Abstract
Rhizoremediation of organic chemicals requires high-level expression of biodegradation genes in bacterial strains that are excellent rhizosphere colonizers. Pseudomonas fluorescens F113 is a biocontrol strain that was shown to be an excellent colonizer of numerous plant rhizospheres, including alfalfa. Although a derivative of F113 expressing polychlorinated biphenyl (PCB) biodegradation genes (F113pcb) has been reported previously, this strain shows a low level of bph gene expression, limiting its rhizoremediation potential. Here, a high-level expression system was designed from rhizobial nod gene regulatory relays. Nod promoters were tested in strain F113 by using β-galactosidase transcriptional fusions. This analysis showed that nodbox 4 from Sinorhizobium meliloti has a high level of expression in F113 that is dependent on an intact nodD1 gene. A transcriptional fusion of a nodbox cassette containing the nodD1 gene and nodbox 4 fused to a gfp gene was expressed in the alfalfa rhizosphere. The bph operon from Burkholderia sp. strain LB400 was cloned under the control of the nodbox cassette and was inserted as a single copy into the genome of F113, generating strain F113L::1180. This new genetically modified strain has a high level of BphC activity and grows on biphenyl as a sole carbon and energy source at a growth rate that is more than three times higher than that of F113pcb. Degradation of PCBs 3, 4, 5, 17, and 25 was also much faster in F113L::1180 than in F113pcb. Finally, the modified strain cometabolized PCB congeners present in Delor103 better than strain LB400, the donor of the bph genes used.
Polychlorinated biphenyls (PCBs) have been widely used for numerous industrial applications due to their thermal and chemical stability. Unfortunately, these physical properties, along with low water solubility and high lipophilicity, have also contributed to the persistence of PCBs in the environment (27). Since numerous toxic effects on different organisms have been documented for PCBs, their persistence causes a major environmental problem (35). The use of PCBs has been abandoned in most countries, but in spite of this, PCBs have been identified in almost every component of the global ecosystem (27). Besides bioaccumulation in higher trophic levels of the food chain, PCBs are especially found in soils and sediments near industrial areas (27). Remediation of PCB-contaminated soil has traditionally been carried out by incineration or burial of the soil in secure landfills. As an alternative and less expensive removal strategy, bioremediation by soil bacteria has been extensively investigated during the last few decades, (21, 41). As part of these investigations, several bacterial strains capable of degrading PCB have been isolated from soils and sediments contaminated with PCBs (6, 21, 22).
Aerobic PCB degradation is catalyzed by the enzymes expressed from the bph genes involved in biphenyl degradation. These genes are conserved and have a high degree of homology among different strains, although there are considerable differences in the congener selectivity and activity of the enzymes expressed by the genes. All enzymes expressed by the bph operon convert PCB to chlorobenzoate and a five-carbon aliphatic acid (2-hydroxypenta-2,4-dienoate); the latter can be further degraded to tricarboxylic acid cycle intermediates. Chlorobenzoates can accumulate and deactivate the ring fission enzymes involved in PCB cometabolism; however, a Pseudomonas fluorescens strain engineered to degrade such compounds (12) shows enhanced removal of these products in the rhizosphere. This upper pathway of PCB degradation generally proceeds by initial introduction of molecular oxygen at the 2,3 position, followed by 1,2-meta cleavage of the molecule. Both reactions occur by dioxygenase attack, normally at the least chlorinated ring (29).
One of the most widely used bacteria in PCB degradation studies is Burkholderia sp. strain LB400 (formerly Pseudomonas sp. strain LB400), a strain originally isolated from PCB-contaminated soil (8). The bph operon in this strain encodes a complete metabolic pathway which allows biphenyl to be used as the sole carbon and energy source and the organism to cometabolically degrade PCBs (21, 22). However, after introduction of such bacteria into soil, a decline in survival and degradation activity has been observed, which has been overcome by periodic reinoculation or by imposing positive selection through continuous addition of a specific substrate, such as biphenyl (5). Since neither of these solutions is realistic for in situ bioremediation, an alternative solution is to insert bph genes into a host that is known to have high survival capability in specific soil compartments. As a natural environment for in situ bioremediation in soil, the rhizosphere seems to be a promising compartment. Not only is the rhizosphere a hot spot for microbial activity (43), but it also enhances the dispersal of the introduced strain in the soil. Thus, expanding the degradation capacities of rhizosphere-competent bacteria might be a useful strategy for generating strains suitable for plant-assisted in situ bioremediation of soil (9, 44), also termed rhizoremediation.
Pseudomonas fluorescens F113 has been isolated from the sugar beet rhizosphere (37) and is an excellent colonizer of several plant rhizospheres, such as those of sugar beet (13, 37), tomato (38), pea (31), and alfalfa (42). A derivative of this strain with potential for rhizoremediation, F113pcb, has been constructed (9). F113pcb contains the bph operon from strain LB400 under the control of its own promoter, which facilitates growth on biphenyl as the sole carbon and energy source. This strain is not affected in sugar beet colonization or biocontrol abilities (9). However, the level of expression of the bph genes is lower than that in the parental strain, which limits the ability of F113pcb to grow on biphenyl and therefore it ability to degrade PCBs.
