Abstract
An Escherichia coli strain capable of producing the potent antibiotic erythromycin C (Ery C) was developed by expressing 17 new heterologous genes in a 6-deoxyerythronolide B (6dEB) producer strain. The megalomicin gene cluster was used as the source for the construction of two artificial operons that contained the genes encoding the deoxysugar biosynthetic and tailoring enzymes necessary to convert 6dEB to Ery C. The reconstructed mycarose operon contained the seven genes coding for the enzymes that convert glucose-1-phosphate (G-1-P) to TDP-l-mycarose, a 6dEB mycarosyl transferase, and a 6dEB 6-hydroxylase. The activity of the pathway was confirmed by demonstrating conversion of exogenous 6dEB to 3-O-α-mycarosylerythronolide B (MEB). The reconstructed desosamine operon contained the six genes necessary to convert TDP-4-keto-6-deoxyglucose, an intermediate formed in the mycarose pathway, to TDP-d-desosamine, a desosamine transferase, a 6dEB 12-hydroxylase, and the rRNA methyltransferase ErmE; the last was required to confer resistance to the host cell upon production of mature macrolide antibiotics. The activity of this pathway was demonstrated by conversion of MEB to Ery C. When the mycarose and desosamine operons were expressed in an E. coli strain engineered to synthesize 6dEB, Ery C and Ery D were produced. The successful production of Ery C in E. coli shows the potentiality of this model microorganism to synthesize novel 6-deoxysugars and to produce bioactive glycosylated compounds and also establishes the basis for the future use of E. coli both in the production of new glycosylated polyketides and for the generation of novel bioactive compounds through combinatorial biosynthesis.
Polyketides are a large family of structurally diverse natural products that are produced by bacteria, fungi, sponges, insects, and plants. They possess a broad range of pharmacological properties that have gained widespread use in human medicine (as antibacterial, antifungal, and anticancer agents and immunosuppressants), veterinary medicine (as antibiotics and anthelmintics), and agriculture (as insecticides). The enormous structural diversity of these compounds is dictated by polyketide synthases (PKS) through a number of programmed events that involve the selection of starter and extender units, carbon chain length, and degree of reduction (13). This natural diversity is expanded through post-PKS biosynthetic steps, such as acylation, alkylation, oxidation, and glycosylation. Many naturally occurring polyketides contain unusual sugars as essential components of the bioactive molecule, examples being the antibiotics erythromycin, oleandomycin, and megalomicin, the antifungal agent nystatin, the antiparasitic agent avermectin, and several anticancer compounds (e.g., doxorubicin and aclacinomycins). All of these polyketides contain deoxysugars attached in a glycosidic linkage to the aglycone core. These sugar components generally participate in the molecular recognition of the cellular target by the bioactive compound, and their presence is usually essential for bioactivity (33).
The need for new antimicrobial drugs is constant due to the inevitable development of resistance that follows the introduction of antibiotics to the clinics (30). Naturally occurring metabolites continue to be an important source for lead compounds in the discovery of novel therapeutic agents for infectious diseases and other illnesses (3). The low success rate during the last decades in finding novel compounds with useful antibiotic activity has moved scientists towards combinatorial biosynthesis in order to discover new natural products for drug development. The modular organization of the type I PKS has allowed the engineering of novel compounds by genetic modification of the catalytic domains that determine the choice or stereochemistry of extender units or degree of reduction at various points in the biosynthesis of the polyketide core (12, 18, 23). Due to the limited genetic tools available in most polyketide-producing organisms, engineering of the PKS genes in more genetically manageable heterologous hosts is preferred.
A significant achievement in this field has been the production of the 6-deoxyerythronolide B (6dEB) aglycone in an engineered strain of Escherichia coli (21). This was followed by the successful biosynthesis of yersiniabactin (22), a polyketide-nonribosomal peptide hybrid, and an ansamycin polyketide precursor (31). These results provide promise for the use of E. coli as a platform, not only for the expression of new clusters of PKS genes but also to greatly enhance the rate at which combinatorial biosynthesis tools are developed and PKS are engineered.
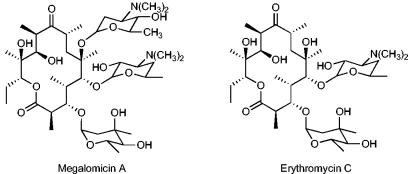
Although advances have been made in the biosynthesis of PKS in E. coli, the challenge of expressing the tailoring enzymes involved in the modification of the polyketide cores in order to produce fully active compounds remains. Megalomicin is a 6-O-megosaminyl derivative of erythromycin C (Ery C) (Fig. 1). The proposed biosynthetic pathway of erythromycin and its relationship to that of megalomicin is identical through the production of Ery C (29), the last common intermediate in the biosynthesis of erythromycin A (Ery A) and megalomicin A. In order to demonstrate the feasibility of employing E. coli to produce bioactive erythromycin analogs, and with the assumption that the enzymes involved in all the common steps of the erythromycin and megalomicin biosynthetic pathways would be similar, we chose the megalomicin (meg) gene cluster from Micromonospora megalomicea (Fig. 2a) as the source of the tailoring genes needed for the biosynthesis of Ery C. Here we describe the expression of 23 foreign genes in E. coli that enable the production of 6dEB and its subsequent conversion to the erythromycin analogs Ery C and Ery D. Although these end products currently represent ca. 12% of the total polyketide formed, the system provides a needed proof of principle that mature erythromycin analogs can be produced in a single heterologous expression system and provides the foundation for further yield improvement by process science and metabolic engineering.
FIG. 1.
Structures of megalomicin A and erythromycin C.
