Abstract
It is well known that biofilm formation by pathogenic staphylococci on implanted medical devices leads to “chronic polymer-associated infections.” Bacteria in these biofilms are more resistant to antibiotics and the immune defense system than their planktonic counterparts, which suggests that the cells in a biofilm have altered metabolic activity. To determine which genes are up-regulated in Staphylococcus aureus biofilm cells, we carried out a comparative transcriptome analysis. Biofilm growth was simulated on dialysis membranes laid on agar plates. Staphylococci were cultivated planktonically in Erlenmeyer flasks with shaking. mRNA was isolated at five time points from cells grown under both conditions and used for hybridization with DNA microarrays. The gene expression patterns of several gene groups differed under the two growth conditions. In biofilm cells, the cell envelope appeared to be a very active compartment since genes encoding binding proteins, proteins involved in the synthesis of murein and glucosaminoglycan polysaccharide intercellular adhesin, and other enzymes involved in cell envelope synthesis and function were significantly up-regulated. In addition, evidence was obtained that formate fermentation, urease activity, the response to oxidative stress, and, as a consequence thereof, acid and ammonium production are up-regulated in a biofilm. These factors might contribute to survival, persistence, and growth in a biofilm environment. Interestingly, toxins and proteases were up-regulated under planktonic growth conditions. Physiological and biochemical tests for the up-regulation of urease, formate dehydrogenase, proteases, and the synthesis of staphyloxanthin confirmed the microarray data.
Numerous reports in the past two decades have shown that especially biofilm-forming staphylococci cause an infection that is best described as a “chronic polymer-associated infection” (46). Staphylococcus aureus and Staphylococcus epidermidis, as commensal inhabitants of the human skin, therefore have easy access to wounds and can reach implanted devices (51), where they frequently cause persistent and chronic wound infections on catheters, shunts, implants, and other implanted devices (6-8, 23, 37-39, 45).
Bacterial biofilm infections are particularly problematic because sessile bacteria can often withstand host immune responses and are generally much more tolerant to antibiotics, biocides, and hydrodynamic shear forces than their planktonic counterparts (36, 48). This makes medical treatment of these infections very difficult, and often the implanted device has to be removed or replaced. It has been estimated that biofilms are associated with 65% of the nosocomial infections in the United States and that the treatment of these biofilm-based infections costs more than one billion dollars annually (2).
The ability to form a biofilm requires at least two properties: adherence of cells to a surface and accumulation to form multilayered cell clusters. A trademark is the production of the slime substance polysaccharide intercellular adhesin (PIA), a polysaccharide composed of β-1,6-linked N-acetylglucosamine with partially deacetylated residues, in which the cells are embedded and protected against the host immune defense and antibiotic treatment. Mutations in the corresponding biosynthesis genes (ica operon) lead to a pleiotropic phenotype; the cells are biofilm and hemagglutination negative, less virulent, and less adhesive on hydrophilic surfaces (22). ica expression is modulated by various environmental conditions. It appears to be controlled by SigB, IcaR, and some unknown activator (29) and can be turned on and off by insertion elements (53).
Very recently, it has been shown that a mutation in the ica genes of a clinical S. aureus isolate has little effect on biofilm formation, whereas a mutation in sarA leads to a biofilm-negative phenotype (3). Particularly in S. aureus, ica genes are only expressed under anoxic conditions; in a routine microtiter plate assay, most of the S. aureus isolates appear to be biofilm negative if they are not cultivated strictly anoxically (10). Several mutants affected in primary adhesion, e.g., atlE (26) or dltA (44) mutants, have been isolated.
S. aureus produces several adhesive compounds (e.g., PIA, biofilm-associated protein [Bap], and other protein adhesins) that enable the bacterial cells to bind to very different surface structures (9, 11, 18, 27, 40). After binding to the surface, the biofilm of S. aureus cells usually becomes multilayered and differentiated (39). The cells are embedded in a slimy matrix, their physiology differs distinctly from that of planktonically grown cells, and they are much more resistant to the host immune response (21, 50). The growth conditions, including the supply of oxygen and nutrients, vary greatly among the various layers of the staphylococcal biofilm, which might promote differential growth and physiology of the cells in the different layers, such as fermentation in the anoxic areas of the biofilm (5, 20). This environmental and physiological diversity might also contribute to the persistence of the biofilm cells, as evidenced by the lowered effect of antibiotics. The cell wall does not act as a diffusion barrier and thus apparently does not play a significant role in general antibiotic resistance, as was assumed previously (42). The higher antibiotic resistance of biofilm cells therefore must have a different explanation.
A major future task is to find new and effective treatments for biofilm-associated infections. Therefore, a better understanding of which genes and proteins are differentially expressed under biofilm and planktonic growth conditions and of how the morphology and physiology of the biofilm cells differ from those of planktonic cells is needed to understand the high persistence and resistance of biofilm cells. As a starting point for further investigations, DNA microarray analysis is a helpful tool to determine differential gene expression patterns under biofilm and planktonic growth conditions. Follow-up investigations on the bacterial proteome, knockout mutants, and enzyme assay development are the next steps. With DNA microarrays covering several times in biofilm development, it might even be possible to determine events or the activity of certain genes that are responsible for establishment and differentiation of biofilms and thereby to gain an understanding of how a biofilm develops over time.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus strain 113 was used for all experiments in this work (28). Cells were grown in BM medium (containing [per liter]10 g peptone 140, 5 g yeast extract, 5 g NaCl, 1 g K2HPO4 · 3H20; pH 7.5) supplemented after autoclaving with 0.67% glucose, which promotes biofilm formation (13, 41). Overnight cultures were inoculated into fresh BM medium and grown to an optical density at 450 nm of 0.2 to 0.3. The culture was diluted to a concentration of 5 × 106 cells/ml and used for further inoculation. For planktonic growth conditions, 30 ml of BM medium was inoculated with 0.2 ml of the cell suspension and incubated at 37°C with shaking at 150 rpm. For biofilm growth conditions, NADIR dialysis membranes (molecular mass cutoff of 10 to 20 kDa; Roth) were cut into circles with a diameter of 8 cm, sterilized in 70% ethanol for 1 h, and aseptically placed onto BM agar plates; the plates were allowed to dry at room temperature overnight. The membranes were inoculated with 0.2 ml of the cell suspension and incubated at 37°C.
RNA isolation.
Biofilm-grown and planktonically grown cells were harvested at five different times (6, 8, 16, 24, and 48 h) and immediately incubated with an appropriate volume of RNAprotect (QIAGEN), vortexed for 5 s, incubated for 5 min at room temperature, and pelleted by centrifugation for 10 min at 4,500 × g. Cell pellets were stored at−70°C until they were used for RNA isolation. Each bacterial pellet was suspended in 920 μl of Tris-EDTA buffer (QIAGEN). Biofilm cells, which adhere very strongly to each other, were separated by a 30-s pulse of sonication. Lysostaphin (320 μl, 0.5 mg/ml) was added to all suspensions, which were then vortexed for 10 s and incubated with shaking for 10 min at 37°C. Sterile glass beads (200 mg) were added, and the cells were vortexed for 30 s, followed by 30 s of incubation on ice. This was repeated three times. RLT buffer (QIAGEN) was added, and the RNA was isolated according to the standard protocol provided with an RNeasy mini kit (QIAGEN). Contaminating DNA in the RNA preparations was removed using “DNA-free” (Ambion). The RNA quality and quantity were determined by agarose gel electrophoresis and by measuring the absorbance at 260 and 280 nm (results not shown). Purified RNA was stored at −20°C. RNA was isolated from four samples for each time and growth condition.
cDNA labeling and DNA microarray hybridization.
cDNA was synthesized from mRNA using a slight modification of the method of the microarray manufacturer (Scienion AG, Berlin, Germany). RNA (3 μg unless indicated otherwise [see below]) from biofilm or planktonic cells was mixed with 750 ng of random hexamer primer (Invitrogen), denatured at 70°C for 10 min, and allowed to cool to room temperature. Since a lower yield of RNA per mg (wet weight) of cells after longer periods of biofilm growth has been observed by other groups (14), larger amounts of RNA from biofilm cells grown for 24 and 48 h (4.5 and 13 μg, respectively) were used to account for degradation.
cDNA was synthesized by mixing the RNA with 400 U of Superscript II reverse transcriptase (Invitrogen) in 1× first-strand buffer containing 10 mM dithiothreitol, low-dT deoxynucleoside triphosphate mixture (500 mM of dGTP, dCTP, and dATP plus 200 mM dTTP), Cy3- or Cy5-labeled dUTPs (300 mM; Amersham), and RNasin (40 U). cDNA derived from planktonic cells was labeled with Cy3, and cDNA derived from biofilm cells was labeled with Cy5. The mixture was incubated for 25 min at 42°C. Then 200 U of Superscript II reverse transcriptase was added, and the mixture was incubated for 35 min at 42°C. The reaction was stopped by adding 5 μl of 500 mM EDTA. NaOH (10 μl, 1 M) was added, and the mixture was incubated for 15 min at 65°C. The mixture was then neutralized with 25 μl of Tris-HCl (1 M, pH 7.5).
The resulting cDNA was purified using a QIAquick PCR purification kit (QIAGEN). The Cy3- and Cy5-labeled cDNAs for the different times were pooled and dried by vacuum centrifugation for 1 h. The cDNAs were then resuspended in 50 μl of prewarmed (42°C) hybridization solution (Scienion) and used for hybridization (60 h at 42°C). The slides were washed and stored according to the recommendations of the manufacturer. For each of the five times, four DNA microarrays were analyzed, using biofilm cDNA as a probe and planktonic cDNA as a reference on one microarray.