A potential way of increasing the biphenyl-degrading activity is to increase the transcription rate of the genes by changing the promoter regions. In this work we tested heterologous rhizobial nodulation promoters (nod boxes) from Sinorhizobium meliloti and its regulatory systems to drive the expression of the bph operon in P. fluorescens F113 derivatives. The nodulation (nod) genes of S. meliloti encode the enzymes required for the synthesis of the lipochitin nodulation factors when they are induced by flavonoids present in alfalfa root exudates (33). The use of one such system allowed us to construct an improved rhizoremediation strain.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. fluorescens strains were routinely grown in low-iron sucrose asparagine medium (36) at 28°C. This medium is semiselective for P. fluorescens and demonstrates production of fluorescence siderophores. To test growth on biphenyl, the strains were grown at 28°C in minimal medium (MM) (9) supplemented with biphenyl crystals (0.5 g/liter). Growth parameters were calculated from triplicate cultures, and optical density at 600 nm was determined. For growth on solid medium, biphenyl crystals were added to a petri dish lid. Escherichia coli strains were grown at 37°C in Luria-Bertani medium (7). Antibiotics were added at the following concentrations when required: 100 μg of rifampin per ml, 70 μg of tetracycline per ml (10 μg/ml for E. coli), 20 μg of kanamycin per ml, and 100 μg of ampicillin per ml.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| P. fluorescens strains | ||
| F113 | Wild-type strain isolated from sugar beet rhizosphere | 37 |
| F113rif | Rifampin-resistant derivative of F113 | 17 |
| F113 LacZY | F113rif::lacZY | 17 |
| F113pcb | F113rif::bph | 9 |
| F113L::1180 | F113LacZY::nodD-nodbox 4bph | This study |
| Plasmids | ||
| pMP220 | IncP Tetr, promoterless lacZ | 39 |
| pHC60 | IncP Tetr, gfp under the control of a constitutive lac promoter | 10 |
| pMP92 | IncP Tetr | 39 |
| pGFP92 | IncP Tetr, promoterless gfp 65S/T | This study |
| pRK2013 | Helper plasmid, Tra+ Kanr | 18 |
| pDDPCB | pUT derivative, miniTn5::bph, Ampr | 16 |
| pCR2.1-TOPO | PCR cloning vector, Ampr Kanr | Invitrogen |
| pBluescript SK+ | Cloning vector, Ampr | Stratagene |
| pBG1075 | pMP220 nodbox 4::lacZ | This study |
| pBG1079 | pMP220 nodD1::lacZ | This study |
| pBG1106 | pMP220 nodD1-nodbox 4::lacZ | This study |
| pBG1093 | pMP220 nodD1-nodbox 1::lacZ | This study |
| pBG1094 | pMP220 nodbox 1::lacZ | This study |
| pBG1110 | pBG1106 ΔnodD1 | This study |
| pBG1157 | pGFP92 nodD1-nodbox 4::gfp | This study |
| pBG1180 | pDDPCB miniTn5::nodD1-nodbox 4::bph | This study |
| pBG1197 | PMP92 nodD1-nodbox 4::bph | This study |
Plant assays.
Alfalfa plants were grown in Leonard jars using perlite as the solid substrate and in soil microcosms as previously described (42). Surface-sterilized seeds were germinated at 4°C, and seedlings were transferred to the Leonard jars or a microcosm system and inoculated with 108 bacterial cells/seedling (three replicates, experiment repeated twice). The plants were grown for 3 weeks in a growth cabinet with a photoperiod consisting of 16 of light and 8 h of darkness and day and night temperatures of 25 and 16°C, respectively. Root colonization ability was tested by colony counting on sucrose asparagine medium and by microscopic inspection as described below.
DNA manipulation.
Standard techniques for cloning and subcloning procedures, plasmid preparation, and agarose gel electrophoresis were used. PCR was performed using the Tth enzyme from Biotools (Madrid, Spain) and standard conditions. The sequences of the primers used in PCR experiments are available on request. Southern blot hybridizations were performed with a nonradioactive kit, using a chemiluminescence method to detect hybridization (Roche Diagnostics, Barcelona, Spain). DNA sequencing was done by the chain termination method using the DyeDeoxy terminator cycle sequencing kit protocol (Applied Biosystems, Madrid, Spain). Sequence analysis was performed using software from the Genetics Computer Group (Madison, Wis.).
Enzymatic activities.
β-Galactosidase activity was determined as described by Miller (28). BphC (2,3-dihydroxybiphenyl-1,2-dioxigenase) activity was determined spectrophotometrically at 434 nm by measuring the appearance of the yellow product 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid from the substrate 2,3-dihydroxybiphenyl (9).
Visualization and colonization dynamics of gfp-marked Pseudomonas strains on Medicago sativa roots.
Strains to be monitored by gfp visualization techniques were tagged with a stable, constitutively produced green fluorescent protein gene in plasmid pHC60 (10) by triparental conjugation. Cultures were examined prior to rhizosphere inoculation by preparing a wet mount of each culture (from a fresh selective plate) and were visualized under blue light (395 nm) using a Nikon E400 epifluorescence microscope equipped with a 100-W mercury short-arc photooptic lamp. The Lucia imaging software (version 4.6) was used to capture and process microscopic images.
An inoculated plant was removed from the growth system (after 13 or 14 days), and any excess soil or perlite was removed by brushing. The roots were stored in distilled water to prevent drying out between sampling and analysis. A root was placed on a microscope slide, and a few drops of distilled water were added. A coverslip was placed over the root, and the root was crushed gently by covering the slide and coverslip with clean absorbent paper and applying gentle pressure. This was done to allow better focusing on the root. The intact root was examined from the top of the root to the root tip at a magnification of ×10 using epifluorescence microscopy. Any colonization by gfp-marked strains was noted before the root was examined at a magnification of ×40 (and a magnification of ×100 in the case of the plants propagated in the nonsterile soil system). The same sample processing procedure was used for confocal laser scanning microscopy, and visualization was performed with Leica TCS NT Scanner equipment.
Resting cell assays to measure PCB degradation.
