Abstract
We have isolated from plant surfaces several bacteria with the ability to catabolize indole-3-acetic acid (IAA). One of them, isolate 1290, was able to utilize IAA as a sole source of carbon, nitrogen, and energy. The strain was identified by its 16S rRNA sequence as Pseudomonas putida. Activity of the enzyme catechol 1,2-dioxygenase was induced during growth on IAA, suggesting that catechol is an intermediate of the IAA catabolic pathway. This was in agreement with the observation that the oxygen uptake by IAA-grown P. putida 1290 cells was elevated in response to the addition of catechol. The inability of a catR mutant of P. putida 1290 to grow at the expense of IAA also suggests a central role for catechol as an intermediate in IAA metabolism. Besides being able to destroy IAA, strain 1290 was also capable of producing IAA in media supplemented with tryptophan. In root elongation assays, P. putida strain 1290 completely abolished the inhibitory effect of exogenous IAA on the elongation of radish roots. In fact, coinoculation of roots with P. putida 1290 and 1 mM concentration of IAA had a positive effect on root development. In coinoculation experiments on radish roots, strain 1290 was only partially able to alleviate the inhibitory effect of bacteria that in culture overproduce IAA. Our findings imply a biological role for strain 1290 as a sink or recycler of IAA in its association with plants and plant-associated bacteria.
One intriguing type of interorganismal interaction is the manipulation by some microbes of their host's hormone system. A good example is given by bacteria of the genus Wolbachia, which convert male woodlice into females, supposedly by suppressing a gland that produces a masculinizing hormone (37). Another striking example is the microbial production of plant hormones such as auxin, cytokinin, and gibberellin, which allows certain bacteria and fungi to direct a plant's physiology toward their own advantage (5, 7, 14, 20).
Indole-3-acetic acid (IAA) is the main auxin in plants, controlling many important physiological processes including cell enlargement and division, tissue differentiation, and responses to light and gravity (45). Bacterial IAA producers (BIPs) have the potential to interfere with any of these processes by input of IAA into the plant's auxin pool. The consequence for the plant is usually a function of (i) the amount of IAA that is produced and (ii) the sensitivity of the plant tissue to changes in IAA concentration. A root, for instance, is one of the plant's organs that is most sensitive to fluctuations in IAA, and its response to increasing amounts of exogenous IAA extends from elongation of the primary root, formation of lateral and adventitious roots, to growth cessation (8).
The existence of BIPs was recognized very early in the research on auxin, and plant-associated BIPs have long been considered a source of contamination in measurements of IAA present in plant tissues (24, 51, 52). Later, BIPs were identified as the cause of symptoms associated with severe plant diseases such as gypsophila gall (26), knot disease of olive and oleander (43), and russet of pear fruit (25), or, in other cases, as benefactors of plants, e.g., nitrogen-fixing Bradyrhizobium japonicum in root nodules (9) and plant-growth-promoting rhizobacteria such as Pseudomonas putida (33). Since the initial discovery of BIPs, an impressive body of scientific information has been established on the biology, ecology, and pathology of these bacteria, and much of the genetics and biochemistry of bacterial IAA production has been elucidated (20, 32).
Although much is known about BIPs, little is known about bacteria that have the converse property, i.e., the ability to destroy IAA. This is surprising given the early realization that, like BIPs, bacterial IAA degraders (BIDs) are abundantly associated with plants (1, 2, 23, 29, 38, 49). Actually, BIDs were also considered a source of contamination, since their ability to destroy IAA obscured true levels of IAA in plant tissues (30, 46). Ironically, there have been more studies dedicated to successfully eliminate BID activity in measurements of plant IAA, e.g., by the use of antibiotics (1, 49) than there have been serious attempts to try to understand the abundance, origin, and significance of these bacteria and their unique ability to destroy IAA.
In one of the few studies on BIDs, Wichner and Libbert reported that bacteria with IAA-destroying activity could readily be isolated from plant surfaces (50). Libbert and Risch (23) described 15 such isolates from the shoots and roots of hydrocultured pea plants. These isolates were identified as Alcaligenes, Pseudomonas, other Pseudomonadaceae, and unidentified strains. More BIDs have been described by Proctor (35), Tsubokura et al. (47), Mino (27), and by the group of Jochimsen (10, 17, 31). For most of these studies, the authors' primary interest was with IAA degradation per se, i.e., elucidation of the degradative pathway and identification of pathway intermediates. We are interested in the ecological role of bacteria that are capable of IAA degradation: what is the selective advantage of IAA degradation, and by what forces is this trait maintained in nature? This report provides an initial description of Pseudomonas putida strain 1290, a bacterial isolate with the unique ability to utilize IAA as a sole source of carbon, nitrogen, and energy. We describe how degradation of IAA supports growth and survival of this bacterium, and explore the significance of this unique trait in its association with plants and other bacteria.
MATERIALS AND METHODS
Strains and growth media.
