Abstract
Mass spectrometry is a potentially attractive means of monitoring the survival and efficacy of bioaugmentation agents, such as the dioxin-mineralizing bacterium Sphingomonas wittichii strain RW1. The biotransformation activity of RW1 phenotypes is determined primarily by the presence and concentration of the dioxin dioxygenase, an enzyme initiating the degradation of both dibenzo-p-dioxin and dibenzofuran (DF). We explored the possibility of identifying and characterizing putative cultures of RW1 by peptide mass fingerprinting (PMF) targeting this characteristic phenotypic biomarker. The proteome from cells of RW1—grown on various media in the presence and absence of DF—was partially purified, tryptically digested, and analyzed using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Mascot online database queries allowed statistically significant identification of RW1 in disrupted, digested cells (P < 0.01 to 0.05) and in digested whole-cell extracts (P < 0.00001 to 0.05) containing hundreds of proteins, as determined by two-dimensional gel electrophoresis. Up to 14 peptide ions of the alpha subunit of the dioxin dioxygenase (43% protein coverage) were detected in individual samples. A minimum of 107 DF-grown cells was required to identify dioxin degradation-enabled phenotypes. The technique hinges on the detection of multiple characteristic peptides of a biomarker that can reveal at once the identity and phenotypic properties of the microbial host expressing the protein. The results demonstrate the power of PMF of minimally processed microbial cultures as a sensitive and specific technique for the positive identification and phenotypic characterization of certain microorganisms used in biotechnology and bioremediation.
The gram-negative bacterium Sphingomonas wittichii strain RW1 is of considerable interest to the field of bioremediation (13, 15) because of its unique ability to mineralize dibenzofuran (DF) and dibenzo-p-dioxin (42) and to biotransform a number of chlorinated diaryl ethers (20, 42). Reactions are initiated by the dioxin dioxygenase (1, 5), a key enzyme whose relaxed substrate range invites the application of dioxin dioxygenase-harboring bacteria as bioaugmentation agents facilitating the accelerated in situ bioremediation of dioxin-contaminated environments (15, 41).
Compared to other environmental pollutants, dioxins are particularly difficult to bioremediate (15, 16). Among the various reasons for this are (i) the need for degradative enzymes having broad substrate ranges for turnover of multiple congeners of the large dioxin family, consisting of 75 chlorinated dioxins and 135 chlorinated dibenzofurans (15); (ii) high mammalian toxicity of dioxins, necessitating very low treatment goals, particularly for dibenzo-p-dioxins and DFs carrying chlorine substituents in the lateral 2, 3, 7, and 8 positions (reviewed in reference 15); (iii) limited bioavailability of dioxins due to their strong sorption to soil and sediment (16, 43); and (iv) the pronounced recalcitrance of dioxins to attack by both aerobic and anaerobic microorganisms indigenous to contaminated soils and sediments (reviewed in reference 15).
Despite these formidable obstacles, a small number of studies have demonstrated the feasibility of removing dioxins from contaminated environments by in situ bioremediation (13, 15, 16). Feasibility studies conducted in the laboratory and in the field demonstrated that for in situ treatment to be successful, the introduction of nonnative microorganisms with dioxin degradation potential is critical. For example, removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soil microcosms was observed only following the addition of a minimum of 4 × 106 CFU of strain RW1 per gram (dry weight) of soil (16). Introduction of larger quantities of cells (108 CFU/g [dry weight] of soil) resulted in complete biotransformation of nonsubstituted diaryl ethers and in a reduction by 50% of 2-chlorodibenzo-p-dioxin, present in soils at an initial concentration of 10 ppm (17).