The cells used for the inoculum in the degradation experiments were grown in MM containing yeast extract and biphenyl (100 mg · liter−1), until they were in the exponential growth phase. The cell suspensions were then centrifuged at 9,500 × g for 5 min and washed twice in MM. The protein was determined using a Bio-Rad DC protein assay kit II according to the manufacturer's instructions, with bovine serum albumin (Sigma) as the standard. Degradation assays were carried out in 8-ml glass vials with Teflon screw caps containing MM (1 ml), the appropriate PCB (see Table 4) in acetone (6), and inoculum to give an optical density of 3.6. Autoclaved cells (121°C for 20 min) were used as controls. All the vials were incubated at 20°C at a 45° angle on a rotary shaker at 250 rpm. At each sampling time, three vials were sacrificed. Degradation was stopped by adding 0.05 ml of 2 M HCl, and residual PCB was extracted by adding 4.0 ml of n-hexane to each vial and shaking the vials horizontally (IKA-Schüttler model MTS 4) at 900 rpm for 5 min. A sample from the n-hexane layer was then transferred to a 2-ml high-performance liquid chromatography (HPLC) vial with a Teflon-lined cap, and the rest of the n-hexane layer was discarded. To confirm complete extraction, a second hexane extraction was performed on the water phase left in the vial by repeating the procedure described above. The extracts were stored at −30°C until they were analyzed.
TABLE 4.
PCB degradation by engineered strains of P. fluorescens F113 determined as zero-order rates during the first 60 min of resting cell assays
| Strain | Degradation of the following substrates (nmol mg of protein−1 min−1):
|
||||
|---|---|---|---|---|---|
| PCB 3 (4-chlorobiphenyl) | PCB 4 (2,2′-dichlorobiphenyl) | PCB 5 (2,3-dichlorobiphenyl) | PCB 17 (2,2′,4-trichlorobiphenyl) | PCB 25 (2,3′,4-trichlorobiphenyl) | |
| F113pcb | 0.38 | 0.14 | 0.24 | 0.12 | 0.01 |
| F113L::1180 | 1.48 | 0.22 | 0.55 | 0.23 | 0.09 |
The PCB in the extracts was determined using HPLC on a Hewlett-Packard series 1100 HPLC with a reverse-phase Zorbax SB C18 column (4.6 by 150 mm). Twenty microliters of extract was injected. The compounds were eluted using the following solvent program: 18% methanol at a flow rate of 1 ml · min−1 from 0 to 2.5 min, followed by a linear gradient for 30 s to reach 82% methanol at a flow rate of 1.5 ml · min−1 for the next 5.5 min and then a linear gradient for 30 s to regain 18% methanol at a flow rate of 1 ml · min−1 from 9 to 15 min. Sodium acetate (0.05 M) was added to the eluant as a buffer (pH 8.3). Peaks were identified and quantified from UV spectra at wavelengths from 200 to 300 nm with a diode array detector, using reference substances at known concentrations.
Cometabolism of PCBs.
A standard PCB mixture, Delor 103 (Chemko, Strázske, former Czechoslovakia), was used in biodegradation experiments. This mixture was commercially produced until 1984, and it contains 59 individual PCB congeners substituted with three to five chlorines per biphenyl molecule (Table 2). Bacteria (5 ml) pregrown on MM with biphenyl was added to 300-ml Erlenmeyer flasks with 50 ml of MM, biphenyl (0.5 g/liter), and Delor 103 (50 mg/liter). Cells were incubated for 14 days at 28°C in a rotary shaker. Abiotic samples were incubated and used as a control.
TABLE 2.
PCB congeners present in Delor 103 and congener content of each chromatographic peak
| Chromatographic peak | PCB no. (IUPAC nomenclature) | Chlorine substitutions |
|---|---|---|
| 1 | 5, 8 | 2,3; 2,4′ |
| 2 | 15, 18 | 4,4′; 2,2′,5 |
| 3 | 17 | 2,2′,4 |
| 4 | 16, 32 | 2,2′,3; 2,4′,6 |
| 5 | 26 | 2,3′,5 |
| 6 | 31 | 2,4′,5 |
| 7 | 28 | 2,4,4′ |
| 8 | 20,33,53 | 2,3,3′; 2′,3,4′; 2,2′,5,6′ |
| 9 | 45 | 2,2′,3,6 |
| 10 | 52, 69 | 2,2′,5,5′; 2,3′,4,6 |
| 11 | 49 | 2,2′,4,5′ |
| 12 | 47, 75 | 2,2′,4,4′; 2,4,4′,6 |
| 13 | 48 | 2,2′,4,5 |
| 14 | 44 | 2,2′,3,5′ |
| 15 | 37,42,59 | 3,4,4′; 2,2′,3,4′; 2,3,3′,6 |
| 16 | 41, 64 | 2,2′,3,4; 2,3,4′,6 |
| 17 | 96 | 2,2′,3,6,6′ |
| 18 | 74 | 2,4,4′,5 |
| 19 | 70 | 2,3′,4′,5 |
| 20 | 66, 88, 95 | 2,3′,4,4′; 2,2′,3,4,6; 2,2′,3,5′,6 |
| 21 | 101 | 2,2′,4,5,5′ |
| 22 | 77, 110 | 3,3′,4,4′; 2,3,3′,4′,6 |
General PCB conversion was evaluated on the basis of the decrease in the initial PCB content (34). At the end of each incubation, after homogenization and subsequent sonication, the whole contents of each flask were extracted with 10 ml of hexane at 20°C on a rotary shaker for 2 h. Following phase separation, the upper, hexane layer was sampled for gas chromatography analysis on a Hewlett-Packard 5890 gas chromatograph with an electron capture detector. A fused silica capillary column (length, 30 m; inside diameter, 0.20 mm) coated with 0.25-μm immobilized phase SE-54 was used with nitrogen as the carrier gas (flow rate, 1 ml/min). The temperature program was 50°C for 1 min, followed by an increase at a rate of 25°C/min until the temperature was 280°C and then isothermal incubation. The injection volume was 2 μl. Using this method, 22 peaks, containing an identified mixture of congeners (Table 2), could be identified. This represented 35 identified congeners from a total of 59 congeners and more than 95% of the PCB content in Delor 103 (3). In each experiment, controls without cells and controls with heat-killed cells were included to eliminate the effects of evaporation and sorption to glass and biomass and to establish that the observed changes in the content of congeners were dependent exclusively on the biological activity of living cells, thus eliminating the abiotic factors involved (15). The accuracy of the results obtained was within 15%, as determined with standards. Results were calculated from the residual amounts of each congener peak of the sample by comparison to the respective peaks of the controls.