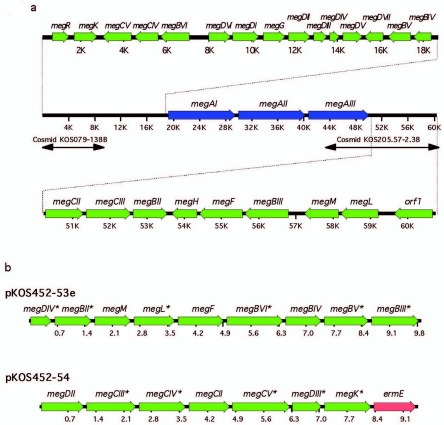
FIG. 2.
Organization of the megalomicin gene cluster. (a) The nucleotide sequences of the left and right hand sides of the meg gene cluster were obtained from cosmids pKOS079-138B (29) and pKOS205.57-2.3B (McDaniel, personal communication). (b) Genetic organization of the mycarose and the desosamine operons. Synthetic operons constructed for expression of the mycarose (pKOS452-53e) and desosamine (pKOS452-54) pathways were cloned under the transcriptional control of the T7 promoter. Asterisks indicate genes with a 5′ translational fusion to a His6-encoding DNA sequence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The relevant plasmids and strains used throughout this study are described in Table 1. Luria-Bertani (LB) medium was used for growth of bacteria. The following antibiotics were added to the media when necessary: kanamycin (50 μg/ml), carbenicillin (100 μg/ml), chloramphenicol (20 μg/ml), streptomycin (25 μg/ml), and phleomycin (50 μg/ml).
TABLE 1.
Relevant plasmids and strains used in this study
| Plasmid or strain | Description or relevant genotypea | Source or reference |
|---|---|---|
| Plasmids | ||
| pET24b | E. coli expression vector, ColE1 ori, kan | Novagen |
| pET28a | E. coli expression vector, ColE1 ori, kan | Novagen |
| pKOS431-54.2 | E. coli expression vector, CloDF13 ori, bleo | S. Mutka, personal communication |
| pKOS431-39.1 | E. coli expression vector, RSF1030 ori, kan | S. Mutka, personal communication |
| pKOS342-96 | ermE in a pET28a background, kan | S. Ward, personal communication |
| pKOS452-53e | megDIV, megBII, megM, megL, megF, megBVI, megBIV, megBV, and megBIII in a pKOS431-39.1 background, kan | This work |
| pKOS452-54 | megDII, megCIII, megCIV, megCII, megCV, megDIII, megK and ermE in a pKOS431-54.2 background, bleo | This work |
| pBP130 | debs2 and debs3 in a pET21b background, bla | 21 |
| pKOS207-129 | debs1 in a pRSF1010 background, str | 19 |
| pGro7 | PBADgroES-groEL in a pACYC184 background, cat | Takara |
| E. coli strains | ||
| DH5α | lacZΔM15 recA1 | Promega |
| BL21 (DE 3) | ompT hsdS (rB− mB−) gal (DE 3) | Novagen |
| K207-3 | F−ompT hsdS (rB− mB−) gal dcm (DE 3) panD::panDS2SAΔprpRBCD::T7 prom-sfp T7 prom-prpE ygfG::T7 prom-accA1-T7 prom-pccB | 19 |
Abbreviations: kan, kanamycin resistance gene; bleo, bleomycin resistance gene; bla, ampicillin resistance gene; str, streptomycin resistance gene; cat, cloramphenicol acetyltransferase gene.
DNA manipulation.
DNA restriction and modifying enzymes were used as recommended by the manufacturer (New England Biolabs). Standard protocols were used for recombinant DNA techniques (Sambrook). DNA fragments were purified from agarose gels with the QIAquick purification kit (QIAGEN). Plasmids were prepared using a QIAprep spin miniprep kit (QIAGEN). Deep Vent DNA polymerase was used in all PCRs according to the supplier's instructions (New England Biolabs).
Plasmid constructions.
Each gene of the mycarose and desosamine biosynthetic pathways, the genes for their specific glycosyltransferases (the megBV and megCIII genes, respectively) and the genes for the P450 C-6 and C-12 hydroxylases (megF and megK, respectively) were individually amplified by PCR from M. megalomicea genomic DNA and sequenced to confirm that they were free of errors. The 5′ primers (Table 2) were designed to have an NdeI site overlapping the translational initiation codon, changing GTG start codons to ATG when necessary. The 3′ primers (Table 2) contained EcoRI and adjacent SpeI sites downstream from the stop codon. PCRs were performed using a DNA thermal cycler 480 (Perkin-Elmer) with the cycling parameters described in Table 2. The PCR products were digested with NdeI and EcoRI and cloned into identical sites of either pET24b or pET28a vectors in order to express proteins with their natural N terminus or as His-tag fusions, respectively. pKOS431-39-1 was derived from pRSF-1b (Novagen) to accept XbaI/EcoRI fragments as follows. (i) The XbaI site at nucleotide 2254 was eliminated by cleavage, filling-in, and blunt end ligation; (ii) the MluI/BlpI fragment of the polylinker was replaced with that from pET26b (S. Mutka, personal communication).
TABLE 2.