DNA microarray analysis.
S. aureus N315 microarrays were purchased from Scienion AG (Berlin, Germany). Microarrays for S. aureus 113 are not available. All the identified up-regulated genes on the S. aureus N315 microarray were compared by BLAST analysis at the DNA level to the known sequences of the genome of S. aureus 8325, the parental strain of S. aureus 113; a level of sequence identity of at least 93% was found.
PCR fragments of the whole genome of S. aureus N315 were spotted on glass slides in the microarray analysis. Each microarray feature was spotted twice. For details, see www.scienion.com. The hybridized microarrays were scanned with an Axon Scanner GenePix 4100. Cy3 and Cy5 were excited at 532 and 635 nm, respectively. Fluorescence was detected with a confocal microscope equipped with the respective optical filters. The hybridization patterns and intensities were quantitatively analyzed using the GenePix Pro software (4.1.1.31 Axon).
A geometric raster was laid over the microarray features to segment the picture and to separate the signals from the background. After localization of the single spots, the spot intensities and the local background were calculated; the features were assigned to the respective accession numbers and annotations. For the annotation, the gene list on the DOGAN web page (database of genomes analyzed in NITE as of 23 January 2004) for S. aureus N315 was used (http://www.bio.nite.go.jp:8080/dogan/MicroTop?GENOME_ID = n315G1).
The fluorescence of each microarray was normalized. The mean of the ratio of medians was used to normalize single signals. This method is appropriate for compensating for variation in hybridization (including labeling efficiency and different target amounts).
Data analysis.
Gene expression data were analyzed using the Acuity software (Axon). The data from GenePix Pro had to be exported into the Acuity software for analysis. The differences in the expression levels of single genes were determined. They were calculated by comparing the intensity of Cy5-labeled hybridized cDNA and Cy3-labeled cDNA with the ratio of medians (635/532). Therefore, a change in expression pattern between biofilm and planktonic cells could be directly calculated with the intensities from one microarray. The values indicate the relative difference in each transcript and were used to identify genes that are differentially expressed under biofilm and planktonic growth conditions.
For biofilm cells, the threshold was set at a 2.5-fold difference in expression; for planktonic cells, the threshold was set at a 3.33-fold difference (data not shown; http://www.uni-tuebingen.de/Mikrobiogen/announce.html)
The values listed in the tables are the calculated means of the differences for four parallel microarrays. The standard deviation (SD) and the coefficient of variation (CV) were calculated using the Acuity program. The SD is the average distance from the mean and indicates whether the values are clustered near the mean. The CV is the standard deviation divided by the mean and is expressed as a percentage (value multiplied by 100), and it is an indicator of relative dispersion. Genes are listed in the tables only if more than one value was detected or included in the calculation and if the SD for the calculated mean was <0.55 or if the CV was <25.
Expression patterns of selected genes or gene groups.
The expression patterns of groups of genes (e.g., genes encoding binding proteins or surface proteins) at the five selected times up to 48 h of growth were determined. This was possible since the differences represented the ratios for the two growth conditions.
Determination of the staphyloxanthin content in cells.
At different times, planktonic and biofilm cells were harvested, the cell densities were adjusted to obtain comparable values, and the cells were pelleted, resuspended in 2 ml of 100% ethanol, and incubated for 30 min at 45°C. The cells were pelleted again, and the absorption of the supernatant at 460 nm, a peak absorption wavelength for staphyloxanthin (52), was measured.
Proteolytic activity.
Biofilm and planktonic cells were harvested at the five times selected. The cells were pelleted, and the supernatants (200 μl) were used to test the proteolytic activity on casein agar (CASO agar; Merck). Proteolytic activity was indicated by halo formation.
Urease activity.
Biofilm and planktonic cells were grown as described above and were harvested after 48 h. The cells were pelleted and resuspended in saline. One urease diagnostic tablet (575-21; Rosco) was added per sample, and the cells were incubated for 4 h at 37°C. The diagnostic tablets contain the indicator phenol red, which turns red when urease catalyzes the hydrolysis of urea to form two molecules of ammonia.
Formate dehydrogenase activity.
Biofilm and planktonic cells were grown as described above and harvested after 24 h. The cells were pelleted by centrifugation for 15 min at 4,500 × g and 4°C, and the pellet was resuspended in 5 ml of buffer containing 5 mM MgCl2, 1 mM EDTA, and 0.8 M Tris-Cl (pH 7.6). Lysostaphin (50 μl, 0.5 mg/ml) was added to lyse the cells; the mixture was incubated for 15 min at 37°C. Two hundred micrograms of glass beads was added, and the mixture was vortexed for 30 s, followed by 30 s of incubation on ice; the vortexing/incubation cycle was repeated three more times. The resulting suspension was centrifuged for 10 min at 15,000 × g and 4°C, and the supernatant was used to measure formate dehydrogenase activity. Buffer containing 10 mM NAD and 20 mM formate was added to 250 μl of cell extract to obtain a final volume of 1 ml. The enzyme activity was determined from the increase in extinction at 340 nm at 30°C, using the extinction coefficient of NADH (ɛ = 6.3 mM−1 cm−1). The enzyme activity was normalized to the protein content determined using the method of Bradford.
RESULTS
Identification of genes expressed at higher levels in biofilm cells.
By comparing gene expression under biofilm and planktonic growth conditions after 6, 8, 12, 24, and 48 h of growth, we were able to identify and statistically validate a number of genes that are differentially expressed under the two growth conditions. Tables 1 to 5 list the up-regulated genes in biofilm cells for each of the five times. The sequences of the identified up-regulated genes of S. aureus N315 (34) were compared to the partially sequenced genome of S. aureus 8325 (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), which is the parental strain of S. aureus 113 (28) and showed identity of at least 93% at the DNA level, which indicated that the identified genes are present in the strain used. For convenience, the open reading frame identification numbers of the previously published S. aureus N315 genome sequence (34) are used in the tables.
TABLE 1.
Genes expressed more highly under biofilm conditions after 6 h of growth
| Type of proteins | N315 open reading frame | Biofilm vs planktonic cells (fold difference) | Name | Product | SD | CV |
|---|---|---|---|---|---|---|
| Cell wall-associated proteins | SA2460 | 8.793 | icaD | Intercellular adhesion protein D | 0.14 | 1.596 |
| SA2462 | 5.569 | icaC | Intercellular adhesion protein C | 0.221 | 3.975 | |
| SA2459 | 4.861 | icaA | Intercellular adhesion protein A | 0.254 | 5.217 | |
| SA0531 | 4.307 | prop | Proline/betaine transporter homologue | 0.127 | 2.95 | |
| Transporter proteins | SA2202 | 3.357 | Protein similar to ABC transporter periplasmic amino acid-binding protein | 0.114 | 3.399 | |
| SA2142 | 3.345 | Protein similar to multidrug resistance protein | 0.096 | 2.883 | ||
| SA1182 | 3.044 | mscL | Large-conductance mechanosensitive channel | 0.153 | 5.036 | |
| SA2200 | 3.033 | Protein similar to ABC transporter ATP binding subunit | 0.077 | 2.542 | ||
| SA2201 | 2.548 | Protein similar to ABC transporter permease protein | 0.12 | 4.72 | ||
| Physiological proteins | SA2414 | 2.962 | Protein similar to glutathione peroxidase | 0.226 | 7.641 | |
| SA2106 | 2.735 | Protein similar to protein of pXO2-46 | 0.128 | 4.685 | ||
| SA1382 | 2.587 | soda | Superoxide dismutase SodA | 0.137 | 5.282 | |
| Ribosomal protein | SA1116 | 2.519 | rpsO | 30S ribosomal protein S15 | 0.095 | 3.764 |
| Other proteins | SA2174 | 3.259 | Protein similar to transcriptional regulator | 0.166 | 5.108 | |
| SA1305 | 2.92 | hu | DNA-binding protein II | 0.099 | 3.375 | |
| SA1949 | 2.814 | Lytic regulatory protein truncated with Tn554 | 0.162 | 5.768 | ||
| Hypothetical proteins | SA0271 | 4.664 | Conserved protein | 0.08 | 1.709 | |
| SA2133 | 3.315 | Conserved protein | 0.072 | 2.18 | ||
| SA0412 | 3.278 | Conserved protein | 0.066 | 2.018 | ||
| SA2143 | 3.148 | Conserved protein | 0.062 | 1.971 | ||
| SA0292 | 3.147 | Hypothetical protein | 0.206 | 6.558 | ||
| SA0509 | 2.906 | Conserved protein | 0.158 | 5.428 | ||
| SA0082 | 2.878 | Conserved protein | 0.142 | 4.949 | ||
| SA2378 | 2.742 | Conserved protein | 0.146 | 5.343 | ||
| SA0739 | 2.729 | Conserved protein | 0.156 | 5.706 |
TABLE 5.
