Abstract
Streptococcus mutans is implicated as a major etiological agent in human dental caries, and one of the important virulence properties of this organism is its ability to form biofilms (dental plaque) on tooth surfaces. We examined the role of autoinducer-2 (AI-2) on S. mutans biofilm formation by constructing a GS-5 luxS-null mutant. Biofilm formation by the luxS mutant in 0.5% sucrose defined medium was found to be markedly attenuated compared to the wild type. Scanning electron microscopy also revealed that biofilms of the luxS mutant formed larger clumps in sucrose medium compared to the parental strain. Therefore, the expression of glucosyltransferase genes was examined and the gtfB and gtfC genes, but not the gtfD gene, in the luxS mutant were upregulated in the mid-log growth phase. Furthermore, we developed a novel two-compartment system to monitor AI-2 production by oral streptococci and periodontopathic bacteria. The biofilm defect of the luxS mutant was complemented by strains of S. gordonii, S. sobrinus, and S. anginosus; however, it was not complemented by S. oralis, S. salivarius, or S. sanguinis. Biofilm formation by the luxS mutant was also complemented by Porphyromonas gingivalis 381 and Actinobacillus actinomycetemcomitans Y4 but not by a P. gingivalis luxS mutant. These results suggest that the regulation of the glucosyltransferase genes required for sucrose-dependent biofilm formation is regulated by AI-2. Furthermore, these results provide further confirmation of previous proposals that quorum sensing via AI-2 may play a significant role in oral biofilm formation.
Quorum sensing (QS) is a process whereby bacteria communicate with one another by means of the secretion of chemical signal molecules called autoinducers (AIs) (3, 4, 35, 38). In the bioluminescent gram-negative marine bacterium Vibrio harveyi, two distinct AIs, AI-1 (6, 9) and AI-2, regulate light emission (36). LuxS is an enzyme involved in the catabolism of S-adenosylmethionine and converts ribose homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione, the precursor of AI-2 (6, 46, 51). This system has been referred to as an interspecies quorum system and may operate as a universal quorum system for many bacteria possessing the characteristic luxS gene (5). The luxS gene is highly conserved across a diverse range of gram-negative and gram-positive bacterial species, and AI-2 is produced by many of these species. QS enables a population of bacteria collectively to regulate gene expression including expression of virulence factors (21, 39), competence for genetic transformation (1, 28, 37), conjugal DNA transfer (20, 52, 56), and the production of antibiotics and secondary metabolites (31, 49), as well as biofilm formation (13). However, more recent investigations have also indicated that AI-2 production is regulated at the level of LuxS substrate availability and not at the level of luxS expression. Consequently, AI-2-dependent signaling can also reflect the metabolic state of the cell and not necessarily cell density (7).
Biofilms are sessile communities of microorganisms attached to a surface (12, 41, 53). It is clear that microorganisms undergo profound changes during their transition from planktonic organisms to cells that are part of a complex, surface-attached community. These changes are reflected in the new phenotypic characteristics developed by biofilm bacteria and occur in response to a variety of environmental signals (22, 40). Formation of these sessile communities and their inherent resistance to antimicrobial agents are important factors in many persistent and chronic bacterial infections (13, 30).
Streptococcus mutans is the principal causative agent of dental caries in humans and its ability to adhere to the tooth surface is paramount for the progression of disease (17, 29). One of the important virulence properties of these organisms is their ability to form biofilms known as dental plaque on tooth surfaces (27). Dental plaque, one of the best-studied biofilms, is a complex community comprising more than 500 bacterial species (23, 24, 43). The early colonizers of the enamel surfaces are predominantly streptococci, which form mixed-species microcolonies during early plaque development (25). To initiate heterogeneous bacterial interactions, diffusible signals may play important roles resulting in dental plaque formation (33).
Among the potential signaling molecules, the competence-stimulating peptides (CSP), approximately 21-mer cationic oligopeptides, regulate the competence pathways of streptococci (1), as well as S. mutans biofilm formation (28, 54). Recently, another diffusible signal molecule, AI-2, was identified in several oral bacteria (8, 11, 14, 24, 34). In the present study, we describe the biofilm phenotype of a S. mutans GS-5 luxS mutant and how AI-2 molecules affect biofilm formation of these organisms. In addition, we have developed a novel assay system for monitoring AI-2 levels using complementation of the S. mutans luxS mutant. Using this assay system, we show that some, but not all, oral streptococci, as well as selected gram-negative oral bacteria, complement the luxS mutation in S. mutans. These results provide a molecular explanation for the dependence of S. mutans sucrose-dependent biofilm formation on AI-2 and further suggest that LuxS-dependent signaling may mediate intra- and interspecies responses among bacteria in oral biofilms as recently proposed (33).