Oligonucleotides used in this study and conditions used for PCR amplification
| Gene | Oligonucleotidesa | Annealing temp (°C)b |
|---|---|---|
| megDIV | Upper, 5′-CCGGGCATATGAGGGTCGAGGAGCTG-3′ | 58 |
| Lower, 5′-GGAATTCACTAGTCCGGGGTCACGTCCGC-3′ | ||
| megBII | Upper, 5′-CCGTCATATGAGCACCGACGCCAC-3′ | 56 |
| Lower, 5′-AGGAATTCACTAGTGCGGGCTCTCACCGTAG-3′ | ||
| desIII | Upper, 5′-GGGGTCATATGAAGGCGCTTGTCCTGTCGG-3′ | 58 |
| Lower, 5′-GGAATTCTTGTGACTAGTCGAGTAGTC-3′ | ||
| desIV | Upper, 5′-GACCTCCATATGACGACTCGACTCCTGGTC-3′ | 54 |
| Lower, 5′-TGAATTCACTAGTCCCTCACACCATCGCCCG-3′ | ||
| megF | Upper, 5′-TGGTCATATGAAACTGCCCGATCTGGAGAG-3′ | 58 |
| Lower, 5′-CGAATTCACTAGTCTCATCCGTTCGGTCGCA-3′ | ||
| megBVI | Upper, 5′-GGCATATGGGGGATCGGGTCAACGGTCATG-3′ | 58 |
| Lower, 5′-GGAATTCACTAGTTTCACGCCGTCGCCCGGTTGAG-3′ | ||
| megBIV | Upper, 5′-GCATATGACAAGACATGTCACACTTCTCGG-3′ | 58 |
| Lower, 5′-CGAATTCACTAGTGTCACTCCTTGGTCGAGATGA-3′ | ||
| megBV | Upper, 5′-TGTACATATGCGGGTCCTGCTCACCTCG-3′ | 58 |
| Lower, 5′-AGAATTCACTAGTCACCTGTCGGCGCGGTGCTG-3′ | ||
| megBIII | Upper, 5′-CAGCATATGCCCGAAACGAGATGCCG-3′ | 56 |
| Lower, 5′-AGAATTCACTAGTTTCATCACACCACTTCCAGG-3′ | ||
| megDII | Upper, 5′-CACATATGACCACCTACGTCTGGTCCTATC-3′ | 58 |
| Lower, 5′-CGAATTCACTAGTCACAGCCCGGTGATGACCTCC-3′ | ||
| megCIII | Upper, 5′-CTCATATGCGCGTCGTCTTCTCCTCCATGGC-3′ | 58 |
| Lower, 5′-TGAATTCACTAGTCATCCGACGGCGGTCCGTTCCC-3′ | ||
| megCIV | Upper, 5′-GACCATATGAAGCGCGTACCGACC-3′ | 58 |
| Lower, 5′-GGAATTCACTAGTCACGAACCGTTGCGTTTCC-3′ | ||
| megCII | Upper, 5′-AGAGCATATGAACACGACCGATCGCG-3′ | 52 |
| Lower, 5′-CGGAATTCACTAGTCAGAGTTCGACCG-3′ | ||
| megCV | Upper, 5′-CGCATATGAAGACCACTCCTACGGCGACCG-3′ | 58 |
| Lower, 5′-TCGAATTCACTAGTTCAGACGGCGCGGATCAGGC-3′ | ||
| megDIII | Upper, 5′-CCGCATATGCCGAACAGCCACTCGACCACG-3′ | 54 |
| Lower, 5′-CGAATTCACTAGTCGACCCTCACCGCCCCGGCTCC-3′ | ||
| megK | Upper, 5′-GGAGACATATGACCACTATCGAACAGATCC-3′ | 58 |
| Lower, 5′-TCGAATTCACTAGTGGTTCAGGCGGACTCG-3′ |
The engineered restriction sites are shown in italics.
The genes were amplified under the following PCR conditions: 30 cycles of 30 s at 94°C, 30 s of annealing at the temperature indicated in the table, and 60 s at 68°C.
pKOS431-54-2 was derived from pCDF-1b (Novagen) to modify the antibiotic resistance and to accept XbaI/EcoRI fragments as follows. (i) The XbaI site at nucleotide 2206 was eliminated by cleavage, filling-in, and blunt end ligation; (ii) the resulting plasmid was digested with BspHI/DrdI, blunted with T4 DNA polymerase, and dephosphorylated with shrimp alkaline phosphatase in order to replace the streptomycin resistance gene by ligation to a blunted SpeI/HindIII bleomycin resistance cassette from pKOS183-112 (B. Julien, personal communication); (iii) the MluI/BlpI fragment of the polylinker was replaced with that from pET26b (S. Mutka, personal communication).
The general strategy for the construction of the mycarose and desosamine operons was carried out as follows. The XbaI/EcoRI fragment from the pET-derived construct harboring the second gene of the operon was cloned into the SpeI/EcoRI site of the pET-derived plasmid already containing the first gene of the final construction. This resulted in an intermediate vector harboring the first and second genes separated by a 43-bp sequence with an appropriately positioned ribosome-binding site (RBS). The process was repeated in a recursive fashion until the entire operon containing all genes in tandem was constructed. The mycarose operon contained in pET28a (pKOS452-25b) was removed from this plasmid as an XbaI/EcoRI fragment and cloned into pKOS431-39.1 to give pKOS452-53e. The desosamine operon harbored in pET24b (pKOS452-8b) was also removed as an XbaI/EcoRI fragment and cloned into pKOS431-54.2 to give pKOS452-54. The final operons are shown in Fig. 2b.
Gene expression.
To analyze expression of the individual sugar biosynthetic genes cloned in pET24b or pET28a, cultures of transformed BL21(DE3) were grown overnight in tubes containing LB medium and appropriate antibiotics. Cultures were diluted 1:50 and grown at 37°C to mid-log phase (0.4 to 0.6 OD600 unit/ml). Gene expression was induced with 0.5 mM IPTG (isopropyl β-d-thiogalactopyranoside), and cultures were grown for 6 h at the same temperature. Total cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or Western blotting as described below.