Bacterial strains that were tested for their ability to degrade IAA were originally isolated from Bartlett pear trees in orchards located near Healdsburg, CA (25). Erwinia herbicola 299R (3) served as a positive control for IAA production and as a negative control for IAA degradation. Escherichia coli DH5α (42) served as a negative control for IAA degradation. Rahnella aquaticus strains CD14 and CD15, as well as Pseudomonas syringae strain CD32, produce high levels of IAA in culture (25) and were used in coinoculation experiments on radish roots. Pseudomonas putida KT2440 (18) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Bacteria were grown at 30°C in Luria broth (LB) or King's B broth (19) or in 1× M9 minimal medium (42) or phosphate buffer (10 mM KPO4), supplemented with 2 mM MgSO4 and 5 or 9 mM IAA or one of the following carbon sources (final concentration): anthranilate (7.15 mM), catechol (8.35 mM), salicylate (7.15 mM), benzoate (8.35 mM), tryptophan (4.54 mM), glucose (0.1%, 5.56 mM), and fructose (0.4%, 22.2 mM). Also tested (at a final concentration of 0.875 mg per ml) were compounds similar to IAA, including indole acetaldehyde (5.50 mM), indole acrylic acid (4.67 mM), indole butyric acid (4.31 mM), indole lactic acid (4.26 mM), indole propionic acid (4.62 mM), indole pyruvic acid (4.31 mM), and naphthalene acetic acid (4.70 mM). Where appropriate, rifampin was added to a final concentration of 40 μg per ml.
Colorimetric assay for determining IAA concentrations.
IAA concentrations in culture supernatants by the method of Salkowski (13). After centrifugation of 1 ml of culture, a 50-μl aliquot of the supernatant was diluted with 450 μl of phosphate buffer. From this, 60 μl was added to 440 μl of phosphate buffer in a glass tube containing 500 μl of reagent R1. Reagent R1 (Salkowski reagent) contains 12 g of FeCl3 per liter of 7.9 M H2SO4. Red color formation was quantified as the absorbance at a wavelength of 540 nm in a Perkin-Elmer Lambda 3A UV/VIS spectrophotometer (Perkin-Elmer, Norwalk, CT). A standard curve was prepared from serial dilutions of a 5 mM IAA stock solution in phosphate buffer. The detection limit of IAA by this method was ca. 50 μM.
Oxygen uptake experiments, preparation of cell extracts, and determination of catechol 1,2-dioxygenase activity.
Oxygen uptake rates were measured with the Oxygraph System (Hansatech, Norfolk, United Kingdom) using 60 μl of a 15× concentrated suspension of washed P. putida 1290R cells (exponentially growing on IAA) in phosphate buffer in a final volume of 2 ml containing 3 mM IAA, catechol, tryptophan, anthranilate, salicylate, or benzoate in phosphate buffer. Cell extracts were prepared, and catechol 1,2 dioxygenase activity measurements were performed as described elsewhere (22).
Radish root assays.
Radish seeds (Cherry Bell; NK Lawn & Garden Co., Chattanooga, TN) were sterilized by agitation for 10 min in 50 ml of 1% sodium hypochlorite containing 100 μl of Tergitol (anionic) per ml. After being rinsed in several volumes of sterile water, 10 seeds were placed in individual growth pouches (Vaughan's Seed Company, Downers Grove, IL) and wet with 10 ml of phosphate buffer or phosphate buffer containing 1 nM, 1 μM, 1 mM, or no IAA, without or with 5 × 109 cells of P. putida 1290R that were prepared by harvesting, washing, and resuspension in phosphate buffer, i.e., a mid-log culture grown on LB. To test the effect of P. putida 1290 on IAA-producing bacteria, radish seeds were inoculated as described above with suspensions (108 cells/ml) of individual IAA-producing bacteria alone or in a mixture with 109 cells of P. putida 1290R/ml. Pouches were wrapped in aluminum foil and placed at room temperature in the dark for 4 days. Individual root lengths were measured, averaged for individual pouches, and tested for significant differences among treatments with SAS (version 6.04; SAS Institute, Inc., Cary, NC).
16S rRNA sequence.
Cells from fresh plate cultures of P. putida 1290 were resuspended in distilled water and used as a template in a standard PCR with primers 6F and 1510R (48) or Pput1290ssu1 (5′-ATTACTGGGCGTAAAGCG-3′) and Pput1290ssu2 (5′-TGTCAAGGCCTGGTAAGG-3′), resulting in amplification products of ∼1.5 kb (nearly complete copy of the 16S rRNA gene) and ∼0.4 kb (central portion of the 16S rRNA gene), respectively. Both were cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and sequenced (BaseClear, Leiden, The Netherlands). DNA sequences were analyzed by using Lasergene software (DNASTAR, Madison, WI). The nearly complete 16S rRNA nucleotide sequence of P. putida 1290R (1,459 bp) is available under accession number AY491973.
RESULTS
Screening of bacterial isolates for their ability to degrade IAA.
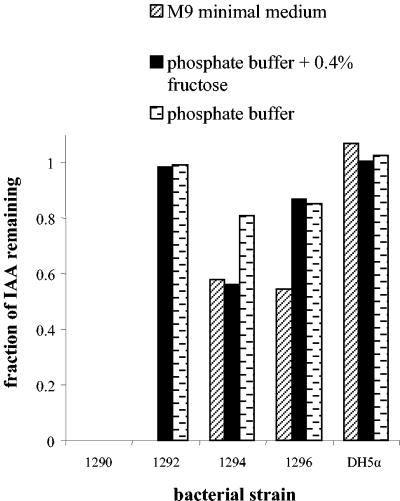
We identified several IAA-degrading isolates that could be assigned to one of four groups based on differences in their ability to catabolize IAA under different circumstances (Fig. 1): (i) complete removal of IAA in M9 minimal medium (which contains ammonium chloride as a nitrogen source), and, independent of the presence of 0.4% fructose, complete removal of IAA in phosphate buffer (which contains no nitrogen); (ii) complete removal of IAA in M9 medium, but not in phosphate buffer, with or without 0.4% fructose; (iii) partial degradation of IAA in M9 medium or phosphate buffer with 0.4% fructose and limited removal of IAA in just phosphate buffer; and (iv) partial degradation of IAA in M9 buffer and, regardless of the presence of 0.4% fructose, limited removal in phosphate buffer. As a control, we used E. coli DH5α, which did not degrade IAA under any condition (Fig. 1). Based on its unique ability to degrade IAA in all of the tested media, one isolate, number 1290, was selected for further study.