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. All streptococcal species were grown anaerobically at 37°C in brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) broth or chemically defined medium (CDM) (54, 55) supplemented with appropriate carbon sources. Transformants of S. mutans were selected after growth on mitis salivarius agar (Difco Laboratories) plates supplemented with erythromycin (10 μg/ml). Porphyromonas gingivalis strains 381 and CW221 (luxS mutant) were grown and maintained anaerobically (10% CO2, 10% H2, 80% N2) in GAM broth (Nissui Medical Co., Tokyo, Japan) supplemented with hemin (5 μg/ml), menadione (1.0 μg/ml), tryptic soy broth (TSB; Difco Laboratories), or enriched tryptic soy agar (40 g of tryptic soy agar [Difco Laboratories], 5 g of BHI, 1 g of cysteine, 5 mg of hemin, and 1 mg of menadione/liter) at 37°C. For the P. gingivalis luxS mutant, erythromycin at a final concentration of 10 μg/ml was added. Actinobacillus actinomycetemcomitans Y4 was cultured at 37°C in a CO2-enriched atmosphere in THY broth (Todd-Hewitt broth [Difco Laboratories] supplemented with 1.0% yeast extract [Difco Laboratories]), diluted THY medium (THY/phosphate-buffered saline ratio, 1:2), or on THY agar plates. Escherichia coli DH5α was grown aerobically in 2× TY (1.6% Bacto Tryptone [Difco Laboratories], 1.0% yeast extract, 0.5% NaCl) medium at 37°C. V. harveyi was cultured at 30°C in Luria-marine (LM) medium (20 g of NaCl, 10 g of Bacto tryptone, and 5 g of yeast extract/liter).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics or distributions | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| GS-5 | Erys; Kans; serotype c human isolate | SUNYaBa |
| ΔluxS | Eryr; GS-5::pAYLS1101; luxS | This study |
| P. gingivalis | ||
| 381 | Type strain | KUb |
| CW221 | Eryr; 381::pVA2198; luxS | Wen and Kuramitsu (SUNYaB) |
| A. actinomycetemcomitans Y4 | Serotype b human isolate | KU |
| S. sobrinus MT8145 | Mutans group oral streptococci, serotype d | KU |
| S. gordonii DL1 | Mitis-sanguinis group oral streptococci | KU |
| S. oralis ATCC10557 | Mitis-sanguinis group oral streptococci | ATCCc |
| S. sanguinis ATCC10556 | Mitis-sanguinis group oral streptococci | ATCC |
| S. salivarius HT9R | Salivarius group oral streptococci | KU |
| S. anginosus FW73 | Aniginosus group oral streptococci | KU |
| V. harveyi | ||
| BB120 | Wild type | 51 |
| BB170 | luxN::Tn5, AI-1 sensor−, AI-2 sensor+, reporter strain | 51 |
| E. coli DH5α | Cloning host | Invitrogen |
| Plasmids | ||
| pBluescript II SK(+) | Ampr; cloning vector | Stratagene |
| pResEmMCS10 | Eryr; integration vector | 47 |
| pAYLS1101 | Ampr Eryr; pBluescript II SK(+) bearing luxS upstream and downstream regions and Eryr gene | This study |
SUNYaB, the culture collection in Department of Oral Biology, State University of New York, Buffalo, N.Y.
KU, the culture collection in Department of Preventive Dentistry, Kyushu University, Fukuoka, Japan.
ATCC, American Type Culture Collection, Manassas, Va.
DNA manipulations.
DNA isolation, endonuclease restriction, ligation, and transformation of competent E. coli cells were carried out as previously described (45). Transformation of S. mutans was accomplished by procedures routinely carried out in this laboratory (44).
Construction of the luxS mutant.
The open reading frame for the luxS gene and the flanking regions were identified in the S. mutans UA159 database from the University of Oklahoma Advanced Center for Genome Technology (http://genome.ou.edu.smutans.html). The luxS-null mutant was constructed by allelic exchange via insertion of an erythromycin resistance (Eryr) determinant into the gene. The plasmid used for disruption of the luxS gene was prepared as follows. The PCR fragments of the upstream and downstream regions of luxS were amplified with the primers luxSUF2883(Sma) and luxSUR3884(Bam), respectively, using chromosomal DNA from S. mutans GS-5 as the template. Initially, PCR products of the upstream region of luxS (luxSU) were cloned into plasmid pBluescript II SK(+) (Stratagene, La Jolla, Calif.). Furthermore, PCR products of the downstream region of luxS, luxSD, were amplified with the primers, luxSDF4347(Bam) and luxSDR5396(Xba) (Table 2), using chromosomal DNA from S. mutans GS-5 and cloned downstream of luxSU. The resultant plasmid was digested with BamHI, and BamHI-digested pResEmMCS10 (47) was inserted (Table 1). The resultant plasmid, pAYLS1101 (Table 1), was linearized with SmaI and resulted in a linear plasmid harboring S. mutans chromosomal DNA flanking the Eryr gene but devoid of the luxS gene, which was used to transform S. mutans GS-5.
TABLE 2.
Oligonucleotide primers and probes
| Primer or probe | Nucleotide sequence | Gene | Size (bp) |
|---|---|---|---|
| Primersa | |||
| luxSUF2883(Sma) | 5′-CCC CCC CCC GGG TCT TCA ATT CGA GCA GGA-3′ | luxS | 1,002 |
| luxSUR3884(Bam) | 5′-CCC CCC GGA TCC TGT CAT AGT AAA CTC CTT-3′ | ||
| luxSDF4347(Bam) | 5′-CCC CCC GGA TCC GTC ATC TAG TGT AAA AT-3′ | luxS | 1,050 |
| luxSDR5396(Xba) | 5′-CCC CCC TCT AGA TAC GGC TTT CTT TGC GG-3′ | ||
| gtfB-F241 | 5′-CAT ACA GTA ACG ACA AGC AGT AGC TCT A-3′ | gtfB | 99 |
| gtfB-R539 | 5′-GTA CGA ACT TTG CCG TTA TTG TCA TA-3′ | ||
| gtfC-F3228 | 5′-GAC AAC ACC TTA CTT CCT AAA TCG-3′ | gtfC | 119 |
| gtfC-R3346 | 5′-GCT TGG TTA CCA CTC GTT GAA TAA-3′ | ||
| gtfD-F4160 | 5′-TGA CAG GTA GCC AAC GCA TTA A-3′ | gtfD | 85 |
| gtfD-R4246 | 5′-GCT CAT CAT AAG CAA CAT CAC CTT-3′ | ||
| recA-F441 | 5′-GCG TGC CTT GAA GTT TTA TTC TTC-3′ | recA | 75 |
| recA-R515R | 5′-TGT TCC CCG GTT CCT TAA ATT-3′ | ||
| TaqMan probesb | |||
| gtfB MGB | 5′-FAM-ATG TTA TTG ATG ATA GCA ATG C-NFQ-MGB-3′ | gtfB | |
| gtfC 3258 | 5′-FAM-AAC CCA AAT CAC GGA ACA AGC AGT TCT G-TAMRA-3′ | gtfC | |
| gtfD 4185 | 5′-FAM-TCA GCG TGT CTT CTT TAC GCG AGA AGG-TAMRA-3′ | gtfD | |
| recA469 | 5′-FAM-CGT CTT GAT GTC CGC GGC AAT ACT C-TAMRA-3′ | recA |
Endonuclease recognition sequences are underlined.