For expression of the mycarose operon, BL21(DE3)/pKOS452-53e cultures were grown at 37°C to mid-log phase (0.4 to 0.6 OD600 unit/ml), induced with 0.5 mM IPTG, and grown for 36 h at 22°C. Volumes of each culture equivalent to 3 OD600 units were resuspended in 1 ml of 20 mM Tris, 150 mM NaCl, pH 7.5, containing 1 tablet of complete EDTA-free protease inhibitor cocktail (Roche) per 50 ml. Cells were disrupted by sonication and centrifuged at 14,000 rpm for 15 min. Pellets were resuspended in 0.5 ml lysis buffer, and soluble and insoluble fractions equivalent to a cell suspension with an OD600 of 0.03 were analyzed by Western blotting.
Protein analysis.
Cell extracts of induced cultures were analyzed by denaturing PAGE (SDS-PAGE) (14a) on Novex 12% Tris-glycine gels (Invitrogen) and visualized by staining with Coomassie blue. Western blotting was carried out as described by Nikolau et al. (20) by transferring proteins to an Invitrolon polyvinylidene difluoride membrane (Invitrogen). His-tagged proteins were identified using a monoclonal anti-His5 antibody (QIAGEN) as a primary antibody and horseradish peroxidase-conjugated goat anti-mouse antibody as a secondary antibody, and the reaction was visualized by enhanced chemiluminescence with ECL reagents (Amersham Pharmacia Biotech).
Bioconversion experiments.
E. coli BL21(DE3) with or without the chaperone expression plasmids and each of the sugar pathway plasmids, pKOS452-53e (mycarose) and pKOS452-54 (desosamine), was cultured overnight at 37°C in LB medium with appropriate antibiotics, subcultured by a 1:50 dilution in the same medium, and grown to mid-log phase (0.4 to 0.6 OD600 unit/ml). Chaperones and sugar gene expression were induced by addition of 2 mg/ml l-arabinose or 1 μg/ml tetracycline and 0.5 mM IPTG, respectively, and cultures were supplemented with 50 μg/ml 6dEB or 3-α-mycarosyl erythronolide B (MEB) as needed. Cultures were grown at 22°C for 2 or 5 days, their pH values were adjusted to 9, and the cultures were then extracted with an equal volume of ethyl acetate. The organic layer was removed, dried under vacuum, dissolved in 5 mM NH4 acetate in 1:1 methanol-H2O, and analyzed by liquid chromatography/mass spectrometry (LC/MS) as described below.
Bioconversion of 6dEB to Ery C was studied in BL21(DE3) containing pKOS452-53e (mycarose) and pKOS452-54 (desosamine) in the presence of pGro7 as a source of the GroEL/GroES chaperone complex. The induction, feeding conditions, and product purification and analysis were carried out as described above.
Erythromycin production.
E. coli K207-3 harboring pBP130, pKOS207-129, pKOS452-53e, pKOS452-54, and the chaperone expression plasmid pGro7 was cultured overnight at 37°C in LB medium with the appropriate antibiotics and then subcultured by a 1:50 dilution to mid-log phase (0.4 to 0.6 OD600 unit/ml). Chaperones, deoxyerythronolide synthase, and sugar gene expression were induced by addition of 0.5 mM IPTG and 2 mg/ml arabinose; cultures were also supplemented with 50 mM glutamate, 50 mM succinate, and 5 mM propionate and then incubated for 5 days at 22°C. The cultures were processed as described for bioconversion experiments.
Polyketide analysis.
Extracts were analyzed using a system consisting of a Gilson 215 sample handler, an Agilent 1100 high-performance liquid chromatography pump, and an Applied Biosystems Mariner time-of-flight mass spectrometer in a positive-ion mode configured with a Turbo-ionspray source.
Samples (20 μl) were chromatographed on a Phenomenex Develosil ODS-UG-3 (3 μm; 2.0 by 150 mm) at 230 μl/min with a linear gradient of 35% 4:1 MeCN-methanol (MeOH) (5 mM NH4 acetyl) to 100% 4:1 MeCN-MeOH (5 mM NH4 acetyl) in H2O over 10 min. The eluate was introduced unseparated into the MS source. Compounds were identified by comparison of their retention times and mass spectra to those of authentic samples. For determination of titers, extracted ion chromatograms were generated from suitable ions of each analyte, and the integrated areas corresponding to each compound were compared to standards. Ery A standards were used to estimate Ery D concentrations, and MEB was used for the demethylated sugar EB derivative.
RESULTS
Megalomicin biosynthetic genes.
A partial sequence of the megalomicin biosynthesis gene cluster has been reported by Volchegursky et al. (29). The putative l-megosamine biosynthetic enzymes were identified and validated by transfer of the megosamine pathway and its cognate glycosyltransferase (MegDI) to an erythromycin-producing strain of Saccharopolyspora erythraea and demonstration of the production of megalomicin. In order to identify the complete set of genes involved in the biosynthesis of this polyketide, we completed the sequencing of the gene cluster and identified by BLAST analysis seven new open reading frames that could be assigned to enzyme activities in megalomicin biosynthesis (Table 3; Fig. 2a).
TABLE 3.
Deduced functions of all the auxiliary genes identified in the megalomicin gene cluster involved in the biosynthesis of Ery C
| Gene | Closest protein match | % Sima | Proposed pathwayb | Proposed function | Reference |
|---|---|---|---|---|---|
| megF | EryF | 91 | C-6 hydroxylase | 32 | |
| megK | EryK | 90 | C-12 hydroxylase | 25 | |
| megL | MtmD | 73 | Myc/Meg/Des | TDP-d-glucose synthase | 16 |
| megM | GilE | 73 | Myc/Meg/Des | TDP-glucose dehydratase | 4 |
| megBVI | EryBVI | 86 | Myc/Meg | 2,3-Dehydratase | 27 |
| megBII | EryBII | 72 | Myc/Meg | 2,3-Reductase | 27 |
| megDIV | EryBVII | 68 | Myc/Meg | 5-Epimerase | 27 |
| megBIII | EryBIII | 89 | Myc | 3-C-Methyltransferase | 27 |
| megBIV | EryBIV | 80 | Myc | 4-Ketoreductase | 27 |
| megBV | EryBV | 85 | Myc | Glycosyltransferase | 27 |
| megCII | EryCII | 78 | Des | 3,4-Isomerase | 27 |
| megCIV | EryCIV | 92 | Des | 2,3-Dehydratase | 27 |
| megCV | EryCV | 87 | Des | 3,4-Enoylreductase | 6 |
| megDIII | EryCVI | 65 | Des/Meg | Dimethyltransferase | 27 |
| megCIII | EryCIII | 90 | Des | Glycosyltransferase | 27 |
| megDII | Med-ORF20 | 83 | Des/Meg | Aminotransferase | 9 |
Sim, similarity. Determined by BLASTX using default parameters.