Genes expressed more highly under biofilm conditions after 48 h of growth
| Type of proteins | N315 open reading frame | Biofilm vs planktonic cells (fold difference) | Name | Product | SD | CV |
|---|---|---|---|---|---|---|
| Cell wall-associated proteins | SA0519 | 7.217 | sdrC | Ser-Asp rich fibrinogen-binding, bone sialoprotein-binding protein | 0.181 | 2.505 |
| SA2431 | 3.317 | isaB | Immunodominant antigen B | 0.256 | 7.727 | |
| SA0572 | 3.643 | Protein similar to esterase/lipase | 0.162 | 4.45 | ||
| SA0742 | 3.068 | clfA | Fibrinogen-binding protein A clumping factor | 0.189 | 6.15 | |
| SA2423 | 2.966 | clfB | Clumping factor B | 0.183 | 6.174 | |
| Transporter proteins | SA2426 | 2.872 | arcD | Arginine/ornithine antiporter | 0.264 | 9.193 |
| SA2203 | 2.784 | Protein similar to multidrug resistance protein | 0.104 | 3.749 | ||
| Physiological proteins | SA0171 | 6.186 | fdh | NAD-dependent formate dehydrogenase | 0.232 | 3.758 |
| SA2425 | 4.657 | arcC | Carbamate kinase | 0.201 | 4.317 | |
| SA2427 | 4.179 | arcB | Ornithine transcarbamoylase | 0.138 | 3.297 | |
| SA2088 | 3.649 | ureD | Urease accessory protein UreD | 0.086 | 2.344 | |
| SA0996 | 3.359 | sdhB | Succinate dehydrogenase iron-sulfur protein subunit | 0.159 | 4.727 | |
| SA2204 | 3.213 | Phosphoglycerate mutase pgm homolog | 0.217 | 6.756 | ||
| SA2007 | 3.143 | Protein similar to alpha-acetolactate decarboxylase | 0.269 | 8.567 | ||
| SA2008 | 3.108 | alsS | Alpha-acetolactate synthase | 0.258 | 8.294 | |
| SA2086 | 2.982 | ureF | Urease accessory protein UreF | 0.12 | 4.022 | |
| SA0218 | 2.867 | pflB | Formate acetyltransferase | 0.128 | 4.458 | |
| SA1531 | 2.738 | ald | Alanine dehydrogenase | 0.013 | 0.463 | |
| SA2428 | 2.527 | arcA | Arginine deiminase | 0.19 | 7.537 | |
| SA0219 | 2.524 | pflA | Formate-acetyltransferase-activating enzyme | 0.351 | 13.911 | |
| Other proteins | SA2424 | 5.58 | Protein similar to transcription regulator Crp/Fnr family protein | 0.313 | 5.615 | |
| SA1941 | 2.818 | dps | General stress protein 20U | 0.149 | 5.272 | |
| Hypothetical proteins | SA2268 | 4.755 | Hypothetical protein | 0.27 | 5.687 | |
| SA0170 | 4.323 | Conserved protein | 0.198 | 4.569 | ||
| SA1937 | 3.34 | Conserved protein | 0.174 | 5.213 | ||
| SA0129 | 3.268 | Hypothetical protein | 0.128 | 3.913 | ||
| SA0271 | 3.198 | Conserved protein | 0.175 | 5.465 | ||
| SA0856 | 3.019 | Conserved protein | 0.224 | 7.43 | ||
| SA2049 | 2.869 | Hypothetical protein | 0.212 | 7.373 | ||
| SA2331 | 2.619 | Hypothetical protein | 0.099 | 3.787 | ||
| SA0292 | 2.561 | Hypothetical protein | 0.218 | 8.512 | ||
| SA1476 | 2.523 | Hypothetical protein | 0.112 | 4.419 | ||
| SA0623 | 2.519 | Hypothetical protein | 0.196 | 7.771 |
Expression of specific genes in biofilm cells.
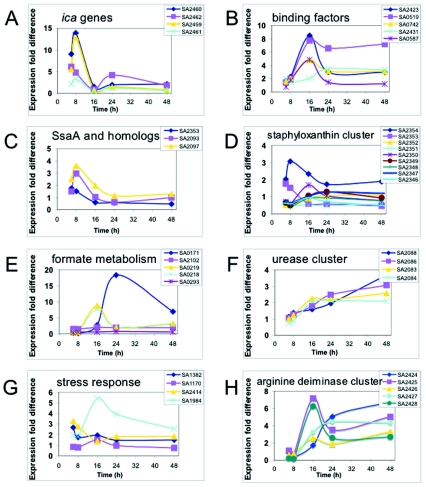
Some cellular functions and proteins, such as slime production and binding proteins, are important for biofilm formation (22). It was therefore of interest to determine whether the encoding genes are expressed at higher levels in the biofilm. The ica genes (9, 27), for example, encode the genes involved in biosynthesis of the glucosamine polymer PIA (40). PIA has been described in various reports to play a crucial role in biofilm formation and virulence. Therefore, the expression pattern for icaADBC (SA2459 to SA2462) was followed during the course of growth (Fig. 1A). The ica genes were primarily expressed under biofilm conditions; after 6 and 8 h of growth, they were expressed 5- to 12-fold more highly under biofilm conditions than under planktonic conditions. The expression level declined at later growth stages.
FIG. 1.
Comparison of the expression profiles of selected gene groups in biofilm cells and planktonic cells. Cells were grown as described in the text, and total RNA was extracted from the cells at the five times indicated and used in DNA microarray analyses. The data indicate the fold differences in expression of selected gene groups in biofilm cells compared to the expression in planktonic cells. (A) Expression pattern of genes encoding the biosynthetic enzymes for PIA, icaADBC (SA2459 to SA2462). (B) Expression pattern of genes encoding the binding proteins clumping factor B (SA2423), Ser-Asp-rich fibrinogen-binding or bone sialoprotein (SA0519), fibrinogen-binding protein A or clumping factor (SA0742), immunodominant antigen B (SA2431), and the lipoprotein streptococcal adhesion PsaA homolog (SA0587). (C) Expression pattern of genes with sequence similarity to genes encoding the extracellular, immune dominant protein staphylococcal secretory antigen A (SsaA), including a hypothetical protein similar to SsaA (SA2353), a secretory antigen precursor SsaA homolog (SA2093), and a hypothetical protein similar to SsaA (SA2097). (D) Expression pattern of genes of the staphyloxanthin biosynthesis cluster, encoding proteins similar to acyltransferase (SA2354), a hypothetical protein (SA2352), phytoene dehydrogenase (SA2351), a conserved hypothetical protein (SA2350), squalene synthase (SA2349), squalene desaturase (SA2348), aspartate aminotransferase (SA2347), and a d-specific d-2-hydroxyacid dehydrogenase ddh homolog (SA2346). (E) Expression pattern of genes involved in formate metabolism, encoding NAD-dependent formate dehydrogenase (SA0171), a formate dehydrogenase homolog (SA2102), formate-acyltransferase-activating enzyme (SA0219), formate acyltransferase (SA0218), and a protein similar to formate transporter NirC (SA0293). (F) Expression pattern of genes involved in urease activity, encoding urease accessory protein UreD (SA2088), urease accessory protein UreF (SA2086), urease beta subunit (SA2083), and urease alpha subunit (SA2084). (G) Expression pattern of genes involved in stress response, encoding superoxide dismutase SodA (SA1382), catalase (SA1170), a protein similar to glutathione peroxidase (SA2414), and alkaline shock protein (SA0194). (H) Expression pattern of genes of the arginine deiminase cluster, encoding a hypothetical protein similar to the transcriptional regulator Crp/Fnr family protein (SA2424), carbamate kinase (SA2425), arginine/ornithine antiporter (SA2426), ornithine transcarbamylase (SA2427), and arginine deiminase (SA2428).
A number of surface-associated proteins are also important for biofilm formation, especially in the initial steps (i.e., adherence) (18, 22). The expression levels of the genes encoding selected binding proteins are shown in Fig. 1B. Expression peaked after 16 h, but the level of expression remained high during the entire biofilm growth period. In particular, the expression level of sdrC (SA0519; Ser-Asp-rich fibrinogen-binding, bone sialoprotein-binding protein) was also high after 24 and 48 h of growth.
It has been postulated that staphylococcal secretory antigen A (SsaA) is involved in biofilm-associated infections since anti-SsaA immunoglobulin G antibody levels are higher in sera of patients with S. epidermidis endocarditis (35). Three genes encoding proteins homologous to SsaA have been identified in the S. aureus genome, and the expression levels of all three genes were higher up to 16 h of growth under biofilm growth conditions than under planktonic growth conditions (Fig. 1C).
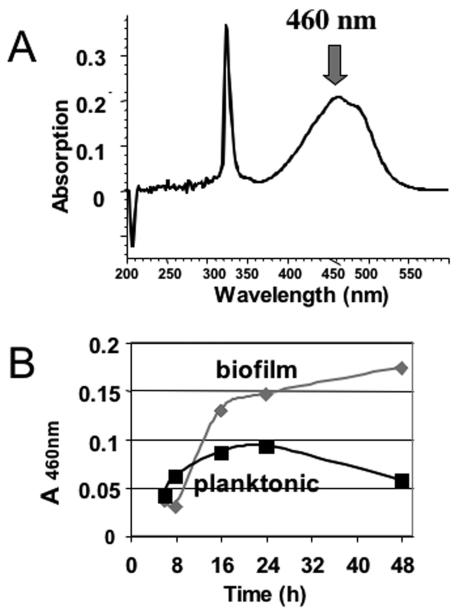
It has been proposed that staphylococci produce pigments to protect themselves from UV radiation and radicals in vivo (43, 52). Therefore, the expression of genes involved in staphyloxanthin biosynthesis was followed (Fig. 1D). In biofilm cells, the corresponding genes were expressed at higher levels than they were expressed in cells grown planktonically. These results were confirmed by direct analysis of staphyloxanthin in ethanol extracts of cells harvested during growth (Fig. 2), which provided physiological verification of the differential gene expression.
FIG. 2.
Production of staphyloxanthin in biofilm and planktonic cultures. Staphyloxanthin was extracted from the culture supernatants with ethanol. After 48 h, the biofilm supernatant was yellow-orange, whereas the planktonic supernatant was almost colorless (not shown). (A) Staphyloxanthin normally shows a typical peak in its spectrum at 460 nm; therefore, the absorption at this wavelength was measured for all samples (B).