FIG. 1.
Complete, partial, or no degradation of IAA by different representative bacterial isolates in various media. Shown is the fraction of IAA that remained in the medium supernatant after a 75-h incubation of bacteria (100-fold diluted from an overnight King’s B-grown culture) with 9 mM IAA at 30°C in either M9 minimal medium, phosphate buffer supplemented with 0.4% fructose, or plain phosphate buffer. Isolate 1290 is the only representative of group 1 (see Results), isolate 1292 and 1294 are the only ones of groups 2 and 3, respectively, and isolate 1296 represents a total of three isolates in group 4.
Phylogeny of IAA-degrading strain 1290 based on 16S rRNA analysis.
The nearly complete nucleotide sequence of the 16S rRNA gene of strain 1290 showed high homology to those from γ-proteobacteria belonging to the genus Pseudomonas. Using RDPII's Sequence Match tool (6), top scores were found with P. putida KT2440 (accession number AE016774, 16S rRNA copies A and B, 99.7% identity in 1,459-bp overlap; AE016775/AE016778, copies C and E, 99.6% identity, 1,459-bp overlap), P. putida ML2 (99.7% identity, 1,459-bp overlap), P. putida PB4 (D37925, 99.6%, 1,328-bp overlap), P. putida mt-2 (D37924, 99.6%, 1,328-bp overlap), P. putida ATCC 17453 (AF094746, 99.6%, 1,459-bp overlap), and Pseudomonas sp. strain WBC-3 (AY040872, 99.6%, 1,459-bp overlap). Based on these results we propose to refer to isolate 1290 as Pseudomonas putida strain 1290. All further experiments described in the present study were done with a spontaneous rifampin-resistant mutant of P. putida 1290, i.e., P. putida 1290R.
Growth of P. putida 1290R on IAA and other compounds in M9 minimal medium or phosphate buffer.
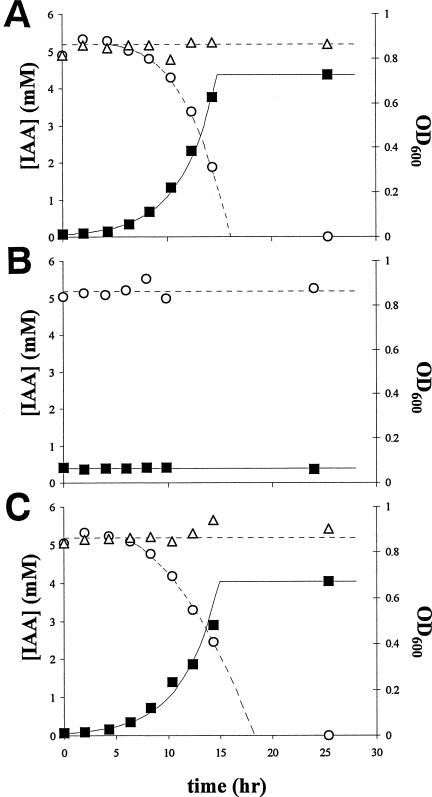
Figure 2A shows as a function of time the growth of P. putida 1290R in M9 minimal medium at the expense of IAA at an initial concentration of 5 mM. The disappearance of IAA was stoichiometrically concomitant with the increase in bacterial biomass. Growth ceased with the depletion of IAA from the medium. The growth rate reached a maximum μmax of 0.39 ± 0.01 h−1, which corresponds to a generation time of 1.8 h. The relative yield -δOD600/δ[IAA] was 0.18, indicating that 1 mmol of IAA supported the formation of 0.18 optical density units at 600 nm (OD600). Since an OD600 of 1 corresponds to ca. 109 cells per ml, 1 g of IAA could theoretically support a population of roughly 1012 bacterial cells. The presence of 0.4% glucose in the medium did not affect the degradation of IAA (not shown). In uninoculated M9 medium, IAA was not degraded. Erwinia herbicola 299R, a known producer of IAA (3), clearly lacked the ability to utilize IAA for growth (Fig. 2B). Also, P. putida KT2440 (18) lacked the ability to grow at the expense of IAA (not shown).
FIG. 2.
Growth of P. putida 1290R at the expense of IAA in M9 minimal medium (A) or phosphate buffer (C). Symbols: ▪, OD600 as a measure for bacterial biomass; ○, IAA concentration in the medium supernatant. Open triangles (▵) show the fate of IAA in the absence of bacteria. (B) No growth or IAA degradation was observed with E. herbicola 299R. The results shown are representative of several repetitions of the experiment.
The growth of P. putida 1290R in minimal medium was also supported by the IAA-like compound indole acetaldehyde but not by indole acrylic acid, indole butyric acid, indole lactic acid, indole propionic acid, indole pyruvic acid, or naphtalene acetic acid (not shown). Furthermore, growth was fast on benzoate and glucose (μmax of 0.24 and 0.56 h−1, respectively), slow on anthranilic acid (μmax of 0.13 h−1), and absent on catechol and tryptophan; on salicylic acid, growth was rapid (μmax of 0.36 h−1) but occurred only after a lag of 3 h (not shown).