Myc, mycarose; Meg, megosamine; Des, desosamine.
We identified four putative biosynthetic genes and a putative regulator of the cluster (megR) in the region upstream of the megDVI gene. MegK showed 90% similarity to EryK and was therefore designated the C-12 P450 hydroxylase. MegCIV and MegCV had 92% and 87% similarity with EryCIV and EryCV, respectively, and were assigned 3,4-dehydratase and 3,4-reductase functions in d-desosamine biosynthesis. Finally, MegBVI, previously named MegT (29), showed 80% similarity to EryBVI, the TDP-4-keto-6-deoxyglucose-2,3-dehydratase required for l-mycarose biosynthesis.
We completed the remaining sequence of the P450 hydroxylase MegF at the other end of the cluster and identified three additional genes that are likely involved in sugar biosynthesis. The open reading frame megBIII encodes a protein that is 89% similar to EryBIII and thus was designated the C-methyltransferase involved in TDP-l-mycarose biosynthesis. The other two genes encode enzymes that are common to all three-sugar pathways. MegL (73% homology to mithramycin MtmD) is a TDP-d-glucose synthase that catalyzes the initial condensation of glucose-1-phosphate (G-1-P) and TTP to form TDP-d-glucose, and MegM (73% homology to gilvocarcin GilE) is the putative TDP-glucose-4,6-dehydratase that catalyzes the conversion of TDP-d-glucose to TDP-4-keto-6-deoxyglucose, the common precursor of the three deoxysugars found in megalomicin. The deduced function for all the auxiliary genes involved in the biosynthesis of megalomicin is presented in Table 3.
The analysis of the complete gene cluster revealed that three of the sugar biosynthetic genes could serve in more than one pathway. megDIV has been proposed to encode the 3,5-epimerase involved in an early stage of l-megosamine biosynthesis (29), but the encoded protein also shows 68% similarity to EryBVII, the epimerase of the l-mycarose pathway in erythromycin biosynthesis (27). Since the epimerization reactions in both pathways are mechanistically similar, and no other epimerase gene was found in the gene cluster, we hypothesized that MegDIV serves as a common enzyme in both l-megosamine and l-mycarose biosynthesis. Likewise, MegDII and MegDIII were proposed to be the aminotransferase and dimethyltransferase activities, respectively, in the last two steps of megosamine biosynthesis (29). Because no homologs were found in the gene cluster, and the corresponding last two steps of l-megosamine and d-desosamine biosynthesis are mechanistically similar, we speculated that MegDII and MegDIII are involved in the biosynthesis of both amino sugars.
Construction and validation of the l-mycarose and d-desosamine operons.
Based on the predicted function of the meg genes, we selected 16 genes encoding all the putative enzymes needed to convert 6dEB to Ery C. To analyze the expression of the 16 genes, each was amplified by PCR using primers that generated a 5′ NdeI site at the start codon, a SpeI site just after the stop codon, and a 3′ terminal EcoRI site. Each of the PCR products was cloned into the NdeI/EcoRI site of pET24b and expressed at 37°C in E. coli BL21(DE3). SDS-PAGE of total cell extracts revealed that only 5 of the 16 genes tested produced sufficient protein to be detected by Coomassie blue staining of gels: megF, megL, megBIV, megCII, and megDII. Because it is known that genes containing rare codons at the N terminus are poorly translated in E. coli (8), 11 of the 16 genes were produced to encode N-terminal His6 fusions. NdeI/EcoRI fragments of the 11 poorly expressed genes were cloned into pET28a to generate N-terminal His6 fusions. Nine of the 11 His-tagged proteins were readily detectable in total cell extracts by Coomassie blue staining of SDS gels; the remaining two, MegBIV and MegD4, were detectable by Western blotting with an anti-His5 monoclonal antibody.
In order to reconstitute the l-mycarose and d-desosamine biosynthetic pathways in E. coli and demonstrate the functionality of the remaining enzymes needed to convert 6dEB to Ery C, we constructed two synthetic operons (Fig. 2b). The mycarose operon, contained in pKOS452-53e, harbored the seven genes needed to convert G-1-P to TDP-l-mycarose, as well as megBV and megF, the genes for the mycarosyltransferase and the C-6 hydroxylase, respectively. For its construction, the XbaI/EcoRI fragment from the pET24b construct containing the second gene of the operon was cloned into the SpeI/EcoRI site of the pET28a construct carrying the first gene. This resulted in a vector containing the first and second genes separated by a 43-bp sequence and containing an appropriately positioned ribosome-binding site. The process was repeated in a recursive fashion until the entire operon containing all the genes in a linear sequence under the transcriptional control of the T7 promoter was constructed in pET28a. The operon was then removed from this plasmid as a XbaI/EcoRI fragment and cloned into pKOS431-39.1, a derivative of pRSF-1b (Novagene), to give pKOS452-53e.