RESULTS
Analysis of nod promoter expression in P. fluorescens F113.
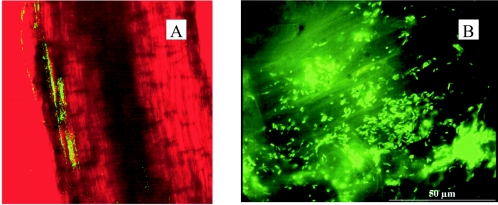
Primers designed from the published sequences of nod promoters (1, 2) were used to amplify these regions from the genome of S. meliloti Rm2011. The nodABC promoter (nodbox 1) was chosen because it is the strongest nod promoter in S. meliloti. The nodM promoter (nodbox 4) was also selected because it has been shown to be expressed in E. coli (2). The nodD1 gene from strain Rm2011, encoding a transcriptional activator of nod promoters, was also amplified. PCR products were cloned in the pCR2.1-TOPO vector, confirmed by sequencing, and subcloned to drive lacZ expression in derivatives of pMP220 (39). The constructs were mobilized into P. fluorescens F113rif by triparental mating, and β-galactosidase activity was determined in exponentially growing cultures in the presence and absence of the nod promoter inducer luteolin. Table 3 shows the expression of these constructs in the P. fluorescens F113 background. We observed that unexpectedly, expression of all constructs was independent of the inducer luteolin. The nodbox 1 promoter did not show activity in F113, and therefore it cannot be used as an expression system in this background. In contrast, the nodbox 4 promoter showed high activity in F113, and the activity was even higher than that in S. meliloti (data not shown). Table 3 also shows that nodbox 4 activity in F113 is dependent on the presence of the nodD1 gene. The presence of this gene increased nodbox 4 activity fivefold, and deletion of the coding region of this gene decreased activity to background levels, even in the presence of the nodD1 promoter which is active in F113 (Table 3). These results prove that the nodbox 4 promoter is highly expressed in P. fluorescens and that its expression depends on NodD1, but it is independent of externally supplied nod inducers. To study the expression of nodbox 4 in strain F113 in the alfalfa rhizosphere, fusions of this promoter to the gfp gene were constructed. The vector used was pGFP92, a derivative of pMP92 in which the gfp-S65T allele was cloned downstream of the multiple cloning site. A DNA fragment containing the nodD1 gene and nodbox 4 was subsequently cloned in the multiple cloning site of pGFP92 to generate pBG1157, which was introduced into F113rif by triparental mating. F113rif(pBG1157) was inoculated onto alfalfa seedlings, and the roots were visualized 2 weeks later by confocal laser scanning microscopy. As shown in Fig. 1A, microcolonies and individual cells of F113rif(pBG1157) produced green fluorescent protein, indicating that nodbox 4 is active in P. fluorescens F113 in the rhizosphere environment.
TABLE 3.
Activity of S. meliloti nodbox promoters in P. fluorescens F113
| Plasmid | Construct | β-Galactosidase activity (Miller units) ona:
|
|
|---|---|---|---|
| Sucrose asparagine medium | Sucrose asparagine medium + luteolin | ||
| None | None | 5 ± 1 | 5 ± 0.8 |
| pMP220 | Promoterless lacZ | 25 ± 2 | 25 ± 3 |
| pBG1079 | nodD1::lacZ | 88 ± 6 | 100 ± 9 |
| pBG1094 | nodbox 1::lacZ | 16 ± 2 | 12 ± 1 |
| pBG1093 | nodD1-nodbox 1::lacZ | 24 ± 2 | 19 ± 1.5 |
| pBG1075 | nodbox 4::lacZ | 343 ± 27 | 373 ± 29 |
| pBG1106 | nodD1-nodbox 4::lacZ | 1,730 ± 135 | 1,663 ± 128 |
| pBG1110 | pBG1106 ΔnodD1 | 142 ± 8 | 134 ± 10 |
Activity of exponentially growing cells. The values are arithmetic means ± standard deviations of two independent experiments performed in triplicate.
FIG. 1.
(A) Confocal laser scanning microscopy image of P. fluorescens F113rif(pBG1157) in the alfalfa rhizosphere in a gnotobiotic system (Leonard jar). Expression of the gfp gene under the nodbox system resulted in green fluorescence visible in individual cells and microcolonies. (B) Epifluorescence image of P. fluorescens F113L::1180(pHC60) in the alfalfa rhizosphere in a PCB-spiked soil microcosm. Expression of the gfp gene from a constitutive lac promoter resulted in green fluorescence observed in bacteria colonizing the root surface (toward the root tip) 14 days after inoculation.