P. putida 1290R grew well on IAA not only in M9 minimal medium but also in phosphate buffer (Fig. 2C). In this case, the bacteria relied on IAA as the sole source of carbon and energy, as well as nitrogen. The growth rate was only slightly lower than in M9 medium (μmax = 0.37 ± 0.01 h−1), as was the relative yield: -δOD600/δ[IAA] = 0.16. Cultures of P. putida 1290R grown on IAA in phosphate buffer consistently turned green fluorescent during the transition from exponential to stationary phase, indicating the production of siderophores.
Metabolic and enzyme activity in P. putida 1290R cells growing on IAA.
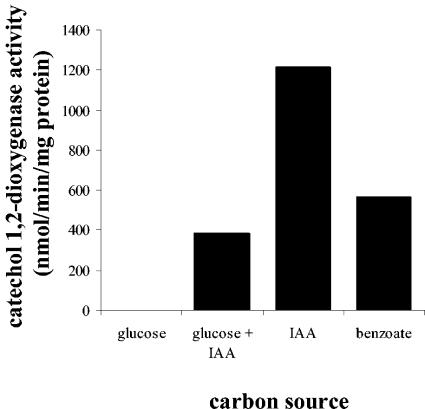
IAA-grown cells of P. putida 1290R showed instantly elevated levels of oxygen uptake upon addition of IAA (not shown), indicating the involvement of oxygenases in the IAA degradative pathway. Oxygen uptake was also elevated after the addition of catechol (not shown), suggesting that catechol is an intermediate of IAA metabolism. Indeed, catechol 1,2-dioxygenase activity could be detected in cell extracts of IAA-grown P. putida 1290R cells (Fig. 3). This activity, albeit to a lower level, was also detected in cells grown either on benzoate or on a mixture of glucose and IAA (Fig. 3). In cell extracts of glucose-grown P. putida 1290R cells no catechol 1,2-dioxygenase could be detected (Fig. 3), which suggests that IAA is an inducer, probably indirectly, of the catechol degradation pathway. No catechol 2,3-dioxygenase activity could be detected in cell extracts of IAA-grown P. putida 1290 cells (not shown), indicating that IAA-derived catechol is channeled exclusively through an ortho- and not a meta-cleavage pathway. The involvement of catechol as an intermediate of IAA metabolism was further confirmed by our finding that a catR mutant of P. putida 1290R, obtained by plasposon mutagenesis (8a) was unable to grow on IAA (not shown). The catR gene product codes for a positive regulator of the cat operon that encodes the degradation of catechol (18). Part of this operon is the catA gene for catechol 1,2-dioxygenase.
FIG. 3.
Catechol 1,2-dioxygenase activity in cell extracts of P. putida 1290R cells grown on M9 minimal medium containing 0.1% glucose, 0.1% glucose, plus 5 mM IAA, 5 mM IAA, or 8.5 mM benzoate.
Production of IAA by P. putida 1290R.
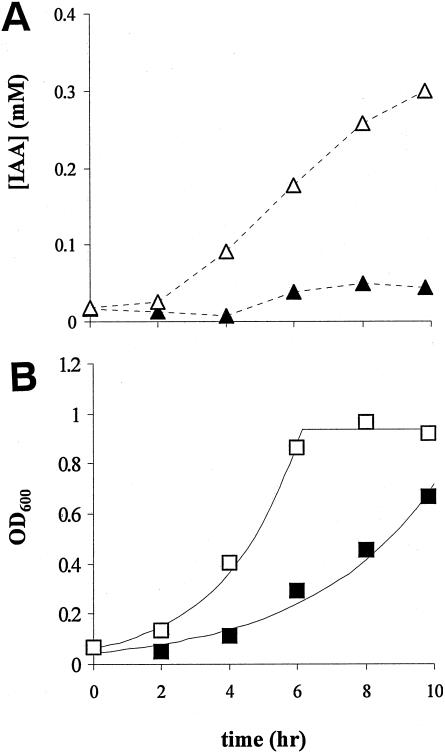
In M9 minimal medium containing glucose and tryptophan, IAA production by P. putida 1290R was obvious but modest compared to the positive control E. herbicola 299R (Fig. 4A). From 4.5 mM tryptophan, only ∼0.05 mM IAA was produced, which represents an ∼1% conversion efficiency, compared to >12% for E. herbicola 299R. We cannot rule out that during this experiment any IAA produced from tryptophan was again degraded by P. putida 1290R, resulting in an underestimation of actual IAA production. An interesting difference between the two strains is that the production of IAA by E. herbicola 299R continued into the stationary-growth phase, whereas IAA production by P. putida 1290R leveled off during the exponential growth phase (compare Fig. 4A and B). Unlike E. herbicola 299R and P. putida 1290R, P. putida KT2440 was not able to produce IAA from tryptophan (not shown).
FIG. 4.
(A) Production of IAA from 4.5 mM tryptophan in M9 minimal medium, supplemented with 0.1% glucose, by P. putida 1290R (▴) or E. herbicola 299R (▵). (B) Corresponding growth curves of P. putida 1290R (▪) and E. herbicola 299R (□). The results shown are representative of several repetitions of the experiment.