To examine the activities of the mycarose pathway and the MegF hydroxylase, we investigated the ability of strain BL21(DE3)/pKOS542-53e to convert exogenous 6dEB to MEB. After induction with IPTG and growth at 22°C for 36 h in the presence of 50 μg/ml 6dEB, LC/MS analysis of the extracts showed a product with a retention time and mass spectrum corresponding to those of authentic MEB. Western blot analysis of total cell extracts using anti-His5 monoclonal antibody showed large amounts of insoluble His-tagged proteins. To improve protein solubility and MEB yields, we examined the effect on the levels of MEB produced by coexpressing the mycarose operon with genes encoding various protein chaperones. Each of four plasmids that encode sets of E. coli chaperones—GroEL/GroES, GroEL/GroES/TIG, GrpE/DnaJ/DnaK, and GroEL/GroES/GrpE/DnaJ/DnaK—were coexpressed with BL21(DE3)/pKOS452-53e and analyzed after 36 h at 22°C for MEB by LC/MS. The best titer was achieved by coexpressing GroEL/GroES (from pGro7), which gave a relative MS peak area of MEB that was sixfold greater than that of the control without chaperones.
To characterize and quantitate the final or intermediate products of the MEB biosynthesis pathway, cultures of BL21(DE3)/pKOS452-53e/pGro7 growing in the presence of 50 μg/ml 6dEB were induced with IPTG and l-arabinose and incubated for 5 days at 22°C. LC/MS analysis of the extracts showed 2% 6dEB, 70% EB, 15% MEB, and 13% of a MEB analog (digitoxosyl- or olivosyl-EB) whose mass spectrum and fragmentation profile indicated the absence of a methyl group in the neutral sugar. The accumulation of the last compound could be explained by an inefficient methyl transfer by MegBIII, although it could also occur through a deficiency of the methyl donor S-adenosylmethionine in the cells. Since approximately 98% of the products were hydroxylated at C-6, a suitable electron donor system for MegF must be present in E. coli. The predominance of EB in recovered products suggests a limitation in the level of TDP-l-mycarose or glycosylation of EB by MegBV.
The desosamine operon was assembled in fashion similar to that of the mycarose operon, giving rise to pKOS452-54 (Fig. 2b). This plasmid harbors the six genes needed to convert TDP-4-keto-6-deoxyglucose to TDP-d-desosamine, genes encoding the glycosyltransferase MegCIII and the C-12 hydroxylase MegK, and ermE, which encodes a ribosomal methyltransferase (28) that dimethylates A2058 of 23S rRNA and confers high-level resistance to Ery A, as well as the ketolide telithromycin (15).
The functionality of the cloned d-desosamine pathway was examined by feeding MEB to BL21(DE3)/pKOS452-54/pGro7 and evaluating extracts for the production of downstream products. The GroEL/GroES complex improved levels of Ery C production in this strain. Cultures were induced with l-arabinose and IPTG and grown at 22°C for 2 days in the presence of 50 μg/ml of MEB. LC/MS analysis revealed approximately 94% MEB, 5% Ery C, and 1% Ery D. The high level of fed MEB recovered may simply reflect inefficient uptake in E. coli (15). Nevertheless, the results clearly demonstrated the functionality of the enzymes involved in the biosynthesis and transfer of d-desosamine to MEB as well as the C-12 hydroxylase that converts Ery D into Ery C in E. coli.
Since genes encoding DesIII and DesIV were not present in the desosamine operon, the needed precursor TDP-4-keto-6-deoxyglucose must be present in the host. It has been documented that this intermediate is formed in E. coli by the G-1-P thymidylyltransferases (RfbB and RffG) and the TDP-d-glucose 4,6-dehydratases (RfbA and RffH) involved in the biosynthesis of TDP-rhamnose and dTDP-4-acetamido-4,6-dideoxygalactose, respectively (17, 26).
Production of Ery C in E. coli from its primary metabolites.
To demonstrate the feasibility of producing Ery C without the requirement of feeding specific intermediates of the pathway, the 6dEB producer strain E. coli K207-3/pBP130/pKOS207-129 (19) was transformed with the expression plasmids pKOS452-53e, pKOS452-54, and pGro7. This new strain contains 23 heterologous genes (approximately 0.5% of the protein genes present in the chromosome of E. coli) all under the transcriptional control of T7 RNA polymerase (Fig. 3). LC/MS analysis of extracts obtained from 5-day cultures demonstrated production of the two erythromycins analogs Ery C (0.4 mg/liter) and Ery D (0.5 mg/liter) (Fig. 4). The sample also contained 6dEB (7 mg/liter), EB (15 mg/liter), MEB (0.14 mg/liter), and 0.28 mg/liter of the analog with one less methyl group in the neutral sugar.
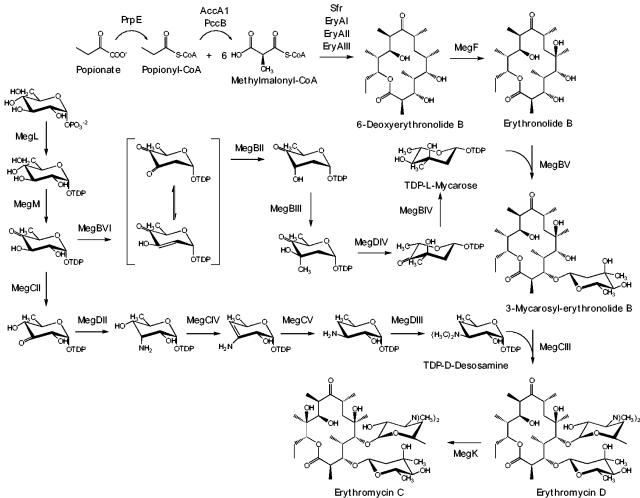
FIG. 3.