Expression of the bph genes under nodbox 4 control in P. fluorescens F113.
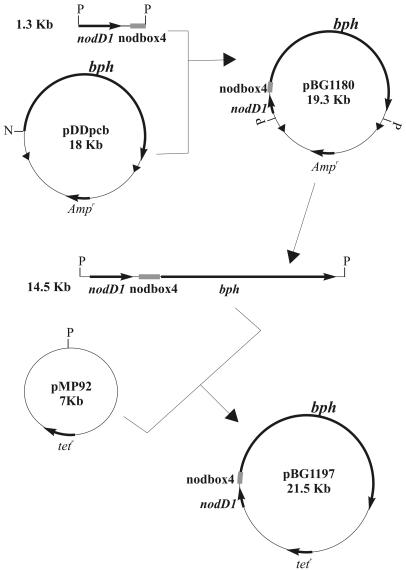
The bph genes from Burkholderia sp. strain LB400 encode a complete pathway for the metabolism of biphenyl (23, 24, 29) and allow the cometabolism of certain PCB congeners (29). To situate bph genes under the control of the nodbox 4 expression system, the DNA fragment containing the nodD1 gene and the nodbox 4 promoter was cloned into the unique NsiI site in plasmid pDDPCB (16) (Fig. 2) to generate pBG1180, in which the nodbox 4 system is located upstream of bphA and in the sense of transcription of the operon. Cloning into this site disrupted ORF0, a putative regulator of the bph operon. The ca. 14-kb fragment from pBG1180 containing the nodbox 4 system and the whole bph operon was then subcloned into pMP92 to generate pBG1197 and was mobilized into F113rif by conjugation. The bph activity was measured by determining the activity of the BphC enzyme in exponentially growing cultures. F113(pBG1197) produced 4,910 ± 230 BphC units, compared to the 1,490 ± 80 BphC units produced by the F113pcb strain. These results show that the activity of the nodbox 4 system is more than three times higher than the activity of the bph promoter in P. fluorescens F113.
FIG. 2.
Construction of nod system-bph fusions introduced into derivatives of P. fluorescens F113 in a replicative vector (pBG1197) or through transposition to a genomic location (pBG1180). The solid triangles represent mini-Tn5 inverted repeats. N, NsiI; P, PstI. Only relevant restriction sites are shown. For a description of the plasmids see Table 1.
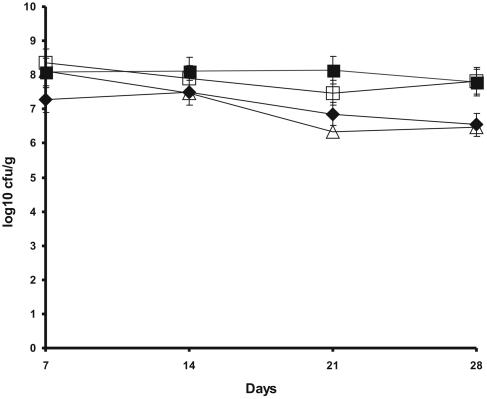
In order to integrate the nodbox-bph construct in the P. fluorescens genome, plasmid pBG1180 (Fig. 2), which contains the construct in a mini Tn5 transposon, was mobilized into F113LacZY (17), a derivative of F113 that contains the lacZY genes integrated via the same transposon. Three transconjugants were obtained, and PCR analysis showed that the three isolates contained the correct insertion (data not shown). Southern blot analysis using an internal fragment of the bphC gene showed that one of the transconjugants, F113L::1180, contained a single insertion, and this transconjugant was selected for further analysis. F113L::1180 was tested for antibiotic resistance (pipericillin resistance is encoded by the vector used in construction), carbon source utilization (using BIOLOG GN plates), and biocontrol ability (as described by Brazil and coworkers [9]), and it was identical to parental strain F113LacZY, indicating that important traits were not inactivated by the transposon insertion (data not shown). We also compared the two strains for rhizosphere colonization in a gnotobiotic system and a soil microcosm (42). Figure 3 shows the colonization dynamics of F113LacZY and F113L::1180 in the alfalfa rhizosphere in the soil microcosm. Although a small decline in rhizosphere counts was observed for F113L::1180 after 21 days, the populations at 28 days were not significantly different (P < 0.001), indicating that the two strains colonized the rhizosphere equally and persisted at similar densities. Figure 1B shows the colonization of the distal part of the root of an alfalfa plant 14 days after inoculation growing in a nonsterile soil microcosm amended with PCB 3 by the gfp-tagged F113L::1180(pHC60). These results clearly show the colonization ability of the genetically modified strain even with PCB contamination.
FIG. 3.
Colonization of M. sativa rhizosphere by P. fluorescens strains. Solid symbols, inoculation with P. fluorescens F113LacZY; open symbols, inoculation with P. fluorescens F113L::1180. Symbols: ♦, F113LacZY counts per gram of root; ▪, total viable counts per gram of soil; Δ, F113L::1180 counts per gram of root; □, total viable counts per gram of soil. All weights are fresh weights. Arithmetic means and standard deviations for three replicates are shown.
The BphC activity of F113L::1180 was 4,980 ± 320 units, indicating that the high activity provided by the nodbox 4 expression system is intrinsic and not related to the copy number of the construct.
Growth of the genetically modified strains on biphenyl.