So far, the cell envelope seems to be a rather dynamic structure in biofilm cells. This was further supported by that the finding that the genes involved in murein synthesis and other genes whose products are involved in the formation of the cell envelope are expressed slightly more highly in biofilm cells (not shown).
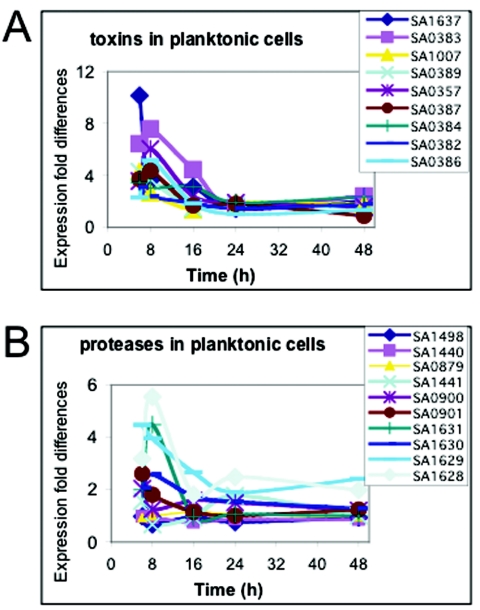
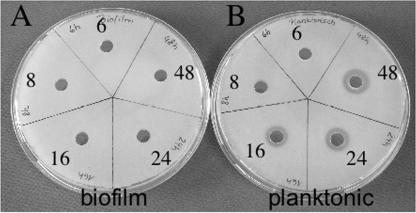
Although this study was mainly geared toward investigation of the expression patterns of genes in biofilm cells, the microarray analysis also provided data on genes expressed at higher levels during planktonic growth. Genes encoding toxins (Fig. 3A), for example, were expressed at much higher levels under planktonic growth conditions, especially during the first 16 h; thereafter, the expression level decreased. No toxin gene was expressed at higher levels in biofilm cells. Genes encoding proteases (Fig. 3B), which also are regarded as virulence factors (15, 47), were also expressed at higher levels under planktonic growth conditions. Physiological verification of the differential gene expression was obtained by testing supernatants of planktonically grown and biofilm-grown cells at all five times for protease activity on casein agar plates. Significant protease activity was found only in the 16-, 24-, and 48-h planktonic cultures (Fig. 4).
FIG. 3.
Comparisonof the expression profiles of genes encoding toxins and proteases in biofilm and planktonic cultures. Microarray analysis was performed and expression levels were determined as described in the legend to Fig. 1. The data indicate the fold differences in expression of genes in planktonic cells compared to the expression in biofilm cells. (A) Expression pattern of genes encoding toxins, including the leukotoxin LukD (SA1637), exotoxin 7 (SA0383), exotoxin 13 (SA0389), exotoxin 8 (SA0384), exotoxin 10 (SA0386), an alpha-hemolysin precursor (SA1007), a protein similar to exotoxin 2 (SA0357), and exotoxin 6 (SA0382). (B) Expression pattern of genes encoding proteases, including protease ClpX (SA1498), a protein similar to protease (SA1440), serine protease HtrA (SA0879), a protein similar to protease (SA1441), a cysteine protease precursor (SA0900), serine protease (SA0901), serine protease SplA (SA1631), serine protease SplB (SA1630), serine protease SplC (SA1629), and serine protease SplD (SA1628).
FIG. 4.
Proteolytic activities of the supernatants of biofilm cells (A) and planktonic cells (B). The supernatants of cells grown for different times were used to test the proteolytic activity on casein agar plates. Proteolytic activity is indicated by halo formation.
The data for all the genes expressed at higher levels in planktonic cells are available at http://www.uni-tuebingen.de/Mikrobiogen/announce.html.
Hypothetical genes.
Unfortunately, more than 200 genes that are up-regulated in biofilm or planktonic cells code for hypothetical proteins with unknown functions. Some of these genes were up-regulated at four times, and one gene (SA0271) was up-regulated at all five times. To obtain information on the possible function of the hypothetical proteins, a BLAST analysis of the derived amino acid sequences was carried out. The results are shown in Table 6. Interestingly, some of the hypothetical proteins up-regulated in biofilm cells show similarities to shock proteins and cell envelope-associated proteins, which supports the general trend found for the expression of genes in biofilm cells.
TABLE 6.
Possible functions of hypothetical genes expressed at higher levels under biofilm growth conditions
| N315 open reading frame | Similarities to identified genes of other organismsa |
|---|---|
| SA0271 | yukE/yfjA Bacillus subtilis family; small heat shock protein; similarity to bacterial protein with unknown function (DUF909) |
| SA2133 | Putative cytochrome (Escherichia coli O157:H7 EDL933); integral membrane protein; bacterial protein with unknown function (DUF805) |
| SA0412 | ybcD (Bacillus subtilis), hypothetical protein |
| SA2143 | yhbJ (Bacillus subtilis), hypothetical protein |
| SA0292 | No similarities found |
| SA0609 | Hypothetical protein (Bacillus subtilis); TNF family signature and profile |
| SA1403 | yqeZ (Bacillus subtilis); protein with unknown function (DUF107) |
| SA2378 | Glyoxylase/bleomycin resistance protein/dioxygenase domain |
| SA2268 | Protein with unknown function (DUF805) |
| SA1985 | No similarities found |
| SA1586 | 6,7-Dimethyl-8-ribityllumazine synthase riboflavin synthase beta chain |
| SA0588 | Membrane protein (Staphylococcus epidermidis) |
| SA1290 | Poly(A) polymerase (Bacillus subtilis); polynucleotide adenyltransferase |
| SA0170 | yrhF (Bacillus subtilis), hypothetical protein |
| SA1937 | Hypothetical protein (Deinococcus radiodurans) |
Similarities were determined by BLAST analysis, DOGAN, COG (1), InterProScan, and EMBL-EBI.
Physiology of biofilms.
The results shown in Tables 1 to 5 and the expression patterns determined for selected genes indicated that several physiological and metabolic pathways could be important for biofilm formation, differentiation, and persistence. Therefore, the expression of genes involved in formate and urea metabolism, the oxidative stress response, and the arginine deaminase cluster was followed over time (Fig. 1E to H). Most of the corresponding genes were expressed at levels that were severalfold higher in biofilm cells than in planktonic cells.
One of the greatest differences in expression was found with the gene encoding NAD-dependent formate dehydrogenase (SA0171). In biofilm cells after 24 h of growth (maximum level), the gene expression level was >17-fold higher than the level in planktonic cells (Fig. 1E). If the gene encoding formate dehydrogenase is expressed at higher levels in biofilm cells, it could be expected that the genes involved in formate synthesis are also up-regulated. Indeed, the expression levels of the S. aureus genes encoding pyruvate formate-lyase, pflA (SA0219) and pflB (SA0218), after 16 (maximum), 24, and 48 h of growth were 2.5- to 8-fold higher in biofilm cells than in planktonic cells (Fig. 1E). In Escherichia coli and related bacteria, the PflA protein is involved in the activation of pyruvate formate-lyase (PflB) under anoxic conditions by generation of an organic free radical, using S-adenosylmethionine and reduced flavodoxin as cosubstrates to produce 5′-deoxyadenosine. To determine whether the gene expression profile has physiological relevance, the specific activity of formate dehydrogenase from biofilm and planktonic cells grown for 24 h was determined. The activity in biofilm cells was 7.5-fold higher than that in planktonic cells (Table 7). This is consistent with the results from the DNA microarrays, in which the gene expression levels were 17-fold higher after 24 h of growth.
TABLE 7.
Specific activities of formate dehydrogenase in biofilm and planktonic cells grown for 24 h
| Cells | n | Formate dehydrogenase sp act (nmol/min/mg)a |
|---|---|---|
| Biofilm | 12 | 29 ± 4.83 |
| Planktonic | 12 | 3.8 ± 3.0 |
Biofilm and planktonic cells were grown for 24 h, and the specific activity of formate dehydrogenase was determined as described in the text. The activity in biofilm cells was 7.5-fold higher than that in planktonic cells.
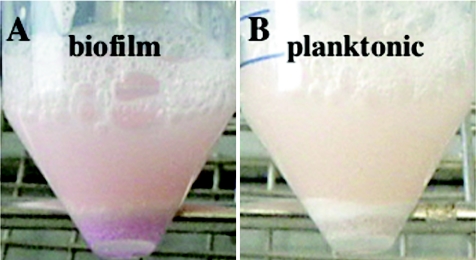
The expression levels of genes encoding urease were also higher in biofilm cells (Fig. 1F). Urease activity was measured using Rosco diagnostic tablets to determine whether the differential gene expression profile has physiological relevance. After 48 h of growth, biofilm cells indeed exhibited much higher urease activity than the corresponding planktonic cells at comparable cell densities (Fig. 5); biofilm cells stained pink-red, which indicated urease activity, whereas planktonic cells showed almost no detectable staining. Although this assay is only semiquantitative, it confirmed the microarray data.
FIG. 5.
Urease activity in biofilm cells (A) and planktonic cells (B). Cells were grown as described in the text and harvested after 48 h of growth. Urease activity was determined using urease diagnostic tablets (575-21; Rosco), which leads to a red color if urea is hydrolyzed by urease to form two molecules of ammonia. The red/purple color of the biofilm cells appeared after 30 min of incubation. The planktonic cells remained nearly white.
It is worth remarking that the expression levels of stress response genes (encoding, for example, superoxide dismutase [SOD], catalase, glutathione peroxidase, and alkaline shock protein) were higher in biofilm cells than in planktonic cells (Fig. 1G). This result suggested that the cells in a biofilm are more exposed to stress factors than cells in a liquid medium.