FAM, 6-carboxyfluorescein; NFQ, nonfluorescent quencher; MGB, minor groove binder; TAMRA, 6-carboxytetramethylrhodamine.
Assay for in vitro biofilms. (i) Quantification of biofilm formation.
Biofilm formation was quantified as previously described (54). Flat-bottom polystyrene microtiter plates (enzyme immunoassay-radioimmunoassay plates, 96-well Easy Wash; Corning, Inc., Corning, N.Y.) containing 100 μl of CDM per well were inoculated with S. mutans GS-5 and its luxS mutant (1.7 × 105 CFU per well) after 24-h growth in BHI. The bacteria grown in BHI were centrifuged and washed with CDM and then suspended with the same amount of CDM. Bacterial cultures were diluted to 1:100 and 100 μl of bacterial solution was added to each well. After 48 h of incubation at 37°C, 25 μl of 1% (wt/vol) crystal violet (CV) solution was added to each well. After 15 min, the wells were rinsed three times with 200 μl of distilled water and air dried. The CV on the abiotic surface was solubilized in 95% ethanol, and the optical density at 570 nm (OD570) was determined. Growth was determined by measuring the turbidities (OD570) of parallel wells after resuspension of the sessile organisms with the planktonic cells.
(ii) Two-compartment system.
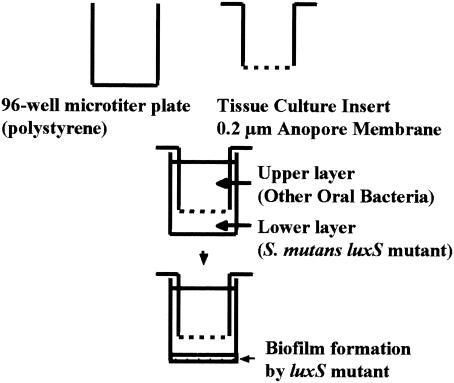
Each well of a 96-well polystyrene plate was separated into two compartments by using 0.2-μm-pore-size Anopore membranes (Nunc tissue culture inserts, 8-well strip; Nalge Nunc International, Naperville, IL.). The S. mutans luxS mutant was inoculated into the lower compartment, and other streptococci or gram-negative bacteria were inoculated into the upper compartment (Fig. 1). After coinoculation of the bacteria in both upper and lower compartments, the membrane strips were removed, and the amount of biofilm in the lower compartment was evaluated as previously described.
FIG. 1.
Two-compartment system for complementation analysis of the S. mutans luxS mutant.
SEM.
Biofilms formed on polystyrene surfaces were examined by scanning electron microscopy (SEM) to verify the quantitative results observed. Biofilms were anaerobically inoculated on 5-by-5-mm polystyrene tips in six-well polystyrene dishes. For the bacterial complementation analysis, the two-compartment assay was carried out by using six-well polystyrene dishes (Corning, Inc.) and 0.2-μm-pore-size Anopore membranes (Nunc 25-mm tissue culture inserts; Nalge Nunc International). The 5-by-5-mm polystyrene tips and the S. mutans luxS mutant were inoculated into the lower compartment, and the indicated streptococci or gram-negative bacteria were added into the upper compartment. After inoculation, biofilms on the polystyrene tips were washed once in distilled water, fixed with formaldehyde, and incubated at 20°C overnight. After dehydration through a graded series of ethanol, the polystyrene tips were air dried and sputter coated with gold. Samples were then examined at ×500 to ×7,500 magnification by using SEM (JEOL JSM-5400LV; JEOL Techniques, Ltd., Tokyo, Japan).
AI-2 bioassay for oral streptococci.
S. mutans strains and other streptococci were cultured in BHI (Difco) broth to stationary phase. The stationary-phase culture was then inoculated into CDM supplemented with 0.5% sucrose until OD570 reached 0.5. The culture supernatant was centrifuged and passed through a 0.2-μm-pore-size filter to remove cells. AI-2 bioassays with V. harveyi BB170 were performed basically as previously described (51). Briefly, V. harveyi BB170 was grown overnight at 30°C in LM medium. The culture was diluted 1:5,000 in fresh AI bioassay medium (AB medium: 0.3 M NaCl, 0.05 M MgSO4, 0.2% Casamino Acids, 1.0% potassium phosphate, 1.0 mM l-arginine, 2% glycerol, 0.01 μg of riboflavin/ml, 1 μg of thiamine HCl/ml), and 90 μl of the diluted cells with 10 μl of the cell-free culture supernatant was added at a 10% (vol/vol) final concentration into wells of 96-well microtiter plates. Positive control wells contained 10 μl of cell-free conditioned medium from V. harveyi BB170, whereas negative control wells contained 10 μl of sterile AB medium. Luminescence was measured every hour with a Luminometer (Fluoroscan Ascent FL; Thermo Labsystems, Vantaa, Finland).
