional Tps protein. This phosphatase is quite specific for trehalose-6-phosphate and does not have significant activity on other phosphorylated substrates (27). The otsB gene could represent a remnant gene without a specific function or it could code for an enzyme with a different activity in vivo (6).
Surprisingly, trehalose did not accumulate in strain HB27 under osmotic stress when it was supplied in the growth medium. Trehalose and maltose are, in fact, taken up from the medium via an ATP binding cassette transporter which was identified and characterized for strain HB27 (28). Moreover, this organism grows in a minimal medium with trehalose as the sole source of carbon and energy. It is possible that strain HB27 lacks an appropriate regulatory system that would induce the accumulation of trehalose under salt stress. The uptake of compatible solutes such as trehalose and their accumulation from the medium to relieve salt stress are very common in prokaryotes (14, 23), but this mechanism does not appear to be important for osmotic adjustment in strain HB27 (28), since compatible solutes other than MG did not accumulate during salt stress in this organism.
T. thermophilus strain CC-16 does not possess mpgS or mpgP. Although strain CC-16 accumulated low levels of trehalose in TD medium containing 1% NaCl, it was unable to grow in medium with 2% NaCl, like the other strains that were unable to synthesize trehalose, because it did accumulate MG. Many prokaryotes accumulate glutamate to relieve low-level osmotic stress and as a counterion to potassium, but as the salt concentration of the growth medium increases, neutral or zwitterionic compatible solutes such as ectoine, trehalose, and glycine betaine become the predominant osmolytes (14, 23). Our results led us to conclude that trehalose is unable to relieve low-level salt stress and that MG appears to have a role in T. thermophilus similar to that of glutamate in low-level osmotic adjustment in other organisms.
This study clearly indicates that the genes for the synthesis of trehalose and mannosylglycerate are necessary for the organisms of the genus Thermus to grow in media with elevated concentrations of salt and that these solutes have a synergistic effect on osmotic adjustments of these thermophilic bacteria.
Acknowledgments
This research was funded by European Commission contracts QLK3-CT-2000-00640 and COOP-CT-2003-508644 and by Fundação para a Ciência e a Tecnologia (FCT), Portugal, and FEDER projects PRAXIS/P/BIO/12082/1998 and POCTI/35715/BIO/2000. S. Alarico acknowledges a Ph.D. scholarship from POCTI SFRH/BD/14140/03. Z. Silva acknowledges a Ph.D. scholarship from PRAXIS XXI (BD/21669/99).
REFERENCES
- 1.Boos, W., U. Ehmann, H. Forkl, W. Klein, M. Rimmele, and P. Postma. 1990. Trehalose transport and metabolism in Escherichia coli. J. Bacteriol. 172:3450-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borges, N., J. D. Marugg, N. Empadinhas, M. S. da Costa, and H. Santos. 2004. Specialized roles of the two pathways for the synthesis of mannosylglycerate in osmoadaptation and thermoadaptation of Rhodothermus marinus. J. Biol. Chem. 279:9892-9898. [DOI] [PubMed] [Google Scholar]
- 3.Bouveng, H., B. Lindberg, and B. Wickberg. 1955. Low-molecular carbohydrates in algae. Structure of the glyceric acid mannoside from red algae. Acta Chem. Scand. 9:807-809. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cashion, P., M. A. Holder-Franklin, J. McCully, and M. Franklin. 1977. A rapid method for the base ratio determination of bacterial DNA. Anal. Biochem. 81:461-466. [DOI] [PubMed] [Google Scholar]
- 6.Cordwell, S. J. 1999. Microbial genomes and “missing” enzymes: redefining biochemical pathways. Arch. Microbiol. 172:269-279. [DOI] [PubMed] [Google Scholar]
- 7.da Costa, M. S., M. F. Nobre, and F. A. Rainey. 2001. The genus Thermus, p. 404-414. In D. R. Boone and R. W. Castenholtz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 8.Degryse, E., N. Glansdorff, and A. Pierard. 1978. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch. Microbiol. 117:189-196. [DOI] [PubMed] [Google Scholar]
- 9.De Smet, K. A. L., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:287-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbein, A. D., Y. T. Pan, I. Pastuszak, and D. Carroll. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17-27. [DOI] [PubMed] [Google Scholar]