In order to compare the growth of the modified strains on biphenyl, we grew F113pcb, F113(pBG1197), and F113L::1180 on minimal medium with biphenyl as the sole carbon and energy source. The strains containing the nodbox expression system had higher growth rates (0.154 h−1 for F113L::1180 and 0.163 h−1 for F113pBG1197) than F113pcb (0.047 h−1), which resulted in a more-than-threefold reduction in the doubling time. No significant differences were observed between the strains containing the nodbox4-bph fusion on a plasmid and as a single genomic insertion. F113(pBG1197) and F113L::1180 grew much faster than F113pcb and reached the stationary phase at 40 h, while F113pcb was still in the mid-exponential phase. These results show that the higher expression of bph genes and BphC activity obtained under nodbox 4 control resulted in a higher growth rate on biphenyl. They also show that the growth of the strains was not carbon limited and that growth could be increased by increasing the bph expression level.
Degradation of PCBs by P. fluorescens F113L::1180.
To investigate the ability of the genetically modified strain to degrade PCBs, we used resting cell assays for PCB 3, PCB 4, PCB 5, PCB 17, and PCB 25. As expected, no PCB degradation was observed with F113 or abiotic controls. One monochlorinated, two dichlorinated, and two trichlorinated PCB congeners were tested (Table 4). All five of these congeners were degraded by both constructs, but the rates were approximately two to five times higher with strain F113L::1180 than with strain F113pcb.
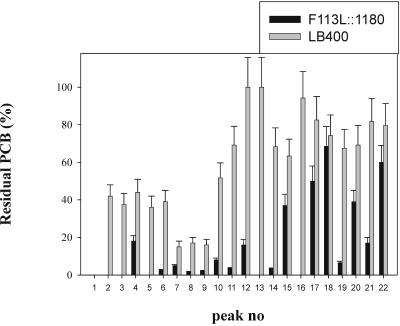
Cometabolism of PCBs was investigated by comparing the Delor 103 biodegradation capability of Burkholderia sp. strain LB400, the source of the bph operon, with that of engineered strain F113L::1180 when the organisms were growing on biphenyl. As shown in Fig. 4, all the congeners degraded by LB400 were also degraded by F113L::1180. Furthermore, the modified strain degraded most congeners to a greater extent.
FIG. 4.
Cometabolism of PCB congeners from Delor 103 by P. fluorescens F113L::1180 and Burkholderia sp. strain LB400. Cells were grown for 14 days at 28°C in minimal medium supplemented with 0.5 g/liter biphenyl and 50 mg/liter Delor 103. After incubation, hexane extracts were analyzed using gas chromatography. The bars indicate the percentages of PCB congeners remaining after incubation, normalized by comparison to abiotic controls. Arithmetic means for three independent experiments analyzed in duplicate and standard errors are shown. The peak numbers refer to congeners, as shown in Table 2.
DISCUSSION
Rhizoremediation (i.e., the breakdown of an organic contaminant in soil through microbial activity that is enhanced by the presence of the root zone) is one of the emerging technologies in soil bioremediation (40, 44). One of the main problems associated with this technology is that usually biodegradation strains are not good rhizosphere colonizers, a condition that has been shown to be essential for biotechnological applications of plant/bacterium systems (11, 26). The fact that multiple bacterial traits are necessary for efficient rhizosphere colonization (26) makes it impractical to improve the colonization ability of good biodegradation strains. An alternative is to introduce biodegradation genes into bacteria that have been shown to be able to efficiently colonize the plant rhizosphere. The strain used in this work, P. fluorescens F113, was isolated from the sugar beet rhizosphere (37) and has been shown to be a good colonizer of the rhizospheres of tomato (38), pea (31), alfalfa (42), grass (Dowling, unpublished results), and willow (Karlson, unpublished results). It has also been shown that F113 does not interfere with beneficial microorganisms, such as the mycorrizal fungus Glomus mosseae (4) or the alfalfa symbiont S. meliloti (42). A derivative of this strain carrying the bph genes that allow PCB degradation has been constructed previously (9), but its degradation activity was low.
A possible strategy to improve degradation activity is to increase the transcriptional expression of the genes coding for the biodegradation enzymes by promoter replacement. This strategy has been successfully employed, and strains with enhanced biphenyl and PCB degradation capability have been obtained through promoter replacement by homologous recombination (32). However, these strains contain antibiotic resistance genes from the cloning procedures and therefore are not suitable for in situ technologies. We tested the nodulation (nod) promoters from S. meliloti to drive gene expression in P. fluorescens. The results for β-galactosidase fusions showed a different behavior for each of the promoters tested. The promoter of the nodD1 gene, which encodes a nodulation transcriptional activator, is expressed constitutively in S. meliloti (19) and E. coli (2). We observed the same expression pattern in P. fluorescens (Table 2), although the expression level is lower than that in S. meliloti but higher than that in E. coli (data not shown). The nodbox 1 promoter, which in S. meliloti depends on NodD1 and a flavonoid inducer (30, 33), is not functional in P. fluorescens under any of the conditions tested. It is likely that this promoter is not recognized by the P. fluorescens transcription system. Conversely, the nodbox 4 promoter, which in S. meliloti has the same expression requirements as nodbox 1 (2), is expressed in P. fluorescens requiring NodD1 but is expressed independently of flavonoid inducers. As NodD1 binds to nodboxes independently of the presence of the inducer (20), it is possible that this complex is enough to initiate transcription in P. fluorescens or that a molecule present in the bacterial cytoplasm could act as an autoinducer. The level of transcription of nodbox 4 in P. fluorescens in the presence of NodD1 is high and therefore useful for overexpressing foreign genes. The use of such heterologous promoters also reduces the possibility of strain variation by homologous recombination and allows uncoupling of gene expression and carbon metabolism, overcoming catabolic repression of the promoter.