Real-time quantitative RT-PCR.
Oligonucleotide primers and probes for the gtf genes, designed by using Primer Express 1.5 software (Applied Biosystems, Foster City, Calif.), are listed in Table 2. The primers for recA, used as an internal control, were also designed by using Primer Express 1.5 software. The fluorescent probes were labeled with a reporter dye (6-caboxyfluorescein) covalently attached at the 5′ end, and a quencher dye (6-carboxytetramethylrhodamine) covalently attached at the 3′ end. Total RNA was isolated from S. mutans GS-5 and the luxS mutant by using TRIzol Reagent (Gibco-BRL, Rockville, Md.) according to the manufacturer's instructions. Single-stranded cDNA was then synthesized in a reaction mixture containing 1.25 U of MultiScribe reverse transcriptase/μl, 0.4 U of RNase inhibitor/μl, 500 mM concentrations of each deoxynucleoside triphosphate, a 200 mM concentration of antisense primer, 1× reverse transcription (RT) buffer, 5.5 mM MgCl2 (TaqMan reverse transcription reagents; Applied Biosystems), and 1.0 μg of total RNA from each phase of culture at 48°C for 30 min. To check for DNA contamination, purified total RNA without reverse transcriptase served as a negative control. The resulting cDNA and negative control were amplified by using the TaqMan Universal PCR Master Mix (Applied Biosystems), which contained deoxynucleoside triphosphates with dUTP, AmpliTaq Gold DNA polymerase, Amperase UNG, optimized buffer, and a passive reference dye. For each PCR, a mixture containing template cDNA, 1× Master Mix, 200 nM concentrations of each forward and reverse primer, and 250 nM TaqMan probe was applied to a 96-well MicroAmp optical reaction plate with optical caps (Applied Biosystems). Amplification and detection of specific products were performed on the ABI Prism 7700 sequence detection system (PE Biosystems) with the following cycle profile: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, 60 cycles at 95°C for 15 s, and 60°C for 1 min. The critical threshold cycle (CT) was defined as the cycle at which the fluorescence becomes detectable above background and is inversely proportional to the logarithm of the initial number of template molecules. A standard curve was plotted for each primer-probe set with CT values obtained from amplification of known quantities of cDNA. To check the linearity of the detection system, a cDNA dilution series (1/10, 1/100, 1/1,000, and 1/10,000) was amplified with primer pairs and probes so that a correlation coefficient could be calculated from the standard curve displaying CT values. The standard curves were used to transform CT values to the relative number of cDNA molecules. The quantities of cDNA for gtfB, gtfC, and gtfD normalized to cDNA synthesized from recA were compared.
RESULTS
Nutrient factors affecting biofilm formation by the S. mutans luxS mutant.
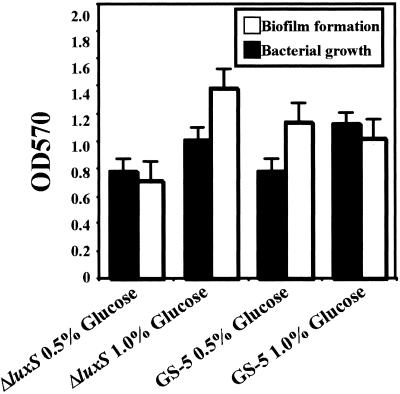
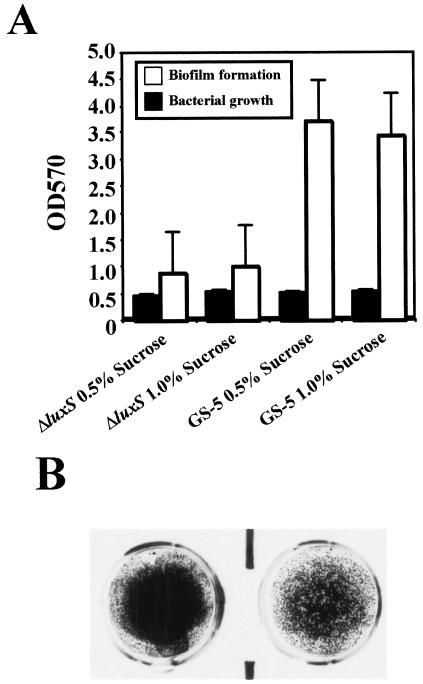
In order to analyze how the difference in the carbon source affects biofilm formation by the S. mutans luxS mutant, we examined glucose- and sucrose-dependent biofilm formation of this mutant. We found that biofilm formation by the luxS mutant in CDM supplemented with 0.5 or 1.0% glucose was not greatly attenuated compared to that of the parent strain, GS-5 (Fig. 2). However, biofilm formation by the luxS mutant in CDM supplemented with 0.5% sucrose was found to be markedly attenuated compared to the parent strain (Fig. 3A and B).
FIG. 2.
Biofilm assays of S. mutans supplemented with glucose. Bacterial growth and biofilm formation of S. mutans GS-5 and luxS mutant were measured. Bacteria were inoculated in CDM supplemented with glucose (0.5 or 1.0%). The data are the averages of three samples, and the standard errors are shown.