The expression levels of the genes of the arginine deaminase cluster were also higher in the biofilm cells than in planktonic cells (Fig. 1H). The highest levels (especially for SA2428 and SA2425) were reached at 16 h of growth, and the levels remained 2.5-fold higher than the levels in planktonic cells even at later times.
DISCUSSION
Here we report a comparative transcriptome analysis of the global gene expression of S. aureus cells grown in a biofilm and in cells grown planktonically in the exponential and stationary growth phases. A number of genes that were expressed at different levels in cells grown under the two conditions were identified.
The expression of the ica genes (9, 19, 22) and thereby the synthesis of PIA (40) are important for adhesion and formation of staphylococcal biofilms. The microarray results reported here showed that the four ica genes are expressed at higher levels after 6 and 8 h of growth in biofilm cells than in planktonic cells (the levels were 5- and 14-fold higher, respectively); the expression level in the biofilm cells decreased thereafter but remained 3-fold higher than that in planktonic cells. Two conclusions can be drawn from these results: the ica genes are up-regulated in an S. aureus biofilm and are necessary for adhesion and the beginning of biofilm formation, and the ica genes are up-regulated only at the beginning of biofilm formation. The gene products have a long half-life (13), and, therefore, the up-regulation of these genes might not be needed once the cells are attached to the surface, biofilm formation has begun, and cell growth is retarded owing to nutrient depletion. Our results corroborated the results of Beenken et al. (3), who also did not observe an increase in ica gene expression at later times in biofilm formation.
As PIA is needed to embed the cells in a slimy matrix and for bacterial adhesion (25, 27), it was of interest to investigate whether other binding factors were also expressed at higher levels in biofilm cells. As Fig. 1B shows, genes for various binding factors were highly expressed in the biofilm cells, and some of these genes were also expressed at later time points. These results could imply that some of these binding factors have shorter half-lives, which would then require permanent synthesis. Furthermore, these factors might be very important for biofilm persistence and differentiation at the later times of biofilm development.
Interestingly, genes involved in murein synthesis, such as all mur genes, were expressed at slightly higher levels in biofilm cells; this has also been shown for Pseudomonas aeruginosa by other groups (17). These genes are involved in the synthesis of peptidoglycan and cell walls. The reasons for their up-regulation in biofilm cells requires further investigation since the cell walls of biofilm and planktonic cells are thought to be similar. Many cell wall-associated binding factors were also up-regulated in biofilm cells, which suggested that the cell envelope is a highly dynamic and active component of biofilm cells. This could explain why biofilm cells are so resistant to shear forces in vivo and why they cannot be easily accessed by the host immune system.
In many patients with chronic polymer-associated infections caused by S. epidermidis, antibodies against SsaA (staphylococcal secretory antigen, a highly immunogenic protein) have been found, which suggests that this factor plays a role in biofilm-associated infections (35). Our microarray results showed that ssaA and homologous genes were expressed at slightly higher levels in biofilm cells than in planktonic cells. Therefore, the expression of ssaA might contribute to the overcoming of the humoral defense system and the persistence of biofilm cells in vivo.
Staphyloxanthin, the major stationary-phase carotenoid, is an orange-red pigment thought to function as a protective antioxidant for staphylococcal cells (43, 52). Its synthesis is regulated by SigB (4). Here, our results showed that the genes involved in the synthesis of staphyloxanthin were expressed at slightly higher levels in biofilm cells than in planktonic cells. This finding was supported by the coloration of ethanol extracts of biofilm cells (not shown) and the absorbance of biofilm cell extracts at 460 nm (Fig. 2A and B), compared to the lack of coloration of ethanol extracts of planktonic cells and their absorbance at 460 nm.
Selected groups of genes were analyzed for up-regulation under planktonic growth conditions. After the initial analysis of the data, it became apparent that especially the genes encoding toxins and other virulence factors (e.g., proteases) were up-regulated in planktonic cells. Various toxins were significantly up-regulated in planktonic cells. No toxin was up-regulated in biofilm cells. The expression of toxins solely in the planktonic cells points toward these cells being much more virulent and being more able to cause acute infections (e.g., sepsis) and wound infections than biofilm cells. The proteases secreted by planktonic cells can also be regarded as virulence factors during the infection process since they can digest host proteins (15, 31, 32, 47). Toxins and proteases, therefore, are probably not factors that promote or contribute to biofilm persistence in the host.
The results of the BLAST analysis have to be interpreted with caution since this is still not a reliable means of identifying the functions of genes; however, the results provide the first clues concerning the function of the hypothetical genes. Here, the results supported the finding that certain gene groups (e.g., genes encoding binding factors) play an important role in biofilm formation. Some of these hypothetical genes (e.g., SA0271 and SA2133) were highly expressed at various times in biofilm cells and therefore should be characterized. The similarity of SA0271 to genes encoding heat shock proteins suggested that various stress factors could play a role in biofilm differentiation.
Our results suggest that not toxins and other virulence factors, but rather processes involved in cell wall synthesis and other distinct physiological activities of the cell, play a crucial role in biofilm persistence. Therefore, it was of interest to determine which metabolic pathways are up-regulated in biofilm cells. One striking example was the gene encoding formate dehydrogenase (SA0171), whose expression level was 17-fold higher in biofilm cells after 24 h of growth than in planktonic cells. This enzyme degrades formate to form CO2 and NADH plus H+. Determination of the formate dehydrogenase specific activity corroborated the microarray data; the specific activity was 7.5-fold higher in biofilm cells than in planktonic cells. The high up-regulation of this gene implies high production of formate in biofilms. Indeed, the genes encoding PflA (formate acetyltransferase-activating enzyme) and PflB (formate acetyltransferase) were more-than-sevenfold up-regulated after 16 h of growth and were still up-regulated after 48 h of growth of biofilm cells.
Cells normally gain energy from the formate pathway, but formic acid is a strong acid (pKa 3.65) and its metabolic products (H+) lead to acidification of the biofilm surroundings. A strong acid concentration in the vicinity of the bacterial cells could lead to necrosis of host tissue and might also affect the host immune response, possibly contributing to the persistence of the staphylococci. In this connection, it would be of interest to investigate the acidity of the tissue surrounding a staphylococcal biofilm in vivo. However, bacterial cells also have to protect themselves from a pH that is too low. Along these lines, it makes sense that the gene encoding urease was up-regulated in our study. The urease accessory protein genes ureD (SA2088) and ureF (SA2086) were expressed at higher levels at the later times. Urease is needed in the urea cycle and in the metabolism of amino acids to degrade urea to form CO2 and NH3. The resulting ammonium and/or ammonia (depending of the pH of the cells) is toxic for the host cells and might accumulate in and outside the bacterial cells. We therefore assume that the urease activity determined (Fig. 5) contributes to the persistence of the bacterial cells in the biofilm by counteracting the low pH values caused by the production of lactic acid, acetic acid, and formic acid. Beenken et al. (3) have also reported up-regulation of the urease operon in 7-day-old biofilms. We therefore believe that urease activity might be an important factor for keeping the biofilm alive. Since excess ammonia would be toxic for the bacterial cells, they should have some mechanism of resistance against this chemical and should also have enzymes or other mechanisms to detoxify this compound. In this context, it is interesting that in biofilm cells the expression of the gene encoding alkaline shock protein Asp23 (SA1984) (33) after 16, 24, and 48 h of growth is 2.5- to 5-fold higher than the expression in planktonic cells (Fig. 1G). The expression of asp23 is regulated by SigB (4).
It has been postulated that oxidative stress could play a role in biofilm differentiation in P. aeruginosa (16, 24, 49). In our study reported here for S. aureus, genes possibly involved in the detoxification of reactive oxygen species (ROS) are expressed at higher levels in biofilm cells. It therefore makes sense that the genes encoding SOD (SA1382) and glutathione peroxidase (SA2414) are up-regulated in biofilm cells, and the findings indicate that ROS, which the bacteria have to detoxify, are present in a biofilm. P. aeruginosa cells are possibly protected from reactive oxygen species by catalase and SOD (49). As the deeper layers of the biofilm become anoxic, the energy metabolism must shift to fermentation. However, the ROS created in the oxic layers or by myeloperoxidase during the host immune response (30) can diffuse into the anoxic layers and damage the cells if insufficient detoxifying enzymes (e.g., SOD and glutathione peroxidase) are produced. The possibility that ROS might play a role in the formation of a biofilm is further supported by the production of staphyloxanthin, which is postulated to protect the bacterial cells against radiation and also organic radicals (52).
Interestingly, we found further similarities with the data of Beenken et al. (3). The genes of the arginine deiminase cluster (arc; in N315: SA2424 to SA2428) are expressed at higher levels in biofilm cells after 16, 24, and 48 h of growth. It can be speculated that these genes are needed for cell metabolism at later times, when some areas of the biofilm become anoxic and the cells can gain some energy in the form of ATP from the conversion of arginine to citrulline (12). Also, the products of the proteins encoded by this gene cluster can be fed into the urea cycle and thereby lead to the generation of ammonia and/or urea. It would be of interest to test whether the resulting ammonia is utilized by the cells to maintain pH homeostasis when they are growing in a biofilm in order to neutralize acids generated by fermentation. Together with a high formate dehydrogenase activity and generation of high levels of acid, this might be a very important finding and might also imply that the urease and arginine deiminase activities are the basis for the survival of cells in a biofilm.
In contrast to Beenken et al. (3), we did not observe a higher level of expression of genes of the potassium-specific transport system (kdp; in N315, SA1879 to SA1881) or of the pyrimidine biosynthesis operon (pyr; in N315, SA1041 to SA1049). However, this might have been due to the different test conditions, such as the age and growth conditions of the biofilm and the growth phase of the planktonic cells.