In order to visualize gene expression directly in the rhizosphere, we constructed a vector (pGFP92) derived from pMP92 (39) that contains a multiple cloning site upstream of a promoterless gfp gene. This plasmid allows in situ inspection of promoter activity. The use of this vector allowed us to show that the nodD1-nodbox 4 expression cassette (nod cassette) is functional in the plant rhizosphere, and therefore, it can be used to drive the expression of genes for rhizoremediation. We cloned the bph operon from Burkholderia sp. strain LB400 under the control of the nod cassette. This operon contains the genes encoding a complete catabolic pathway in a single transcriptional unit (16). A P. fluorescens F113 strain containing this construct in a low-copy-number vector showed enhanced enzymatic activity (dihydroxybiphenyl dioxygenase) compared with the previously reported strain F113pcb (9). It was also superior for growth on biphenyl as the sole carbon and energy source, having a higher growth rate and reaching the stationary phase in 40 h, compared with the 80 h required by F113pcb. These results show that increasing the transcription rate of bph genes results in an increase in the biodegradation ability of P. fluorescens and that growth is not limited by carbon availability. To our knowledge, this is the first use of nod promoters to induce foreign genes in a heterologous host.
The bph genes under the control of the nod cassette were also stably integrated into the P. fluorescens genome through a minitransposon system (14). The resulting strain (F113L::1180) does not contain antibiotic resistance genes from the vector and is suitable for in situ applications. Furthermore, the strain construction process (random insertion) generated hybrid DNA sequences that were unique and could be used to design molecular tools to monitor strain performance, such as those designed for strain F113LacZY (25). F113L::1180 showed no differences in rhizosphere colonization, enzymatic activity, or growth on biphenyl compared with the strain that carried the construct in a plasmid, indicating that the higher level of expression of the bph promoters was intrinsic to the expression system and not a copy number effect. F113L::1180 was also superior to F113pcb in PCB degradation, as tested with five different PCB congeners. As only the promoter and not the structural genes were changed with respect to the LB400 strain, it was predicted that a similar range of congeners can be degraded by F113L::1180. We confirmed this hypothesis by showing that F113L::1180 can degrade a wide number of PCB congeners present in a complex mixture, such as Delor 103 (similar to Aroclor 1242). Our results show not only that F113L::1180 can degrade the same congeners as the LB400 strain (the source of the bph genes) but also that it can do this to a greater extent. On the other hand, the modular structure of the nod cassette could be used to enhance biodegradation or rhizoremediation of other xenobiotics by promoter replacement in P. fluorescens.
Acknowledgments
This work was funded by European Commission grants BIO4-CT97-2227 and QLK3-CT-2001-00101. D.N.D. was also funded in part by the HEA PRTLI TSR Strand 3 and SFI programs, Ireland. M. Martín was also funded by the Spanish Ministry for Science and Technology (grant BIO2003-03412) and is the recipient of a Ramón y Cajal contract from MCYT. R.I.O. is the recipient of a predoctoral fellowship from the Basque Country Government.
REFERENCES
- 1.Baev, N., M. Amar, R. Defez, and M. Iaccarino. 1992. The expression of the nodD and nodABC genes of Rhizobium leguminosarum is not regulated in response to combined nitrogen. FEMS Microbiol. Lett. 97:205-208. [Google Scholar]
- 2.Baev, N., G. Endre, G. Petrovics, Z. Banfalvi, and A. Kondorosi. 1991. 6 nodulation genes of nod box locus-4 in Rhizobium meliloti are involved in nodulation signal production—nodM codes for d-glucosamine synthetase. Mol. Gen. Genet. 228:113-124. [DOI] [PubMed] [Google Scholar]
- 3.Ballschmiter, K., and M. Zell. 1980. Analysis of polychlorinated biphenyls (PCB) by glass-capillary gas chromatography—composition of technical Aroclor-PCB and Clophen-PCB mixtures. Fresenius Z. Anal. Chem. 302:20-31. [Google Scholar]
- 4.Barea, J. M., G. Andrade, V. Bianciotto, D. Dowling, S. Lohrke, P. Bonfante, F. O'Gara, and C. Azcon-Aguilar. 1998. Impact on arbuscular mycorrhiza formation of Pseudomonas strains used as inoculants for biocontrol of soil-borne fungal plant pathogens. Appl. Environ. Microbiol. 64:2304-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barriault, D., and M. Sylvestre. 1993. Factors affecting PCB degradation by an implanted bacterial strain in soil microcosms. Can. J. Microbiol. 39:594-602. [DOI] [PubMed] [Google Scholar]
- 6.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 51:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 9.Brazil, G. M., L. Kenefick, M. Callanan, A. Haro, V. De Lorenzo, D. N. Dowling, and F. O'Gara. 1995. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol. 61:1946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin-a-Woeng, T. F. C., G. V. Bloemberg, I. H. M. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 12.Crowley, D. E., M. Brennerova, C. Irwin, V. Brenner, and D. D. Focht. 1996. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol. Ecol. 20:79-89. [Google Scholar]
- 13.Delany, I. R., U. F. Walsh, I. Ross, A. M. Fenton, D. M. Corkery, and F. O'Gara. 2001. Enhancing the biocontrol efficacy of Pseudomonas fluorescens F113 by altering the regulation and production of 2,4-diacetylphloroglucinol—improved pseudomonas biocontrol inoculants. Plant Soil 232:195-205. [Google Scholar]