FIG. 3.
Biofilm assays of S. mutans supplemented with sucrose. (A) Bacterial growth and biofilm formation of S. mutans GS-5 and luxS mutant in CDM supplemented with sucrose (0.5 and 1.0%). The data are the averages of three samples, and the standard errors are shown. (B) Biofilm phenotypes of S. mutans GS-5 (left) and luxS mutant (right). S. mutans was inoculated in CDM supplemented with 0.5% sucrose on polystyrene surfaces.
SEM analysis of wild-type and luxS mutant biofilms.
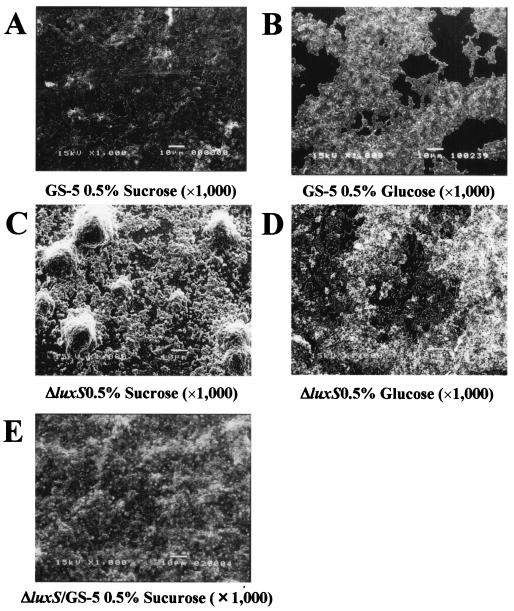
In addition, to further assess the biofilm phenotype of the luxS mutant in medium supplemented with different carbon sources, we also used SEM analysis (Fig. 4). The luxS mutant showed no significant qualitative difference in the biofilm phenotypes compared to GS-5 in the media supplemented with 0.5% glucose (Fig. 4A and B). In contrast, the luxS mutant formed biofilms that markedly differed from GS-5 in morphology when grown with 0.5% sucrose (Fig. 4C and D). The luxS mutant exclusively formed very large clumps in medium supplemented with sucrose compared to the biofilms formed by parental GS-5 strain (Fig. 4C).
FIG. 4.
SEM images of S. mutans GS-5 and luxS mutant biofilms on polystyrene surfaces. Bacteria were incubated on polystyrene tips after 48 h of incubation. S. mutans GS-5 inoculated in CDM supplemented with 0.5% sucrose (A) and glucose (B). Both images were obtained at a ×1,000 magnification. luxS mutant inoculated in CDM supplemented with 0.5% sucrose (C) and glucose (D). Images were obtained at a ×1,000 magnification. SEM analysis of the S. mutans luxS mutant complemented by GS-5 (E). Bacteria were incubated in the 0.5% sucrose-CDM for 48 h. Images were obtained at ×1,000 magnification.
Quantification of glucosyltransferase gene transcription in the S. mutans luxS mutant.
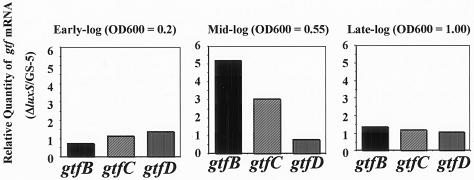
The increased sucrose-dependent colonization of hard surfaces by the luxS sessile cells could result from increased insoluble glucan synthesis catalyzed by the glucosyltransferases (Gtfs) of the mutant. In order to assess gtf gene expression in the S. mutans luxS mutant, real-time PCR assays were used for the quantification of the transcript levels of the gtfB, gtfC, and gtfD genes using an equal amount of total RNA from different growth stages in CDM supplemented with 0.5% sucrose. Initially, we evaluated the transcription levels of the recA gene in the luxS mutant and GS-5 in each culture phase. We observed no significant differences in the expression of the recA gene from samples in each culture phase (data not shown). The gtfBCD mRNA levels were almost the same in the early-log and late-log phases within each strain. However, the mRNA expression levels of the gtfB and gtfC genes, coding for Gtfs involved in insoluble glucan synthesis, in the luxS mutant were almost 5.5- and 3-fold higher, respectively, compared to GS-5 in the mid-log phase (Fig. 5). On the other hand, gtfD, coding for a Gtf synthesizing soluble glucans, expression in the luxS mutant was slightly reduced in the mid-log phase compared to that of GS-5 (Fig. 5).
FIG. 5.
Real-time quantitative RT-PCR analysis of gtfB, gtfC, and gtfD gene expression at different growth stages of S. mutans GS-5 and the luxS mutant. The relative quantities of each gtf cDNA from S. mutans GS-5 and the luxS mutant were assessed by TaqMan assays. The amounts of gtf cDNA were normalized to the amount of cDNA of recA after RT of the total RNA isolated from S. mutans GS-5 and the luxS mutant. The data are expressed as the means of four experiments. The results shown are representative of those obtained from two independently isolated RNA preparations.
Complementation analysis of the luxS mutant by other oral bacteria by using the two-compartment assay system.