Over 160 genes have been identified as genes that are expressed at significantly higher levels in biofilm cells. These genes include those involved in the synthesis of binding factors, peptidoglycan, and PIA and in the detoxification of formate, urea, and ROS. All these activities might contribute to the observed persistence and resistance of cells in a biofilm. On the other hand, in planktonic cells, genes encoding toxins and proteases were expressed at significantly higher levels. As many of the gene products are serious pathogenicity factors, one would expect planktonic cells to be more aggressive with respect to virulence and to have a higher tendency to spread. Some of these genes reflect specific growth phases, nutrition, or oxygen conditions rather than sessile growth in a biofilm per se. Also, the total number of up-regulated genes in the biofilm decreased with time, which could be explained by decelerated growth rates, by dormant states in biofilm cells, and particularly by reduced metabolism due to depletion of nutrients, unfavorable oxygen concentrations (5, 20), or acidification of the medium due to increased fermentation activity. We assume that particularly the genes that are expressed at significantly higher levels in biofilm cells after 24 and 48 h (Tables 4 and 5) are important for the perpetuation of the biofilm and the survival of cells in this dense and nutrient-poor community. By comparative transcriptome analysis of biofilm versus planktonic cells, we demonstrated that biofilm cells show distinct metabolic activity.
TABLE 4.
Genes expressed more highly under biofilm conditions after 24 h of growth
| Type of proteins | N315 open reading frame | Biofilm vs planktonic cells (fold difference) | Name | Product | SD | CV |
|---|---|---|---|---|---|---|
| Cell wall-associated proteins | SA0572 | 8.007 | Protein similar to esterase/lipase | 0.21 | 13.992 | |
| SA0519 | 6.559 | sdrC | Ser-Asp-rich fibrinogen-binding bone sialoprotein-binding protein | 0.089 | 9.777 | |
| SA2431 | 3.51 | isaB | Immunodominant antigen B | 0.406 | 14.286 | |
| SA2462 | 3.236 | icaC | Intercellular adhesion protein C | 0.212 | 15.07 | |
| SA0742 | 3.131 | clfA | Fibrinogen-binding protein A clumping factor | 0.19 | 12.987 | |
| SA2423 | 2.976 | clfB | Clumping factor B | 0.235 | 13.086 | |
| Transporter protein | SA2203 | 2.62 | Hypothetical protein similar to multidrug resistance protein | 0.107 | 6.323 | |
| Physiological proteins | SA0171 | 16.161 | fdh | NAD-dependent formate dehydrogenase | 0.246 | 1.522 |
| SA0996 | 4.37 | sdhB | Succinate dehydrogenase iron-sulfur protein subunit | 0.171 | 6.357 | |
| SA2427 | 3.965 | arcB | Ornithine transcarbamoylase | 0.088 | 1.774 | |
| SA2204 | 3.842 | Phosphoglycerate mutase pgm homolog | 0.15 | 11.092 | ||
| SA1244 | 3.706 | odhB | Dihydrolipoamide succinyltransferase | 0.095 | 5.37 | |
| SA2007 | 3.274 | Protein similar to alpha-acetolactate decarboxylase | 0.096 | 10.403 | ||
| SA0995 | 2.694 | sdhA | Succinate dehydrogenase flavoprotein subunit | 0.113 | 5.445 | |
| SA1245 | 2.634 | odhA | 2-Oxoglutarate dehydrogenase E1 | 0.21 | 5.676 | |
| Other proteins | SA2424 | 4.967 | Protein similar to transcription regulator Crp/Fnr family protein | 0.112 | 6.372 | |
| SA1984 | 4.624 | asp23 | Alkaline shock protein 23 (ASP23) | 0.062 | 8.395 | |
| SA1941 | 4.123 | dps | General stress protein 20U | 0.19 | 8.038 | |
| SA2335 | 2.671 | adaB | Probable methylated DNA-protein cysteine methyltransferase | 0.081 | 7.988 | |
| SA0456 | 2.659 | spoVG | Stage V sporulation protein G homologue | 0.105 | 9.873 | |
| Hypothetical proteins | SA0170 | 7.154 | Conserved protein | 0.271 | 3.795 | |
| SA2268 | 4.487 | Hypothetical protein | 0.09 | 8.851 | ||
| SA1985 | 4.194 | Hypothetical protein | 0.07 | 5.94 | ||
| SA1986 | 4.086 | Hypothetical protein | 0.238 | 5.148 | ||
| SA0856 | 4.055 | Conserved protein | 0.061 | 8.327 | ||
| SA1634 | 3.495 | Truncated protein | 0.067 | 7.195 | ||
| SA0623 | 3.789 | Hypothetical protein | 0.091 | 9.284 | ||
| SA0292 | 3.395 | Hypothetical protein | 0.092 | 2.721 | ||
| SA0585 | 3.354 | Conserved protein | 0.141 | 11.972 | ||
| SA2049 | 3.339 | Hypothetical protein | 0.174 | 14.838 | ||
| SA1937 | 3.283 | Conserved protein | 0.045 | 6.515 | ||
| SA0772 | 3.25 | Conserved protein | 0.089 | 8.143 | ||
| SA0406 | 3.015 | Hypothetical protein | 0.182 | 30.65 | ||
| SA0271 | 2.979 | Conserved protein | 0.067 | 2.247 | ||
| SA0129 | 2.955 | Hypothetical protein | 0.206 | 6.968 | ||
| SA2133 | 2.684 | Conserved protein | 0.072 | 8.619 | ||
| SA0752 | 2.675 | Hypothetical protein | 0.067 | 10.286 | ||
| SA0570 | 2.61 | Hypothetical protein | 0.176 | 14.459 | ||
| SA1476 | 2.564 | Hypothetical protein | 0.121 | 11.113 |
TABLE 2.
Genes expressed more highly under biofilm conditions after 8 h of growth
| Type of proteins | N315 open reading frame | Biofilm vs planktonic cells (fold difference) | Name | Product | SD | CV |
|---|---|---|---|---|---|---|
| Secreted proteins | SA2097 | 3.521 | Protein similar to secretory antigen precursor SsaA | 0.117 | 3.31 | |
| SA2093 | 2.728 | ssaA | Secretory antigen precursor SsaA homolog | 0.215 | 7.874 | |
| SA0620 | 2.534 | Secretory antigen SsaA homologue | 0.125 | 4.913 | ||
| Cell wall-associated proteins | SA2460 | 12.276 | icaD | Intercellular adhesion protein D | 0.275 | 2.236 |
| SA2459 | 8.238 | icaA | Intercellular adhesion protein A | 0.514 | 6.237 | |
| SA2462 | 3.584 | icaC | Intercellular adhesion protein C | 0.381 | 10.633 | |
| SA2356 | 3.12 | isaA | Immunodominant antigen A | 0.098 | 3.154 | |
| SA2461 | 3.039 | icaB | Intercellular adhesion protein B | 0.233 | 7.656 | |
| SA1893 | 2.604 | Lipoprotein precursor | 0.182 | 6.999 | ||
| Transporter proteins | SA0589 | 6.344 | Protein similar to ABC transporter ATP-binding protein | 0.106 | 1.673 | |
| SA0531 | 5.629 | prop | Proline/betaine transporter homologue | 0.294 | 5.222 | |
| SA2142 | 5.002 | Protein similar to multidrug resistance protein | 0.061 | 1.221 | ||
| SA2200 | 3.714 | Protein similar to ABC transporter ATP-binding subunit | 0.081 | 2.169 | ||
| SA1183 | 3.878 | opuD | Glycine betaine transporter | 0.086 | 2.228 | |
| SA2201 | 3.447 | Protein, similar to ABC transporter permease protein | 0.061 | 1.762 | ||
| SA1519 | 3.367 | aapA | d-Serine/d-alanine/glycine transporter | 0.082 | 2.443 | |
| SA0682 | 3.299 | Protein similar to di-tripeptide ABC transporter | 0.08 | 2.425 | ||
| SA1547 | 2.98 | ptaA | Phosphotransferase system N-acetylglucosamine-specific II ABC component | 0.24 | 8.051 | |
| SA2202 | 2.7 | Protein similar to ABC transporter periplasmic amino acid-binding protein | 0.225 | 8.327 | ||
| SA0813 | 2.67 | mnhA | Na+/H+ antiporter subunit | 0.118 | 4.434 | |
| SA1699 | 2.633 | Protein similar to transporter | 0.351 | 13.339 | ||
| SA0793 | 2.615 | dltA | d-Alanine-d-alanyl carrier protein ligase | 0.103 | 3.937 | |
| SA2172 | 2.59 | gltT | Proton/sodium-glutamate symport protein | 0.151 | 5.849 | |
| SA0733 | 2.575 | secG | Probable protein export membrane protein | 0.12 | 4.679 | |
| SA0493 | 2.52 | secE | Preprotein translocase subunit | 0.148 | 5.868 | |
| Physiological proteins | SA0912 | 4.061 | qoxB | Quinol oxidase polypeptide I QoxB | 0.14 | 3.445 |
| SA0913 | 3.513 | Protein similar to quinol oxidase polypeptide II QoxA | 0.199 | 5.672 | ||
| SA0911 | 3.229 | qoxC | Quinol oxidase polypeptide III QoxC | 0.153 | 4.745 | |
| SA0910 | 3.218 | Protein similar to quinol oxidase polypeptide IV QoxD | 0.108 | 3.353 | ||
| SA0505 | 3.554 | fus | Translational elongation factor G | 0.234 | 6.585 | |
| SA0502 | 3.337 | Protein similar to ribosomal protein | 0.156 | 4.677 | ||
| SA1075 | 3.284 | hmrB | HmrB protein | 0.081 | 2.457 | |
| SA0842 | 3.107 | FabH | 3-Oxoacyl-(acyl carrier protein) synthase homologue | 0.186 | 5.991 | |