- 14.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demnerova, K., H. Stiborova, M. B. Leigh, D. Pieper, J. Pazlarova, V. Brenner, T. Macek, and M. Mackova. 2003. Degradation of PCBs and CBs by indigenous bacteria isolated from contaminated soil. Water Air Soil Pollut. 3:47-55. [Google Scholar]
- 16.Dowling, D. N., R. Pipke, and D. F. Dwyer. 1993. A DNA module encoding bph genes for the degradation of polychlorinated-biphenyls (PCBs). FEMS Microbiol. Lett. 113:149-154. [DOI] [PubMed] [Google Scholar]
- 17.Fedi, S., D. Brazil, D. N. Dowling, and F. O'Gara. 1996. Construction of a modified mini-Tn5 LacZY non-antibiotic marker cassette: ecological evaluation of a LacZY marked Pseudomonas strain in the sugarbeet rhizosphere. FEMS Microbiol. Lett. 135:251-257. [DOI] [PubMed] [Google Scholar]
- 18.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher, R. F., H. L. Brierley, J. T. Mulligan, and S. R. Long. 1987. Transcription of Rhizobium meliloti nodulation genes—identification of a nodD transcription initiation site in vitro and in vivo. J. Biol. Chem. 262:6849-6855. [PubMed] [Google Scholar]
- 20.Fisher, R. F., T. T. Egelhoff, J. T. Mulligan, and S. R. Long. 1988. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 2:282-293. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa, K., and F. Matsumura. 1976. Microbial metabolism of polychlorinated biphenyls—studies on relative degradability of polychlorinated biphenyl components by Alcaligenes sp. J. Agric. Food Chem. 24:251-256. [DOI] [PubMed] [Google Scholar]
- 22.Guilbeault, B., M. Sondossi, D. Ahmad, and M. Sylvestre. 1994. Factors affecting the enhancement of PCB degradative ability of soil microbial populations. Int. Biodeterior. Biodegrad. 33:73-91. [Google Scholar]
- 23.Hofer, B., S. Backhaus, and K. N. Timmis. 1994. The biphenyl polychlorinated biphenyl degradation locus (bph) of Pseudomonas sp. LB400 encodes 4 additional metabolic enzymes. Gene 144:9-16. [DOI] [PubMed] [Google Scholar]
- 24.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 25.Hogan, J., O. Sherlock, D. Ryan, C. Whelan, S. Francesconi, R. Rivilla, and D. N. Dowling. 2004. Fluorescence resonance energy transfer (FRET) based molecular detection of a genetically modified PCB degrader in soil. FEMS Microbiol. Lett. 236:349-357. [DOI] [PubMed] [Google Scholar]
- 26.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 27.McFarland, V. A., and J. U. Clarke. 1989. Environmental occurrence, abundance, and potential toxicity of polychlorinated biphenyl congeners: considerations for a congener-specific analysis. Environ. Health Perspect. 81:225-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulligan, J. T., and S. R. Long. 1985. Induction of Rhizobium meliloti nodC expression by plant exudate requires NodD. Proc. Natl. Acad. Sci. USA 82:6609-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naseby, D. C., and J. M. Lynch. 1999. Effects of Pseudomonas fluorescens on ecological functions in the pea rhizosphere are dependent on pH. Microb. Ecol. 37:248-256. [DOI] [PubMed] [Google Scholar]
- 32.Ohtsubo, Y., M. Shimura, M. Delawary, K. Kimbara, M. Takagi, T. Kudo, A. Ohta, and Y. Nagata. 2003. Novel approach to the improvement of biphenyl and polychlorinated biphenyl degradation activity: promoter implantation by homologous recombination. Appl. Environ. Microbiol. 69:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, N. K., J. W. Frost, and S. R. Long. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977-980. [DOI] [PubMed] [Google Scholar]
- 34.Ryslava, E., Z. Krejčík, T. Macek, H. Novakova, and M. Mackova. 2003. Study of PCB biodegradation in real contaminated soil. Fresenius Environ. Bull. 12:296-301. [Google Scholar]
- 35.Safe, S. H. 1994. Polychlorinated-biphenyls (PCBs)—environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit. Rev. Toxicol. 24:87-149. [DOI] [PubMed] [Google Scholar]
- 36.Scher, F. M., and R. Baker. 1982. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 72:1567-1573. [Google Scholar]
- 37.Shanahan, P., D. J. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simons, M., A. J. Vanderbij, I. Brand, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1996. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant-Microbe Interact 9:600-607. [DOI] [PubMed] [Google Scholar]
- 39.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid PRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 40.Trapp, S., and U. Karlson. 2001. Aspects of phytoremediation of organic pollutants. J. Soils Sediments 1:37-43. [Google Scholar]
- 41.Unterman, R., D. L. Bedard, M. J. Brennan, L. H. Bopp, F. J. Mondello, R. E. Brooks, D. P. Mobley, J. B. McDermott, C. C. Schwartz, and D. K. Dietrich. 1988. Biological approaches for polychlorinated biphenyl degradation, p. 253-269. In G. S. Omenn (ed.), Environmental biotechnology: reducing risks from environmental chemicals through biotechnology. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 42.Villacieros, M., B. Power, M. Sanchez-Contreras, J. Lloret, R. I. Oruezabal, M. Martin, F. Fernandez-Pinas, I. Bonilla, C. Whelan, D. N. Dowling, and R. Rivilla. 2003. Colonization behaviour of Pseudomonas fluorescens and Sinorhizobium meliloti in the alfalfa (Medicago sativa) rhizosphere. Plant Soil 251:47-54. [Google Scholar]
- 43.Walton, B. T., and T. A. Anderson. 1990. Microbial degradation of trichloroethylene in the rhizospheres. Potential application to biological remediation of waste sites. Appl. Environ. Microbiol. 56:1012-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yee, D. C., J. A. Maynard, and T. K. Wood. 1998. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monoooxygenase constitutively. Appl. Environ. Microbiol. 64:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]