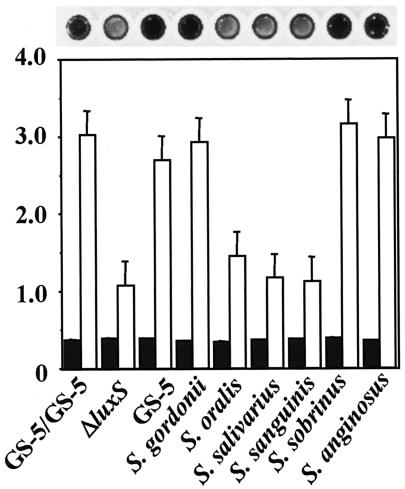
In order to determine whether other oral bacteria can communicate with S. mutans via AI-2 signaling, we determined whether other bacteria could complement the luxS mutation of strain GS-5. For the bacterial complementation analysis, we used a novel two-compartment biofilm assay system (Fig. 1). Initially, we performed complementation analysis of the GS-5 luxS mutant by using oral streptococci in the CDM supplemented with 0.5% sucrose. Biofilm formation by the luxS mutant was restored to wild-type GS-5 levels in the presence of S. gordonii, S. sobrinus, and S. anginosus in the upper compartments (Fig. 6). However, no significant restoration was observed with S. oralis, S. salivarius, and S. sanguinis (Fig. 6). That this complementation was dependent upon AI-2 was demonstrated by the observation that GS-5, but not its luxS mutant, grown in the upper compartment complemented biofilm formation by the luxS mutant in the bottom compartment (Fig. 6). The phenotypic alteration of the S. mutans luxS mutant complemented by GS-5 was also analyzed by SEM. Compared to the large clumps of cells formed by the luxS mutant in the presence of sucrose (Fig. 4C), much smaller aggregates were detected when the mutant was complemented by the parental strain GS-5 (Fig. 4E), a result similar to that observed with the parental strain alone (Fig. 4A).
FIG. 6.
Complementation analysis of S. mutans luxS mutant by other oral streptococci. Bacteria were inoculated in the upper and lower wells in CDM supplemented with 0.5% sucrose for 48 h. Bacterial growth and biofilm formation of the S. mutans luxS mutant in the lower wells were measured. The data are the averages of three samples, and the standard errors are shown. Bars: □, biofilm formation; ▪, bacterial growth. The CV-stained biofilms of the S. mutans luxS mutant complemented by other streptococci on polystyrene surface are shown above the graph. GS-5/GS-5, GS-5 complemented by GS-5 (positive control); ΔluxS, luxS mutant complemented by luxS mutant (negative control); GS-5, luxS mutant complemented by GS-5, S. gordonii, luxS mutant complemented by S. gordonii DL1; S. oralis, luxS mutant complemented by S. oralis ATCC 10557; S. salivarius, luxS mutant complemented by S. salivarius HT9R; S. sanguinis, luxS mutant complemented by S. sanguinis ATCC 10556; S. sobrinus, luxS mutant complemented by S. sobrinus MT8145; S. anginosus, luxS mutant complemented by S. anginosus FW73.
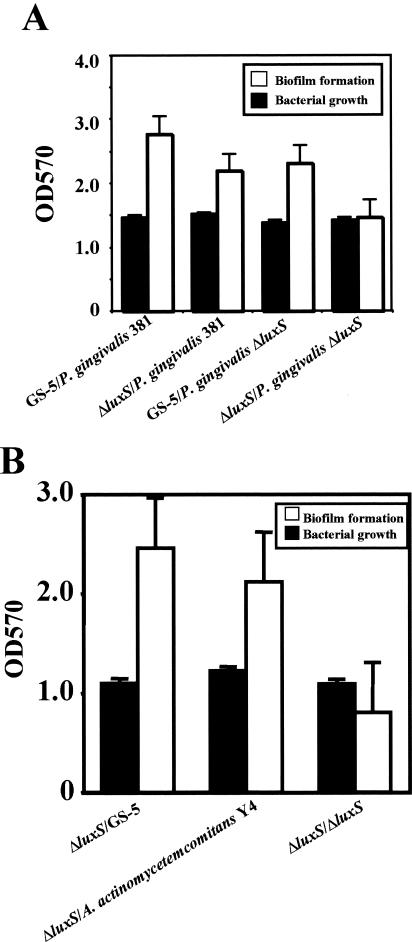
Furthermore, we performed complementation analysis with the gram-negative periodontopathic bacteria P. gingivalis and A. actinomycetemcomitans. Both organisms were recently demonstrated to secrete AI-2 (11, 14). P. gingivalis 381 and its luxS mutant were inoculated into the diluted TSB medium supplemented with 0.5% sucrose. Each strain was inoculated into the upper wells of the two-compartment system. Biofilm formation by the S. mutans luxS mutant was restored to parental levels when the luxS mutant was coinoculated with the P. gingivalis 381 parental strain. However, biofilm formation by the luxS mutant was not restored when coinoculated with the P. gingivalis luxS mutant (CW221) (Fig. 7A). In addition, biofilm formation by the S. mutans luxS mutant was restored to GS-5 levels when cocultured with A. actinomycetemcomitans Y4 (Fig. 7B).
FIG. 7.
Complementation analysis of S. mutans luxS mutant by periodontopathic bacteria. The S. mutans luxS mutant was inoculated into the diluted TSB medium (for P. gingivalis) or diluted THY medium (for A. actinomycetemcomitans) supplemented with 0.5% sucrose in the lower-wells of the two-compartment system. (A) P. gingivalis 381, P. gingivalis luxS mutant (CW221), and (B) A. actinomycetemcomitans Y4 were inoculated into the diluted TSB medium or diluted THY medium supplemented with 0.5% sucrose in the upper wells. The bacterial growth and biofilm formation of the S. mutans luxS mutant in the lower wells were measured. The data are the averages of three samples, and the standard errors are shown. (A) GS-5/P. gingivalis 381, GS-5 complemented by P. gingivalis 381 (positive control); ΔluxS/P. gingivalis 381, S. mutans luxS mutant complemented by P. gingivalis 381; GS-5/P. gingivalis ΔluxS, GS-5 complemented by P. gingivalis luxS mutant; ΔluxS/P. gingivalis ΔluxS, S. mutans luxS mutant complemented by P. gingivalis luxS mutant. (B) ΔluxS/GS-5, S. mutans luxS mutant complemented by GS-5; ΔluxS/A. actinomycetemcomitans Y4, S. mutans luxS mutant complemented by A. actinomycetemcomitans Y4.