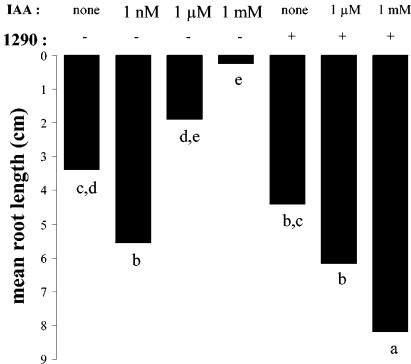
P. putida 1290R as a sink of IAA in plant root development.
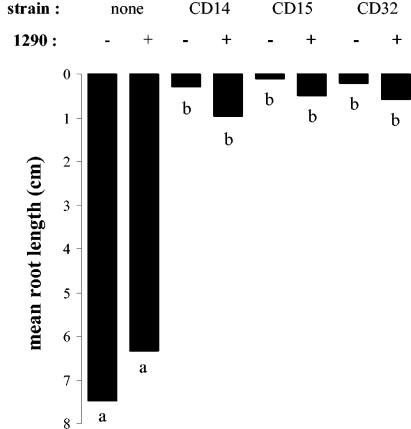
To assign a possible biological role to bacterial IAA degradation, cells of P. putida 1290R were tested for their ability to partially or completely abolish the effect of exogenously added IAA on the development of radish roots (Fig. 5). The effect of IAA by itself on root development was clearly concentration dependent. At 1 mM, roots were completely stunted, whereas at 1 nM the root growth was significantly stimulated compared to the control treatment. Although the addition of bacteria alone had no significant effect on root development, in combination with 1 μM or 1 mM IAA, the bacteria completely abolished the negative effects of IAA. In fact, compared to the control treatment, root development was significantly stimulated by the addition of mixtures of IAA and bacteria (Fig. 5). P. putida 1290R was also examined for its ability to interfere in root assays with bacteria that are known to overproduce IAA in culture (Fig. 6). Roots inoculated with Rahnella aquaticus strains CD14 or CD15 or with Pseudomonas syringae CD32 were severely stunted compared to roots that were untreated or treated with P. putida 1290R. In coinoculation with P. putida 1290R, the stunting caused by these strains was reduced, on average, by a factor 3 to 4. Although this effect was statistically not significant, we consistently observed this trend in several repetitions of the experiment (not shown).
FIG. 5.
Effect of IAA, P. putida 1290R, and combinations thereof on the elongation of radish roots in growth pouches. Different letters indicate significantly different (P < 0.05) root lengths. −, No bacteria were added initially; +, addition of 5 × 108 cells of P. putida 1290R per ml.
FIG. 6.
Effect of coinoculation of P. putida 1290R and bacterial IAA producers CD14, CD15, and CD32 on radish root elongation. Different letters indicate significantly different (P < 0.05) root lengths. Inoculation in the absence (−) or the presence (+) of P. putida 1290R was as indicated.
DISCUSSION
The ability to degrade IAA may serve P. putida strain 1290 in two, not mutually exclusive ways. First, IAA is a substrate for growth. This aspect of bacterial IAA degradation has often been overlooked in other studies, as demonstrated by the fact that we are, to the best of our knowledge, the first to report a quantitative correlation between increase in bacterial biomass and IAA degradation (Fig. 2A and C). A second function of IAA degradation is the manipulation of IAA concentrations in its immediate surroundings. We will address both functions in more detail below and elaborate on whether and how these functions apply to other BIDs.
It is interesting that IAA is close to what bacteria such as P. putida 1290 might consider an ideal food source. It conveniently carries the two most abundant elements of a bacterial cell (C and N) in a single molecule, with a C/N ratio of 8.6. If we suppose that half of the carbon in IAA is used for the generation of energy, the remaining C/N ratio is 4.3, which is close to the C/N of 3.6 for a typical bacterium (28). Another interesting observation is that, under optimal conditions (Fig. 2), the growth rates of P. putida 1290 on IAA are surprisingly high compared to those reported for bacteria growing aerobically on similar N-heterocyclic aromatic compounds (40, 44). This combination of high nutritional value and easy digestibility make IAA, in principle, a potent selector for the presence and expansion of P. putida 1290 populations in locales where IAA is present.
Some BIDs degrade IAA only partially or only under certain conditions. For example, while strain 1292 degraded and grew on IAA in M9 medium, showing that it can use IAA as a source of carbon and energy, it did not degrade or grow on IAA in phosphate buffer (Fig. 1). This may be explained by the inability of this bacterium to release nitrogen from IAA. It would suggest that degradation and therefore growth utilization of IAA by this strain is only partial, which would account for the reduced yield of this strain on IAA in M9 medium (not shown). Compared to strain 1290, strain 1292 probably uses either a different pathway or one that is similar but incomplete; this remains to be investigated. Another factor influencing the degradation and growth utilization of IAA is catabolite repression. Although the presence of fructose or glucose did not interfere with the ability of P. putida 1290 to degrade IAA, Libbert and Risch reported that degradation of IAA by their isolates was reduced in the presence of 0.2% glucose in phosphate buffer and even abolished with 5% glucose or 1% malic acid (23). In both the rhizosphere and on leaves, sugars such as glucose and fructose are abundantly available (16, 21), suggesting that in such environments strains such as the ones described by Libbert and Rische would be less able to utilize available IAA and therefore would have a disadvantage over strains such as P. putida 1290.