| SA2354 | 2.956 | Protein similar to acyltransferase | 0.135 | 4.552 | ||
| SA0945 | 2.946 | pdhC | Dihydrolipoamide S-acetyltransferase component of pyruvate dehydrogenase complex E2 | 0.198 | 6.734 | |
| SA0731 | 2.874 | eno | Enolase | 0.168 | 5.836 | |
| SA2204 | 2.862 | Phosphoglycerate mutase pgm homolog | 0.211 | 7.38 | ||
| SA1717 | 2.861 | Glutamyl-tRNAGln amidotransferase subunit C | 0.272 | 9.502 | ||
| SA2191 | 2.833 | Protein similar to NirC | 0.215 | 7.605 | ||
| SA0802 | 2.832 | Protein similar to NADH dehydrogenase | 0.296 | 10.437 | ||
| SA0843 | 2.621 | fab 3-Oxoacylsynthase | 0.16 | 6.093 | ||
| SA2077 | 2.614 | Protein similar to biotin biosynthesis protein | 0.193 | 7.402 | ||
| SA2027 | 2.557 | adk | Adenylate kinase | 0.293 | 11.443 | |
| SA1548 | 2.516 | Protein similar to acylglycerol-3-phosphate O-acyltransferase homolog | 0.134 | 5.332 | ||
| Ribosomal proteins | SA2016 | 3.324 | rpsI | 30S ribosomal protein S9 | 0.128 | 3.856 |
| SA2017 | 3.241 | rplM | 50S ribosomal protein L13 | 0.111 | 3.43 | |
| SA1471 | 3.173 | rpmA | 50S ribosomal protein L27 | 0.142 | 4.466 | |
| SA2030 | 3.109 | rpmD | 50S ribosomal protein L30 | 0.15 | 4.818 | |
| SA2045 | 3.058 | rplW | 50S ribosomal protein L23 | 0.224 | 7.318 | |
| SA1081 | 3.039 | rpsP | 30S ribosomal protein S16 | 0.079 | 2.605 | |
| SA2029 | 2.874 | rplO | 50S ribosomal protein L15 | 0.162 | 5.653 | |
| SA0504 | 2.836 | rpsG | 30S ribosomal protein S7 | 0.245 | 8.648 | |
| SA0495 | 2.834 | rpsK | 50S ribosomal protein L11 | 0.262 | 9.254 | |
| SA1473 | 2.79 | rplU | 50S ribosomal protein L21 | 0.223 | 7.978 | |
| SA0354 | 2.772 | rpsR | 30S ribosomal protein S18 | 0.071 | 2.557 | |
| SA0503 | 2.757 | rpsL | 30S ribosomal protein S12 | 0.191 | 6.919 | |
| SA2031 | 2.75 | rpsE | 30S ribosomal protein S5 | 0.271 | 9.837 | |
| SA0497 | 2.737 | rplJ | 50S ribosomal protein L10 | 0.136 | 4.973 | |
| SA1116 | 2.736 | rpsO | 30S ribosomal protein S15 | 0.085 | 3.097 | |
| SA2032 | 2.662 | rplR | 50S ribosomal protein L18 | 0.248 | 9.305 | |
| SA0496 | 2.629 | rplA | 50S ribosomal protein L1 | 0.147 | 5.605 | |
| SA2048 | 2.617 | rpsJ | 30S ribosomal protein S10 | 0.205 | 7.846 | |
| SA2035 | 2.611 | rplE | 50S ribosomal protein L5 | 0.302 | 11.562 | |
| SAS079 | 2.587 | rpsN | 30S ribosomal protein S14 | 0.244 | 9.43 | |
| Other proteins | SA2106 | 5.192 | Protein similar to protein of pXO2-46 | 0.271 | 5.22 | |
| SA1949 | 4.484 | Lytic regulatory protein truncated with Tn554 | 0.177 | 3.942 | ||
| SA1956 | 3.783 | Lytic regulatory protein truncated with Tn554 | 0.139 | 3.681 | ||
| SA0469 | 3.215 | ftsH | Cell division protein | 0.119 | 3.712 | |
| SA1023 | 3.174 | ftsL | Cell division protein | 0.025 | 0.796 | |
| SA0353 | 2.964 | ssb | Single-strand DNA-binding protein of phage φPVL | 0.191 | 6.461 | |
| SA1305 | 2.77 | hu | DNA-binding protein II | 0.135 | 4.89 | |
| Hypothetical proteins | SA0271 | 7.686 | Conserved protein | 0.104 | 1.347 | |
| SA0292 | 4.76 | Conserved protein | 0.173 | 3.639 | ||
| SA0609 | 3.935 | Conserved protein | 0.195 | 4.945 | ||
| SA0890 | 3.847 | Conserved protein | 0.289 | 7.516 | ||
| SA1403 | 3.677 | Conserved protein | 0.142 | 3.868 | ||
| SA0588 | 3.593 | Conserved protein | 0.132 | 3.67 | ||
| SA1053 | 3.231 | Conserved protein | 0.106 | 3.272 | ||
| SA1472 | 3.155 | Conserved protein | 0.173 | 5.497 | ||
| SA1419 | 3.084 | Conserved protein | 0.125 | 4.068 | ||
| SA2133 | 3.001 | Conserved protein | 0.248 | 8.261 | ||
| SA1944 | 2.909 | Hypothetical protein | 0.245 | 8.414 | ||
| SA1056 | 2.819 | Hypothetical protein | 0.262 | 9.292 | ||
| SA0412 | 2.815 | Conserved protein | 0.116 | 4.121 | ||
| SA1402 | 2.805 | Conserved protein | 0.117 | 4.164 | ||
| SA0291 | 2.802 | Hypothetical protein | 0.068 | 2.412 | ||
| SA1971 | 2.762 | Hypothetical protein | 0.161 | 5.841 | ||
| SAS048 | 2.658 | Hypothetical protein | 0.17 | 6.389 | ||
| SA2378 | 2.643 | Conserved protein | 0.149 | 5.635 | ||
| SA2143 | 2.629 | Conserved protein | 0.16 | 6.079 | ||
| SA1293 | 2.612 | Conserved protein | 0.219 | 8.378 | ||
| SA1912 | 2.511 | Hypothetical protein | 0.069 | 2.742 | ||
| SA0975 | 2.502 | Conserved protein | 0.253 | 10.124 |
TABLE 3.
Genes expressed more highly under biofilm conditions after 16 h of growth
| Type of proteins | N315 open reading frame | Biofilm vs planktonic cells (fold difference) | Name | Product | SD | CV |
|---|---|---|---|---|---|---|
| Cell wall-associated proteins | SA2423 | 8.485 | clfB | Clumping factor B | 0.099 | 1.167 |
| SA0519 | 7.747 | sdrC | Ser-Asp-rich fibrinogen-binding bone sialoprotein-binding protein | 0.157 | 2.027 | |
| SA0742 | 4.81 | clfA | Fibrinogen-binding protein A clumping factor | 0.189 | 3.931 | |
| SA0587 | 4.297 | Lipoprotein streptococcal adhesin PsaA homologue | 0.279 | 6.501 | ||
| SA1893 | 2.589 | Lipoprotein precursor | 0.096 | 3.717 | ||
| Transporter proteins | SA0589 | 11.177 | Protein similar to ABC transporter ATP-binding protein | 0.178 | 1.592 | |
| SA1519 | 2.899 | aapA | d-Serine/d-alanine/glycine transporter | 0.057 | 1.958 | |
| SA2203 | 2.813 | Protein similar to multidrug resistance protein | 0.308 | 10.95 | ||
| SA0928 | 2.577 | Protein similar to cation ABC transporter | 0.291 | 11.285 | ||
| SA2426 | 2.555 | arcD | Arginine/ornithine antiporter | 0.077 | 3.027 | |
| Physiological proteins | SA2204 | 8.233 | Phosphoglycerate mutase pgm homolog | 0.048 | 0.585 | |
| SA0219 | 8.033 | pflA | Formate-acetyltransferase-activating enzyme | 0.219 | 2.728 | |
| SA0218 | 7.14 | pflB | Formate acetyltransferase | 0.182 | 2.555 | |
| SA2425 | 6.033 | arcC | Carbamate kinase | 0.343 | 5.678 | |
| SA2428 | 5.122 | arcA | Arginine deiminase | 0.348 | 6.798 | |
| SA0225 | 4.386 | Protein similar to glutaryl-coenzyme A dehydrogenase | 0.306 | 6.967 | ||
| SA0994 | 4.293 | sdhC | Succinate dehydrogenase cytochrome b-558 | 0.185 | 4.303 | |
| SA0232 | 4.067 | lctC | l-Lactate dehydrogenase | 0.216 | 5.311 | |
| SA1553 | 3.258 | fhs | Formyltetrahydrofolate synthetase | 0.195 | 5.974 | |
| SA0913 | 3.195 | Protein similar to quinol oxidase polypeptide II QoxA | 0.133 | 4.157 | ||
| SA1531 | 3.146 | ald | Alanine dehydrogenase | 0.182 | 5.776 | |
| SA2427 | 3.088 | arcB | Ornithine transcarbamoylase | 0.182 | 5.878 | |
| SA0133 | 3.079 | dra | Deoxyribose-phosphate aldolase | 0.14 | 4.543 | |
| SA1906 | 2.993 | atpG | ATP synthase gamma chain | 0.173 | 5.767 | |
| SA1939 | 2.988 | Deoxyribose phosphate aldolase | 0.058 | 1.945 | ||
| SA2008 | 2.924 | alsS | Alpha-acetolactate synthase | 0.234 | 8.015 | |
| SA0911 | 2.889 | qoxC | Quinol oxidase polypeptide III QoxC | 0.038 | 1.307 | |
| SA1910 | 2.823 | atpE | ATP synthase C chain | 0.228 | 8.065 | |
| SA0231 | 2.822 | Protein similar to flavohemoprotein | 0.139 | 4.942 | ||
| SA1088 | 2.794 | sucC | Succinyl-coenzyme A synthetase | 0.23 | 8.221 | |
| SA0912 | 2.781 | qoxB | Quinol oxidase polypeptide I QoxB | 0.063 | 2.268 | |
| SA1245 | 2.741 | odhA | 2-Oxoglutarate dehydrogenase E1 | 0.152 | 5.53 | |
| SA0171 | 2.651 | fdh | NAD-dependent formate dehydrogenase | 0.089 | 3.358 | |
| SA1911 | 2.613 | atpB | ATP synthase A chain | 0.109 | 4.186 | |
| SA1561 | 2.575 | murC | UDP-N-acerylmuramate-alanine ligase | 0.076 | 2.961 | |
| Ribosomal protein | SA2035 | 2.504 | rplE | 50S ribosomal protein L5 | 0.138 | 5.518 |
| Other proteins | SA1984 | 5.321 | asp23 | Alkaline shock protein 23 (ASP23) | 0.096 | 1.805 |
| SA0452 | 2.612 | veg | VEG protein homologue | 0.047 | 1.799 | |
| SA1949 | 2.595 | Lytic regulatory protein truncated with Tn554 | 0.087 | 3.357 | ||
| Hypothetical proteins | SA2268 | 8.134 | Hypothetical protein | 0.269 | 3.302 | |
| SA0588 | 6.614 | Conserved protein | 0.322 | 4.876 | ||
| SA1985 | 5.151 | Hypothetical protein | 0.13 | 2.516 | ||