AI-2 production in oral bacteria.
To confirm that the various streptococci, P. gingivalis, and A. actinomycetemcomitans produce AI-2, we examined the AI-2 levels with V. harveyi BB170 (luxN::Tn5, AI-1 sensor −, AI-2 sensor +) as a reporter strain. Cell-free supernatants of the bacteria were added to V. harveyi BB170, and the luminescence induced by the culture supernatant of these oral bacteria was measured. The luminescence level of BB170 induced by supernatant from S. mutans GS-5 was ca. 8% of that stimulated by V. harveyi BB120 (Table 3). In contrast, the luminescence of BB170 was significantly decreased by supernatants prepared from the GS-5 luxS mutant compared to GS-5 (Table 3). The enhanced luminescence of BB170 was equivalent to GS-5 when stimulated by S. gordonii DL1 or S. sobrinus MT8145. S. salivarius HT9R and S. anginosus FW73 stimulated the luminescence of BB170 approximately twofold higher compared to S. mutans GS-5. However, the luminescence of BB170 strain was decreased by S. oralis ATCC 10557 and S. sanguinis ATCC 10556 compared to GS-5 (Table 3).
TABLE 3.
AI-2 bioassay of oral streptococci
| Organism with LuxS | AI-2 activity (% luminescence relative to V. harveyi BB120)a ± SD |
|---|---|
| S. mutans GS-5 | 7.91 ± 1.82 |
| S. mutans ΔluxS | 0.00 ± 0.01 |
| S. gordonii DL1 | 9.10 ± 1.50 |
| S. oralis ATCC 10557 | 2.90 ± 0.77 |
| S. sanguinis ATCC 10556 | 3.36 ± 1.08 |
| S. salivarius HT9R | 16.71 ± 2.71 |
| S. sobrinus MT8145 | 8.89 ± 2.41 |
| S. anginosus FW73 | 14.11 ± 1.15 |
n = 3.
DISCUSSION
AI-2 is a novel bacterial signal produced by both gram-negative and gram-positive genera, and the LuxS-dependent QS circuit, originally identified in V. harveyi, was identified in many gram-negative and gram-positive bacteria (6). A recent study of AI-2 secretion by periodontal bacteria showed that P. gingivalis, Prevotella intermedia, Fusobacterium nucleatum, and A. actinomycetemcomitans secrete AI-2-like signals that can induce luminescence in V. harveyi (11, 14, 15). LuxS-based signaling is thought to represent an important means of intergeneric communication, especially in biofilms (32). Furthermore, individual species regulate different aspects of metabolism and virulence factor expression in response to AI-2. Recently, the luxS gene has also been isolated from P. gingivalis and appears to be important for regulating aspects of iron acquisition by this organism (11).
The nutrient content of the medium was found to regulate the development of biofilms in several organisms (10, 16, 42). For cariogenic dental pathogens, identification of the luxS gene for S. mutans GS-5 was recently reported (34). Therefore, we initially examined the effects of carbohydrates on biofilm formation by the S. mutans luxS mutant and the parental strain. Biofilm formation by these strains was equivalent in glucose-CDM. However, there was a marked difference in sucrose-mediated biofilm formation between these strains. Therefore, we hypothesized that the extracellular glucan synthesis genes are involved in luxS regulation of S. mutans sucrose-dependent biofilm formation.
For the confirmation of this hypothesis, we compared biofilm formation by these strains by using SEM analysis. In CDM supplemented with 0.5% glucose, the luxS mutant and GS-5 parental strain did not exhibit phenotypic differences. However, phenotypic differences between the luxS mutant and GS-5 were apparent in biofilms grown in CDM supplemented with 0.5% sucrose. In the sucrose-CDM, the luxS mutant formed many large clumps of cells on the surface of polystyrene (Fig. 4C). A recent report by Merritt et al. also revealed that the S. mutans luxS mutant forms altered biofilm structures compared by using dark-field microscopy to the wild-type strain on glass coverslips in BHI broth supplemented with 1% sucrose (34). Furthermore, these authors reported a noticeable difference in biofilm structure of the luxS mutant compared to that of the wild type based on visual inspection. Merritt et al. indicated that the luxS mutant had a very rough texture, and our present results confirm this at a higher magnification.