Three different pathways have been proposed for bacterial degradation of IAA. One pathway, suggested by Proctor for a Pseudomonas strain isolated from soil (35), involves the intermediates skatole (3-methyl indole), indoxyl (3-hydroxy indole), salicylic acid, and catechol. Another pathway, proposed by Tsubokura et al. for a bacterium isolated from air (47) and by Egebo et al. for Bradyrhizobium japonicum (10), assumes degradation via o-formaminobenzoylacetic acid, o-aminobenzoylacetic acid, and anthranilic acid. A third pathway, described by Olesen and Jochimsen, also for Bradyrhizobium japonicum (31), assumes the assimilation of IAA via dioxindole-3-acetic acid, dioxindole, isatin, α-aminophenyl glyoxylic acid (isatinic acid), and anthranilic acid. Which of these pathways, if any, is operational in P. putida 1290 remains to be elucidated. Besides catechol, none of the intermediates are known. The addition of salicylic acid but not anthranilic acid to cell suspensions of IAA-grown P. putida 1290 cells resulted in a slight increase in oxygen uptake (not shown), suggesting that perhaps salicylic acid, not anthranilic acid, is a pathway intermediate, similar to Proctor's soil Pseudomonas strain. The genetic basis for IAA metabolism in any of these IAA degraders, including P. putida 1290, remains to be elucidated.
There are two apparent sources of IAA that maintain and explain the existence of BIDs and their ability to use IAA as a food substrate, namely, plants and BIPs (or other microbial producers of IAA). In a study by Libbert et al., hardly any IAA could be extracted from sterile pea and corn plants, whereas nonsterile plants contained considerable amounts of IAA; the latter finding was attributed to the presence of IAA-producing bacteria (24). This BIP-derived IAA would be readily available to BIDs, as long as BIPs and BIDs are in close association with each other. Such an association is very likely, since BIPs and BIDs are both found abundantly on plant surfaces such as roots, shoots, and leaves (23). Brandl and Lindow (4) estimated, based on the extent of root growth inhibition by BIPs, that bacteria such as E. herbicola 299R could produce IAA concentrations in the rhizosphere as high as ca. 10 μM. Such levels of IAA production would yield 1.75 μg of IAA per ml of water solution in equilibrium with the plant. Since as much as 1 ml of liquid might commonly be associated with a root, we can estimate from the data presented here that BIP-derived IAA in these environments would sustain substantial BID populations of 106 cells or more.
It is not clear whether and in what quantities plants leak or secrete IAA and how much of it could be used by BIDs. As estimated from the growth experiments presented here, little IAA would be needed to establish a substantial population of IAA degraders: roughly 1 pg of IAA suffices for the generation of one bacterial cell. This provokes the intriguing but untried hypothesis that a plant would have the potential to specifically enrich for IAA degraders by the secretion of IAA. Thus, IAA could represent a means for the plant to select for a highly specific microbial population to cover its surfaces. It needs further investigation to see whether and how such a selection for BIDs is beneficial to plants. Possibly, BIDs produce plant growth stimulating substances or carry properties that are antagonistic to plant pathogens such as by the production of siderophores (34) or secondary metabolites with antimicrobial activity. Another intriguing possibility is that bacteria such as P. putida 1290R protect plants from fungal infections by keeping plant surfaces free of IAA: fungal pathogens may use IAA as a chemical cue that signals plant presence and induces mechanisms for invasion (36).
It is clear from the present study that by its IAA-degrading ability P. putida 1290 has the potential to manipulate IAA concentrations in its interaction with plants. We were able to show that P. putida 1290R abolished the harmful effects of the exogenously added hormone at micro- and millimolar concentrations (Fig. 5). It will be necessary to assess whether P. putida 1290R also has the capacity to lower IAA concentrations inside the plant by acting as a sink for IAA on the outside of the plant. A very similar phenomenon has been reported for a number of rhizobacteria that express the enzyme 1-aminocyclopropane-1-carboxylate deaminase and are capable of lowering the plant endogenous levels of another plant hormone, ethylene (12). An interesting observation from the coinoculation of P. putida 1290 and 1 mM IAA was that it seemed to stimulate radish root elongation more than compared to untreated roots. This synergistic effect, which implicates the potential use of P. putida 1290 as a seed inoculant to stimulate plant growth, can be explained by a number of reasons. Perhaps IAA was degraded to concentrations at which it became unavailable to the bacteria but at which it still exerted a positive effect on the roots. Another possibility is that the stimulation is a secondary consequence of the enrichment for P. putida 1290, whereby the roots are stimulated by an unknown bacterial factor. It can also not be ruled out that a metabolite of IAA degradation was released by P. putida 1290R and enhanced root development.
Under the conditions tested, P. putida 1290R was not very efficient in protecting radish roots from damage by IAA-producing bacteria (Fig. 6). One reason could be that IAA is not the main cause of the inhibitory effect of these bacteria on root development. In that case, we would not have expected P. putida 1290, as an IAA degrader, to exert a large effect. Another possibility is that P. putida 1290 and each of the IAA-producing strains occupied different niches on the plant root, so that any IAA from the latter would not be available to IAA degrader P. putida 1290R. As an IAA producer, P. putida 1290R did not have the same deleterious effect on radish roots as did the other IAA-producing strains. We suspect that this is a consequence of its ability to both produce and destroy IAA. A dual BID/BIP status is not unique to P. putida 1290R. For example, 2 of 15 of the IAA degraders reported by Libbert and Risch (23) share this trait. Two other examples are Bradyrhizobium japonicum and Pseudomonas savastanoi. For both, it has been suggested that the dual status enables these bacteria to finely control the level of IAA in interaction with their respective host plants. Symbiotic nitrogen fixation by nodules of B. japonicum on soybean apparently requires a carefully balanced level of IAA (41), which depends on the ability of the bacterium to degrade IAA (10, 17, 31). For optimal infection and colonization of olive and oleander, P. savastanoi requires a functional synthetase that converts IAA into the inactive IAA-lysine (15, 39); knockout mutants showed accumulation of IAA, attenuation of virulence, and reduced growth in planta (11). P. putida 1290 most likely would exist in a nearly commensalistic association with plants since its ability to degrade IAA may function in homeostasis, i.e., balancing IAA-based interactions away from extremes that might lead to a parasitic relationship, but one that would benefit the bacterium. A systematic inventory of the IAA-degrading capacity among bacteria, whether they produce IAA or not, will expose whether, to what extent, and under what circumstances a bacterial IAA-degradative phenotype is exploited for the purpose of manipulating plant physiology.