- 12.Empadinhas, N., J. D. Marugg, N. Borges, H. Santos, and M. S. da Costa. 2001. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemical and genetic characterization of key enzymes. J. Biol. Chem. 276:43580-43588. [DOI] [PubMed] [Google Scholar]
- 13.Empadinhas, N., L. Albuquerque, A. Henne, H. Santos, and M. S. da Costa. 2003. The bacterium Thermus thermophilus, like hyperthermophilic archaea, uses a two-step pathway for the synthesis of mannosylglycerate. Appl. Environ. Microbiol. 69:3272-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinski, E. A. 1995. Osmoadaptation in bacteria. Adv. Microbiol. Physiol. 37:272-328. [PubMed] [Google Scholar]
- 15.Henne, A., H. Brüggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Denker, R. Huber, H.-P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H.-J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 16.Koh, S., J. Kim, H.-J. Shin, D. Lee, J. Bae, D. Kim, and D.-S. Lee. 2003. Mechanistic study of intramolecular conversion of maltose to trehalose by Thermus caldophilus GK24 trehalose synthase. Carbohydr. Res. 338:1339-1343. [DOI] [PubMed] [Google Scholar]
- 17.Martins, L. O., N. Empadinhas, J. D. Marugg, C. Miguel, M. S. da Costa, and H. Santos. 1999. Biosynthetic pathway for mannosylglycerate in Rhodothermus marinus. Characterization, cloning and expression of mannosylglycerate synthase. J. Biol. Chem. 274:35407-35414. [DOI] [PubMed] [Google Scholar]
- 18.Martins, L. O., R. Huber, H. Huber, K. O. Stetter, M. S. da Costa, and H. Santos. 1997. Organic solutes in hyperthermophilic Archaea. Appl. Environ. Microbiol. 63:896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruta, K., K. Hattori, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1291:177-181. [DOI] [PubMed] [Google Scholar]
- 20.Maruta, K., K. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Rhizobium sp. M-11. Biosci. Biotechnol. Biochim. 60:717-720. [DOI] [PubMed] [Google Scholar]
- 21.Nunes, O. C., C. M. Manaia, M. S. da Costa, and H. Santos. 1995. Compatible solutes in the thermophilic bacteria Rhodothermus marinus and “Thermus thermophilus”. Appl. Environ. Microbiol. 61:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 24.Sharp, R. J., and R. A. D. Williams. 1988. Properties of Thermus ruber strains isolated from Icelandic hot springs and DNA-DNA homology of Thermus ruber and Thermus aquaticus. Appl. Environ. Microbiol. 54:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva, Z., N. Borges, L. O. Martins, R. Wait, M. S. da Costa, and H. Santos. 1999. Combined effect of the growth temperature and the salinity of the medium on the accumulation of compatible solutes by Rhodothermus marinus and Rhodothermus obamensis. Extremophiles 3:163-172. [DOI] [PubMed] [Google Scholar]
- 26.Silva, Z., S. Alarico, A. Nobre, R. Horlacher, J. Marugg, W. Boos, A. I. Mingote, and M. S. da Costa. 2003. Osmotic adaptation of Thermus thermophilus RQ-1: a lesson from a mutant deficient in the synthesis of trehalose. J. Bacteriol. 185:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva, Z., S. Alarico, and M. S. da Costa. 2005. Trehalose biosynthesis in Thermus thermophilus RQ-1: biochemical properties of the trehalose-6-phosphate synthase and trehalose-6-phoshate phosphatase. Extremophiles 9:29-36. [DOI] [PubMed] [Google Scholar]
- 28.Silva, Z., M. M. Sampaio, A. Henne, A. Böhm, R. Gutzat, W. Boos, M. S. da Costa, and H. Santos. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J. Bacteriol. 187:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strøm, A. R., and I. Kaasen. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205-210. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi, H., M. Hamaoki, H. Matsuzawa, and T. Ohta. 1983. Heat-stable extracellular proteolytic enzyme produced by Thermus caldophilus strain GK24, an extremely thermophilic bacterium. J. Biochem. 93:7-13. [DOI] [PubMed] [Google Scholar]
- 31.Tsusaki, K., T. Nishimoto, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1997. Cloning and sequencing of trehalose synthase gene from Thermus aquaticus ATCC 33923. Biochim. Biophys. Acta 1334:28-32. [DOI] [PubMed] [Google Scholar]
- 32.Xavier, K. B., L. O. Martins, R. Peist, M. Kossmann, W. Boos, and H. Santos. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]