In contrast, biofilm formation by the previously attached luxS mutant cells was enhanced relative to the parental strain in the presence of sucrose. This result is compatible with our observation that the luxS mutant exhibited enhanced insoluble glucan synthesis and therefore autoaggregation relative to the strain GS-5 (data not shown). This would also lead to the attenuation of biofilm formation when the luxS mutant was initially inoculated in the presence of sucrose. The resulting large aggregates would not be able to attach and form biofilms as well as the parental strain. S. mutans synthesizes sucrose-derived glucans by Gtf enzymes (26) encoded by the gtfB (2, 48), gtfC (18), and gtfD (19) genes. Therefore, we analyzed gtf expression in relation to the luxS mutation. Real-time RT-PCR in the present study revealed that gtfB and gtfC genes, coding for the Gtfs synthesizing insoluble glucans, were induced in mid-log phase compared to those of the parental strain. However, the gtfD gene coding for the Gtf synthesizing water-soluble glucans was not induced under these conditions. The gtfBCD transcript levels of both strains were almost the same in the early-log and stationary growth phases. Therefore, the luxS mutant would be expected to synthesize more adherent insoluble glucans than the wild-type strain during growth. The laboratory of S. D. Goodman indicated that there is increased GtfB activity in the luxS mutants (34). Our results confirm this at the transcriptional level. Fong et al. reported that A. actinomycetemcomitans AI-2 activity was maximal in early- and mid-log phases, which influences the expression of the A. actinomycetemcomitans leukotoxin. Leukotoxin levels increased severalfold in early-log-phase cells after exposure to conditioned medium from recombinant E. coli cultures expressing luxS (14). On the other hand, in Salmonella enterica serovar Typhimurium the expression of the luxS gene is controlled by environmental factors, and AI production and signaling activity increase during the mid-to-late-exponential-growth phases (50). Thus, the observation that gtfB and gtfC expression of the S. mutans luxS mutant was enhanced relative to the parental strain suggests that QS may also regulate insoluble glucan synthesis by S. mutans.
The widespread distribution of the luxS gene and the observation that AI-2 is capable of inducing a response in heterologous organisms suggests that the AI-2 signal system transcends species barriers and may function to signal the total bacterial cell community and/or influence its the metabolic potential (3, 51). The LuxS-based signaling system may be relevant for organisms in mixed oral biofilms such as dental plaque. Fong et al. (14) recently reported that A. actinomycetemcomitans AI-2 complemented a P. gingivalis luxS mutant (14). In the present study, we used a two-compartment system to examine complementation analysis of the S. mutans luxS mutant. We confirmed this system was useful for complementation analysis between two organisms. In addition, we examined whether bacterial culture supernatants restore biofilm formation of the luxS mutant by using the two-compartment assay system. We confirmed that bacterial supernatants restore biofilm formation by the luxS mutant using the two-compartment assay system (data not shown). Although this method is not as sensitive as the V. harveyi system (5), it is convenient and does not require a luminometer. Using the complementation assay system, biofilm formation by the S. mutans luxS mutant was restored to the same levels as the parental strain by S. gordonii, S. sobrinus, and S. anginosus strains. We confirmed the existence of the luxS gene in these streptococci by PCR, and all strains used in the present study possessed the luxS homologue (data not shown). Therefore, the noncomplementing streptococci may produce factors that antagonize AI-2 activities, and this is currently under investigation. We also confirmed the complementation of S. mutans luxS mutant biofilm formation by using the periodotopathic bacteria P. gingivalis and A. actinomycetemcomitans. However, the P. gingivalis luxS mutant did not restore biofilm formation by the S. mutans luxS mutant. Furthermore, we performed reporter assays with these oral bacteria and V. harveyi to confirm AI-2 production by the oral streptococci. The levels of AI-2 produced by oral streptococci and biofilm formation by the luxS mutant complemented by these bacteria were generally directly associated (Fig. 6 and Table 3). However, it is not clear why this was not the case with S. salivarius. This may suggest that additional factors are also expressed by some streptococci which also modulate AI-2 activity. These results provide additional support for the previous suggestion that LuxS-dependent intercellular signaling may modulate interspecies communication in oral biofilms. However, the functional significance of such interactions still remains to be determined.
Acknowledgments
This investigation was supported in part by NIH grant DE03258 (to H.K.K.) and by Grant-in-Aid for the Encouragement of Scientists 13771265 (to T.T.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., T. Shiroza, M. Hayakawa, S. Sato, and H. K. Kuramitsu. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 53:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 7.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 10.Carlsson, J. 2000. Growth and nutrition as ecological factors, p. 67-130. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymondham, United Kingdom.
- 11.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 14.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 71:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanada, N., and H. K. Kuramitsu. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect. Immun. 56:1999-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada, N., and H. K. Kuramitsu. 1989. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect. Immun. 57:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjelleberg, S., and S. Molin. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5:254-258. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 24.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolenbrander, P. E., N. Ganeshkumar, F. J. Cassels, and C. V. Hughes. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 7:406-413. [DOI] [PubMed] [Google Scholar]
- 26.Kuramitsu, H. K. 2000. Streptococcus mutans: molecular genetic analysis, p. 280-286. In R. P. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 27.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 31.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 32.McLean, R. J., M. Whiteley, D. J. Stickler, and W. C. Fuqua. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259-263. [DOI] [PubMed] [Google Scholar]
- 33.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 36.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison, D. A., and M. S. Lee. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res. Microbiol. 151:445-451. [DOI] [PubMed] [Google Scholar]
- 38.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 43.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry, D., L. M. Wondrack, and H. K. Kuramitsu. 1983. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect. Immun. 41:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 47.Shiroza, T., and H. K. Kuramitsu. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiroza, T., S. Ueda, and H. K. Kuramitsu. 1987. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 169:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 50.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 51.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tun-Garrido, C., P. Bustos, V. Gonzalez, and S. Brom. 2003. Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J. Bacteriol. 185:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida, A., and H. K. Kuramitsu. 2002. Streptococcus mutans biofilm formation: utilization of a gtfB promoter-green fluorescent protein (PgtfB::gfp) construct to monitor development. Microbiology 148:3385-3394. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]