Erythromycin C biosynthesis pathway. All of the heterologous proteins present in E. coli K207/pBP130/pKOS207-129/pKOS452-53e/pKOS452-54 that are involved in the bisosynthesis of Ery C are shown. The common intermediates of the l-mycarose and d-desosamine pathways are indicated.
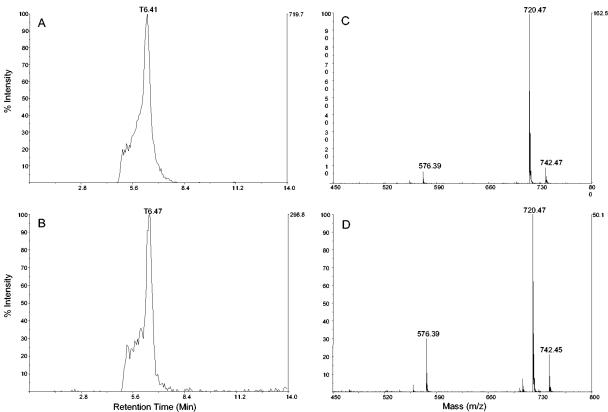
FIG. 4.
Analysis of erythromycin C produced by E. coli. (A and B) LC/MS-extracted ion chromatogram (m/z 720.5 ± 0.2) of the erythromycin C standard (A) and of an extract from a fermentation broth of E. coli K207/pBP130, pKOS207-129, pKOS452-53e, pKOS452-54, and pGro7 (B). (C and D) LC/MS of the erythromycin C standard (C) and of the peak corresponding to erythromycin C in the extract from a fermentation broth of E. coli K207/pBP130, pKOS207-129, pKOS452-53e, pKOS452-54, and pGro7 (D).
Interestingly, we did not find a 3-desmethyl-Ery C derivative that should result from the desosaminylation of the desmethyl mycarosyl-EB compound. However, a 3-desmethyl-Ery A has been reported in eryBIII mutants of S. erythraea (7).
DISCUSSION
In this study we have succeeded for the first time in the complete biosynthesis and export of a potent macrolide antibiotic in a genetically modified strain of E. coli. Although this microorganism presents a new environment for PKS expression (there are no PKS found in E. coli) and polyketide biosynthesis, we have demonstrated the utility and promise of the biosynthetic capabilities of this bacterium for producing glycosylated polyketides.
The development of new erythromycin analogs with altered aglycones (5, 10, 14) has been greatly facilitated by heterologous expression of the deoxyerythronolide synthase in Streptomyces coelicolor (11) and, more recently, in a metabolically engineered strain of E. coli (21). The aglycones thus obtained are purified and subsequently modified into promising new bioactive compounds by an erythromycin-producing strain of S. erythraea containing the necessary tailoring enzymes but in which the erythromycin PKS had been rendered inactive (2). Although the two-fermentation system is highly refined, it would clearly be advantageous to produce mature erythromycin analogs in a single organism. While the final titer for Ery C in our system is not yet useful for broader application, improved production of this compound can be approached by rational optimization of putative rate-limiting steps or by the empirical processes of titer development.
Interestingly, the distribution of products in the full producer strain does not correlate with that observed in the feeding experiments using the mycarose or the desosamine pathways and suggests the presence of differential amounts of substrates or enzyme activities. In other work, we have observed unpredictable differential expression of genes when multiple T7 promoters were used, presumably due to competition for systems needed in transcription/translation (unpublished results). The producer strain contains five different plasmids and probably hundreds of copies of the strong φ10 promoter from T7 phage, which might produce a heavy burden on the metabolism of the microorganism after its induction. We also demonstrated that E. coli provided the electron donor system, probably by the ferredoxin (flavodoxin) NADP+ oxidoreductase encoded by fpr (1), which is needed for the P450 hydroxylases MegF and MegK. However, the complete conversion of 6dEB to EB and the accumulation of Ery D in the final producing strain might indicate that the electron donor system is optimal for MegF but not for MegK. Expression of a more specific redox pair (ferredoxin/ferredoxin reductase) for MegK might also be necessary to optimize this production system.
Changing the sugar moieties in natural products might be an important contribution to the generation of novel derivatives with potentially novel pharmacological properties. Recently, a plasmid containing the genes involved in the biosynthesis of l-oleandrose was developed in Streptomyces antibioticus (24). By replacement of oleU with different 4-ketoreductase genes, and by using the “sugar-flexible” glycosyltransferase ElmGT, Rodriguez et al. were able to produce novel glycosylated compounds. Although this example is an important achievement towards the generation of combinatorial libraries, the restricted genetic tools available for Streptomyces and the relatively low growth rate of these bacteria limit the system. The idea of converting E. coli into a model system for generating combinatorial libraries, where the biosynthesis of different PKS aglycones could be combined with the expression of different sugar pathways and glycosyltransferases of relaxed substrate specificity, has gained growing interest. The results presented here represent a step towards the development of such a model system.
Acknowledgments
We thank Chau Tran for assistance with the MS, Daniel Kurth for the construction of some of the intermediate plasmids, and Mark Burlingame for facilitating production of the 6dEB and MEB. We also thank Dan Santi, David Hopwood, Sarah Kodumal, Gary Ashley, and Leonard Katz for their comments on the manuscript.