Acknowledgments
We thank the Niyogi lab for use of the Oxygraph system and Maria Marco and Wietse de Boer for valuable commentary on the manuscript. We also thank Renée Koutsoukis for help with root elongation assays.
This research was funded in large part by U.S. Department of Agriculture National Research Initiative grant 96-35303-3377 and U.S. Department of Energy grant DE-FG03-86ER13518 and by The Netherlands Organization for Scientific Research (NWO) Innovational Research Incentive Grant VENI 863.03.005.
Footnotes
Publication 3454 of the The Netherlands Institute of Ecology (NIOO-KNAW).
REFERENCES
- 1.Andreae, W. A., and M. W. H. van Ysselstein. 1960. Studies on 3-indoleacetic acid metabolism. VI. 3-Indoleacetic acid uptake and metabolism by pea roots and epicotyls. Plant Physiol. 35:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouillenne-Walrand, M., C. Leyh, and T. Gaspar. 1963. Mise en évidence d'un protecteur de l'acide beta indol-acétique dans un extrait de feuilles de Zea mays L. Bull. Soc. R. Sci. Liege 4:262-268. [Google Scholar]
- 3.Brandl, M. T., and S. E. Lindow. 1996. Cloning and characterization of a locus encoding an indole pyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl. Environ. Microbiol. 62:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl, M. T., and S. E. Lindow. 1998. Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Appl. Environ. Microbiol. 64:3256-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruckner, B., and D. Blechschmidt. 1991. The gibberellin fermentation. Crit. Rev. Biotechnol. 11:163-192. [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costacurta, A., and J. Vanderleyden. 1995. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 21:1-18. [DOI] [PubMed] [Google Scholar]
- 8.Davies, P. J. 1995. The plant hormone concept: concentration, sensitivity, and transport, p. 13-18. In P. J. Davies (ed.), Plant hormones: physiology, biochemistry, and molecular biology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 8a.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analyses of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dullaart, J. 1970. The bioproduction of indole-3-acetic acid and related compounds in root nodules and roots of Lupinus luteus and by its microbial symbiont. Acta Bot. Neerl. 19:573-615. [Google Scholar]
- 10.Egebo, L. A., S. V. S. Nielsen, and B. U. Jochimsen. 1991. Oxygen-dependent catabolism of indole-3-acetic acid in Bradyrhizobium japonicum. J. Bacteriol. 173:4897-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, N. L., and T. Kosuge. 1988. Role of indoleacetic acid lysine synthetase in regulation of indole-acetic-acid pool size and virulence of Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 170:2367-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glick, B. R., D. M. Penrose, and J. P. Li. 1998. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190:63-68. [DOI] [PubMed] [Google Scholar]
- 13.Glickmann, E., and Y. Dessaux. 1995. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 61:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland, M. A. 1997. Occam's razor applied to hormonology: are cytokinins produced by plants? Plant Physiol. 115:865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutzinger, O., and T. Kosuge. 1968. Microbial synthesis and degradation of indole 3-acetic acid. III. The isolation and characterization of indole-3-acetyl-epsilon-l-lysine. Biochemistry 7:601-605. [DOI] [PubMed] [Google Scholar]
- 16.Jaeger, C. H., S. E. Lindow, S. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, J. B., H. Egsgaard, H. Vanonckelen, and B. U. Jochimsen. 1995. Catabolism of indole-3-acetic acid and 4-chloroindole-3-acetic and 5-chloroindole-3-acetic acid in Bradyrhizobium japonicum. J. Bacteriol. 177:5762-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez, J. I., B. Minambres, J. L. Garcia, and E. Diaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824-841. [DOI] [PubMed] [Google Scholar]
- 19.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 20.Lambrecht, M., Y. Okon, A. Vande Broek, and J. Vanderleyden. 2000. Indole-3-acetic acid: a reciprocal signalling molecule in bacteria-plant interactions. Trends Microbiol. 8:298-300. [DOI] [PubMed] [Google Scholar]
- 21.Leveau, J., and S. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 6:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leveau, J. H. J., and J. R. van der Meer. 1996. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 178:6824-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libbert, E., and H. Risch. 1969. Interactions between plants and epiphytic bacteria regarding their auxin metabolism. V. Isolation and identification of the IAA-producing and -destroying bacteria from pea plants. Physiol. Plantarum 22:51-58. [Google Scholar]
- 24.Libbert, E., S. Wichner, U. Schiewer, H. Risch, and W. Kaiser. 1966. The influence of epiphytic bacteria on auxin metabolism. Planta 68:327-344. [DOI] [PubMed] [Google Scholar]
- 25.Lindow, S. E., C. Desurmont, R. Elkins, G. McGourty, E. Clark, and M. T. Brandl. 1998. Occurrence of indole-3-acetic acid-producing bacteria on pear trees and their association with fruit russet. Phytopathology 88:1149-1157. [DOI] [PubMed] [Google Scholar]
- 26.Manulis, S., and I. Barash. 2003. Pantoea agglomerans pvs. gypsophilae and betae, recently evolved pathogens? Mol. Plant Pathol. 4:307-314. [DOI] [PubMed] [Google Scholar]
- 27.Mino, Y. 1970. Studies on the destruction of indole-3-acetic acid by a species of Arthrobacter. IV. Decomposition products. Plant Cell Physiol. 11:129-138. [Google Scholar]
- 28.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 29.Nielsen, S. V. S., L. A. Egebo, and B. Jochimsen. 1988. Bradyrhizobial indoleacetic acid metabolism and its significance for root nodule development, p. 151-152. In R. Palacios and D. P. S. Verma (ed.), Molecular genetics of plant-microbe interactions. APS Press, St. Paul, Minn.