| SA0271 | 4.887 | Conserved protein | 0.123 | 2.519 | ||
| SA1986 | 4.65 | Hypothetical protein | 0.238 | 5.121 | ||
| SA0227 | 3.765 | Conserved protein | 0.091 | 2.418 | ||
| SA0929 | 3.547 | Conserved protein | 0.195 | 5.507 | ||
| SA1403 | 3.155 | Conserved protein | 0.229 | 7.248 | ||
| SA0412 | 2.991 | Conserved protein | 0.054 | 1.793 | ||
| SA2133 | 2.945 | Conserved protein | 0.182 | 6.177 | ||
| SA1912 | 2.771 | Hypothetical protein | 0.197 | 7.125 | ||
| SA0371 | 2.707 | Hypothetical protein | 0.231 | 8.524 | ||
| SA1925 | 2.702 | Conserved protein | 0.112 | 4.138 | ||
| SA1402 | 2.69 | Conserved protein | 0.056 | 2.068 | ||
| SA0292 | 2.58 | Hypothetical protein | 0.131 | 5.084 | ||
| SA1916 | 2.532 | Conserved protein | 0.12 | 4.745 | ||
| SA0609 | 2.5 | Conserved protein | 0.059 | 2.378 |
Acknowledgments
This work was supported by a grant from the Friedrich-Ebert-Stiftung to A. Resch, by the BMBF Kompetenznetz PathoGenoMik (grant 031U213B), by the DFG “Graduate College Infection Biology,” and by the Landesstiftung Baden-Württemberg.
Special thanks go to Karen Brune for editing the manuscript.
REFERENCES
- 1.Ahmed, K., and M. N. Jones. 2003. The effect of shear on the desorption of liposomes adsorbed to bacterial biofilms. J. Liposome Res. 13:187-197. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, L. K., and R. P. Gaynes. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect. Dis. Clin N. Am. 11:245-255. [DOI] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, M. R., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-780. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, G. D., L. Baldassarri, and W. A. Simpson. 1994. Colonization of medical devices by coagulase-negative staphylococci, p. 45-78. In A. L. Bisno and F. A. Waldvogel (ed.), Infections associated with indwelling medical devices. ASM Press, Washington, D.C.
- 7.Christensen, G. D., A. L. Bisno, J. T. Parisi, M. G. McLaughlin, G. M. Hester, and R. W. Luther. 1982. Nosocomial septicemia due to multiple-resistant Staphylococcus epidermidis. Ann. Intern. Med. 96:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1983. Experimental foreign body infections in mice challenged with slime-producing Staphylococcus epidermidis. Infect. Immun. 40:407-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella, C., M. A. Tormo, E. Knecht, B. Amorena, I. Lasa, T. J. Foster, and J. R. Penades. 2002. Expression of the biofilm-associated protein interferes with host protein receptors of Staphylococcus aureus and alters the infective process. Infect. Immun. 70:3180-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobinsky, S., and D. Mack. 2001. Efficient RNA isolation method for analysis of transcription in sessile Staphylococcus epidermidis biofilm cultures. Methods Enzymol. 336:255-262. [DOI] [PubMed] [Google Scholar]
- 15.Dubin, G. 2002. Extracellular proteases of Staphylococcus spp. Biol. Chem. 383:1075-1086. [DOI] [PubMed] [Google Scholar]
- 16.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Zoeiby, A., F. Sanschagrin, P. C. Havugimana, A. Garnier, and R. C. Levesque. 2001. In vitro reconstruction of the biosynthetic pathway of peptidoglycan cytoplasmic precursor in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 201:229-235. [DOI] [PubMed] [Google Scholar]
- 18.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 19.Gerke, C., A. Kraft, R. Süssmuth, O. Schweitzer, and F. Götz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, P., P. J. Collier, and M. R. Brown. 1990. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmann, D. A., and G. B. Pier. 1993. Pathogenesis of infections related to intravascular catheterization. Clin. Microbiol. Rev. 6:176-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 23.Gray, E. D., G. Peters, M. Verstegen, and W. E. Regelmann. 1984. Effect of extracellular slime substance from Staphylococcus epidermidis on the human cellular immune response. Lancet i:365-367. [DOI] [PubMed] [Google Scholar]
- 24.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 25.Heilmann, C., and F. Gotz. 1998. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl. Bakteriol. 287:69-83. [DOI] [PubMed] [Google Scholar]
- 26.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 27.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 28.Iordanescu, S. 1976. Temperature-sensitive mutant of a tetracycline resistance staphylococcal plasmid. Arch. Roum. Pathol. Exp. Microbiol. 35:257-264. [PubMed] [Google Scholar]
- 29.Jefferson, K. K., S. E. Cramton, F. Götz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 30.Jesaitis, A. J., M. J. Franklin, D. Berglund, M. Sasaki, C. I. Lord, J. B. Bleazard, J. E. Duffy, H. Beyenal, and Z. Lewandowski. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171:4329-4339. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroda, M., T. Ohta, and H. Hayashi. 1995. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 207:978-984. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 35.Lang, S., M. A. Livesley, P. A. Lambert, W. A. Littler, and T. S. Elliott. 2000. Identification of a novel antigen from Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 29:213-220. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locci, R., G. Peters, and G. Pulverer. 1981a. Microbial colonization of prosthetic devices. III. Adhesion of staphylococci to lumina of intravenous catheters perfused with bacterial suspensions. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. B 173:300-307. [PubMed] [Google Scholar]
- 38.Ludwicka, A., R. Locci, B. Jansen, G. Peters, and G. Pulverer. 1983. Microbial colonization of prosthetic devices. V. Attachment of coagulase-negative staphylococci and “slime”-production on chemically pure synthetic polymers. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. B 177:527-532. [PubMed] [Google Scholar]
- 39.Ludwicka, A., G. Uhlenbruck, G. Peters, P. N. Seng, E. D. Gray, J. Jeljaszewicz, and G. Pulverer. 1984. Investigation on extracellular slime substance produced by Staphylococcus epidermidis. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 258:256-267. [DOI] [PubMed] [Google Scholar]
- 40.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34-39. [DOI] [PubMed] [Google Scholar]
- 43.Marshall, J. H., and G. J. Wilmoth. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Götz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 45.Peters, G., R. Locci, and G. Pulverer. 1982. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J. Infect. Dis. 146:479-482. [DOI] [PubMed] [Google Scholar]
- 46.Peters, G., and G. Pulverer. 1984. Pathogenesis and management of Staphylococcus epidermidis ‘plastic’ foreign body infections. J. Antimicrob. Chemother 14(Suppl. D):67-71. [DOI] [PubMed] [Google Scholar]
- 47.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 48.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 49.Stewart, P. S., F. Roe, J. Rayner, J. G. Elkins, Z. Lewandowski, U. A. Ochsner, and D. J. Hassett. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 66:836-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tojo, M., N. Yamashita, D. A. Goldmann, and G. B. Pier. 1988. Isolation and characterization of a capsular polysaccharide adhesin from Staphylococcus epidermidis. J. Infect. Dis. 157:713-722. [DOI] [PubMed] [Google Scholar]
- 51.von Eiff, C., C. Heilmann, M. Herrmann, and G. Peters. 1999. Basic aspects of the pathogenesis of staphylococcal polymer-associated infections. Infection 27(Suppl. 1):S7-S10. [DOI] [PubMed] [Google Scholar]
- 52.Wieland, B., C. Feil, E. Gloria-Maercker, G. Thumm, M. Lechner, J. M. Bravo, K. Poralla, and F. Götz. 1994. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176:7719-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lößner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]