REFERENCES
- 1.Bianchi, V., P. Reichard, R. Eliasson, E. Pontis, M. Krook, H. Jörnvall, and E. Haggard-Ljungquist. 1993. Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J. Bacteriol. 175:1590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carreras, C., S. Frykman, S. Ou, L. Cadapan, S. Zavala, E. Woo, T. Leaf, J. Carney, M. Burlingame, S. Patel, G. Ashley, and P. Licari. 2002. Saccharopolyspora erythraea-catalyzed bioconversion of 6-deoxyerythronolide B analogs for production of novel erythromycins. J. Biotechnol. 92:217-228. [DOI] [PubMed] [Google Scholar]
- 3.Cragg, G. M., D. J. Newman, and K. M. Snader. 1997. Natural products in drug discovery and development. J. Nat. Prod. 60:52-60. [DOI] [PubMed] [Google Scholar]
- 4.Fischer, C., F. Lipata, and J. Rohr. 2003. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J. Am. Chem. Soc. 125:7818-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frykman, S., T. Leaf, C. Carreras, and P. Licari. 2001. Precursor-directed production of erythromycin analogs by Saccharopolyspora erythraea. Biotechnol. Bioeng. 76:303-310. [DOI] [PubMed] [Google Scholar]
- 6.Gaisser, S., G. A. Böhm, J. Cortés, and P. F. Leadlay. 1997. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 256:239-251. [DOI] [PubMed] [Google Scholar]
- 7.Gaisser, S., G. A. Bohm, M. Doumith, M. C. Raynal, N. Dhillon, J. Cortes, and P. F. Leadlay. 1998. Analysis of eryBI, eryBIII and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 258:78-88. [DOI] [PubMed] [Google Scholar]
- 8.Gramajo, H. C., J. White, C. R. Hutchinson, and M. J. Bibb. 1991. Overproduction and localization of components of the polyketide synthase of Streptomyces glaucescens involved in the production of the antibiotic tetracenomycin C. J. Bacteriol. 173:6475-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichinose, K., M. Ozawa, K. Itou, K. Kunieda, and Y. Ebizuka. 2003. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 149:1633-1645. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen, J. R., C. R. Hutchinson, D. E. Cane, and C. Khosla. 1997. Precursor-directed biosynthesis of erythromycin analogs by an engineered polyketide synthase. Science 277:367-369. [DOI] [PubMed] [Google Scholar]
- 11.Kao, C. M., L. Katz, and C. Khosla. 1994. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science 265:509-512. [DOI] [PubMed] [Google Scholar]
- 12.Katz, L. 1997. Manipulation of modular polyketide synthases. Chem. Rev. 97:2557-2576. [DOI] [PubMed] [Google Scholar]
- 13.Katz, L. 2002. Polyketide diversity, p. 157-175. In W. K. Shounfeld and H. A. (ed.), Macrolide antibiotics. Birkhauser Verlag, Basel, Switzerland.
- 14.Kennedy, J., S. Murli, and J. T. Kealey. 2003. 6-Deoxyerythronolide B analogue production in Escherichia coli through metabolic pathway engineering. Biochemistry 42:14342-14348. [DOI] [PubMed] [Google Scholar]
- 14a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Liu, M., and S. Douthwaite. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombo, F., K. Siems, A. F. Brana, C. Mendez, K. Bindseil, and J. A. Salas. 1997. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J. Bacteriol. 179:3354-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 177:5539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel, R., A. Thamchaipenet, C. Gustafsson, H. Fu, M. Betlach, M. Betlach, and G. Ashley. 1999. Multiple genetic modifications of the erythromycin gene cluster to produce a library of novel “unnatural” natural products. Proc. Natl. Acad. Sci. USA 96:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murli, S., J. Kennedy, L. C. Dayem, J. R. Carney, and J. T. Kealey. 2003. Metabolic engineering of Escherichia coli for improved 6-deoxyerythronolide B production. J. Ind. Microbiol. Biotechnol. 30:500-509. [DOI] [PubMed] [Google Scholar]
- 20.Nikolau, B. J., E. S. Wurtele, and P. K. Stumpf. 1985. Use of streptavidin to detect biotin-containing proteins in plants. Anal. Biochem. 149:448-453. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer, B. A., S. J. Admiraal, H. Gramajo, D. E. Cane, and C. Khosla. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790-1792. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer, B. A., C. C. Wang, C. T. Walsh, and C. Khosla. 2003. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 69:6698-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez, E., and R. McDaniel. 2001. Combinatorial biosynthesis of antimicrobials and other natural products. Curr. Opin. Microbiol. 4:526-534. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez, L., I. Aguirrezabalaga, N. Allende, A. F. Brana, C. Mendez, and J. A. Salas. 2002. Engineering deoxysugar biosynthetic pathways from antibiotic-producing microorganisms. A tool to produce novel glycosylated bioactive compounds. Chem. Biol. 9:721-729. [DOI] [PubMed] [Google Scholar]
- 25.Stassi, D., S. Donadio, M. J. Staver, and L. Katz. 1993. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J. Bacteriol. 175:182-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers, R. G., S. Donadio, M. J. Staver, E. Wendt-Pienkowski, C. R. Hutchinson, and L. Katz. 1997. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology 143:3251-3262. [DOI] [PubMed] [Google Scholar]
- 28.Vester, B., and S. Douthwaite. 1994. Domain V of 23S rRNA contains all the structural elements necessary for recognition by the ErmE methyltransferase. J. Bacteriol. 176:6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volchegursky, Y., Z. Hu, L. Katz, and R. McDaniel. 2000. Biosynthesis of the anti-parasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. Mol. Microbiol. 37:752-762. [DOI] [PubMed] [Google Scholar]
- 30.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, K., M. A. Rude, C. T. Walsh, and C. Khosla. 2003. Engineered biosynthesis of an ansamycin polyketide precursor in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:9774-9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber, J. M., J. O. Leung, S. J. Swanson, K. B. Idler, and J. B. McAlpine. 1991. An erythromycin derivative produced by targeted gene disruption in Saccharopolyspora erythraea. Science 252:114-117. [DOI] [PubMed] [Google Scholar]
- 33.Weymouth-Wilson, A. C. 1997. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14:99-110. [DOI] [PubMed] [Google Scholar]