- 30.Nissl, D., and M. H. Zenk. 1969. Evidence against induction of protein synthesis during auxin-induced initial elongation of Avena coleoptiles. Planta 89:323-341. [DOI] [PubMed] [Google Scholar]
- 31.Olesen, M. R., and B. U. Jochimsen. 1996. Identification of enzymes involved in indole-3-acetic acid degradation. Plant Soil 186:143-149. [Google Scholar]
- 32.Patten, C. L., and B. R. Glick. 1996. Bacterial biosynthesis on indole-3-acetic acid. Can. J. Microbiol. 42:207-220. [DOI] [PubMed] [Google Scholar]
- 33.Patten, C. L., and B. R. Glick. 2002. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68:3795-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persello-Cartieaux, F., L. Nussaume, and C. Robaglia. 2003. Tales from the underground: molecular plant-rhizobacteria interactions. Plant Cell Environ. 26:189-199. [Google Scholar]
- 35.Proctor, M. H. 1958. Bacterial dissimilation of indoleacetic acid: a new route of breakdown of the indole nucleus. Nature 181:1345. [DOI] [PubMed] [Google Scholar]
- 36.Prusty, R., P. Grisafi, and G. R. Fink. 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigaud, T., C. Soutygrosset, R. Raimond, J. P. Mocquard, and P. Juchault. 1991. Feminizing endocytobiosis in the terrestrial crustacean Armadillidium vulgare Latr (Isopoda): recent acquisitions. Endocytobiosis Cell Res. 7:259-273. [Google Scholar]
- 38.Riviere, J., and B. Berthier. 1964. Action des microorganismes de la rhizosphère sur la croissance du blé. III. Isolement et identification des bacteries dégradant l'acide indole-3-acétique. Ann. Inst. Pasteur 3:250-256. [Google Scholar]
- 39.Roberto, F. F., H. Klee, F. White, R. Nordeen, and T. Kosuge. 1990. Expression and fine-structure of the gene encoding N-epsilon-(indole-3-acetyl)-l-lysine synthetase from Pseudomonas savastanoi. Proc. Natl. Acad. Sci. USA 87:5797-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronen, Z., A. Abeliovich, and A. Nejidat. 1997. Biodegradation of alkylpyridines by bacteria isolated from a polluted subsurface. Biodegradation 8:357-361. [DOI] [PubMed] [Google Scholar]
- 41.Rosendahl, L., and B. U. Jochimsen. 1995. In vitro indole-3-acetic acid uptake in symbiosomes from soybean (Glycine max L) root nodules. Symbiosis 19:99-110. [Google Scholar]
- 42.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Silverstone, S. E., D. G. Gilchrist, R. M. Bostock, and T. Kosuge. 1993. The 73-kb pIAA plasmid increases competitive fitness of Pseudomonas syringae subspecies savastanoi in oleander. Can. J. Microbiol. 39:659-664. [DOI] [PubMed] [Google Scholar]
- 44.Sutton, S. D., S. L. Pfaller, J. R. Shann, D. Warshawsky, B. K. Kinkle, and J. R. Vestal. 1996. Aerobic biodegradation of 4-methylquinoline by a soil bacterium. Appl. Environ. Microbiol. 62:2910-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taiz, L., and E. Zeiger. 1998. Plant physiology, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 46.Tomaszewski, M., and K. V. Thimann. 1966. Interactions of phenolic acids, metallic ions, and chelating agents on auxin-induced growth. Plant Physiol. 41:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsubokura, S., Y. Sakamoto, and K. Ichihara. 1961. The bacterial decomposition of indoleacetic acid. J. Biochem. 49:38-42. [Google Scholar]
- 48.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wichner, S. 1968. Beziehungen zwischen Pflanzen und epiphytischen Bakterien hinsichtlich ihres Auxinstoffwechsels. IV. IES-Zerstoerung und Produktion von Inhibitoren der IES-Oxydase durch epiphytische Bakterien. Flora Abt. A 159:141-166. [Google Scholar]
- 50.Wichner, S., and E. Libbert. 1964. Hinweise zur Exoenzym-Natur einer IES-Oxydase. Naturwissenschaften 51:268. [Google Scholar]
- 51.Wichner, S., and E. Libbert. 1968. Interactions between plants and epiphytic bacteria regarding their auxin metabolism. I. Detection of IAA-producing epiphytic bacteria and their role in long duration experiments on tryptophan metabolism in plant homogenates. Physiol. Plant 21:227-241. [Google Scholar]
- 52.Wichner, S., and E. Libbert. 1968. Interactions between plants and epiphytic bacteria regarding their auxin metabolism. II. Influence of IAA-producing epiphytic bacteria on short-term IAA production from tryptophan in plant extracts. Physiol. Plant 21:500-509. [Google Scholar]