Successful bioaugmentation strategies employing bacteria harboring the dioxin dioxygenase will require monitoring of both bacterial survival and expression of the genes coding for the biodegradative function. Commonly employed genotypic analyses, such as targeted amplification of either 16S rRNA genes or functional gene sequences, are highly sensitive and specific but do not inform about biotransformation activity. Similarly, the use of monoclonal antibodies in conjunction with epifluorescence microscopy can reveal the presence of strain RW1 (38) and other specific bacteria in environmental samples but typically fails to yield any additional information on the biodegradative activity of the detected microorganism. Analysis of mRNA can suggest—but does not confirm—the presence of a functional enzyme of interest. Therefore, short of directly assaying the biochemical activity of cells, the detection of pollutant-transforming enzymes is the most definitive technique for confirming the presence of catalysis-enabled phenotypes of pollutant-degrading microorganisms.
We explored the use of peptide mass fingerprinting (PMF) (22, 34) using vacuum matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for monitoring the dioxin dioxygenase as a biomarker of phenotypes of RW1 that are physiologically able to biotransform dioxin-related compounds. The mass spectrometric target of our investigation was the dioxin dioxygenase of S. wittichii strain RW1, a heteromer consisting of two alpha and two beta subunits (National Center for Biotechnology Information [NCBI] accession numbers gi|3426122 and gi|3426121, respectively) (1). We hypothesized that the enzyme complex would represent an excellent target for mass spectrometric analysis because (i) it has been thoroughly characterized (1, 5), (ii) its two protein subunits are contained in searchable online databases (e.g., NCBI), (iii) the respective DNA sequences have only weak similarity (40%) to other three-component dioxygenases (1), (iv) the genes are stable, (v) it is indicative of dioxin degradation activity (1), and, last but not least, (vi) it is unique to the dioxin-degrading organism of interest, strain RW1.
The specific aims of our study were (i) to identify a predetermined strain-specific proteinaceous biomarker, the dioxin dioxygenase, in minimally processed microbial pure cultures of RW1 by PMF using vacuum MALDI-TOF MS; (ii) to determine the minimal number of cells required for statistically significant (P < 0.05) identification of putative cultures of the bioremediation agent; and (iii) to determine the effects of different sample preparation techniques and growth substrates on the detectability of the dioxin-degrading bacterium.
MATERIALS AND METHODS
Culturing of strain RW1.
Liquid cultures of S. wittichii strain RW1 (DSMZ 6014) were grown at 30°C in a water bath shaker in M9 phosphate-buffered minimal medium (37) supplemented with (i) DF crystals (Sigma-Aldrich, Milwaukee, WI), (ii) 50 mM glucose, or (iii) both. Saturated DF medium contained approximately 3 to 5 mg liter−1 of the binuclear aromatic compound in the dissolved phase. Turbidity of the cultures was monitored using a DR/4000U spectrophotometer (Hach, Loveland, CO) at a wavelength of 560 nm. Viable bacteria were enumerated by plate counts using M9 medium supplemented with 1.5% agar (Difco, Franklin Lakes, NJ) and 5 mM sodium benzoate. Negative control samples composed of cells of RW1 lacking the dioxin dioxygenase were obtained via growth of the bacterium on nonselective Luria-Bertani broth, a complex medium that represses dioxin dioxygenase expression (17).
Microorganisms serving as negative controls.
More than 20 different Proteobacteria served as negative controls throughout this study. Most of these represented poorly characterized environmental monocultures and mixed cultures that had been obtained via selective enrichment using dioxin-like compounds as sole sources of carbon and energy. Pseudomonas putida KT2440 (DSMZ 6125) was the only negative control strain for which the complete genome was available in searchable online databases. All cultures were grown under selective conditions on aromatic substrates to maximize the expression of aromatic-ring dioxygenases.
Sample preparation.
Four different types of cell preparations were furnished for MALDI-TOF MS. Cells growing in the early, mid-, and late exponential phase were harvested by centrifugation (3,000 × g; 30 min; 4°C), washed, and resuspended in 50 mM NH4HCO3 (fraction 1; undisrupted cells). Biomass was disrupted on ice using a Sonic Dismembrator (Fisher Scientific, Pittsburgh, PA) on low setting for three bursts of 10 s, with cooling periods of 30 s between bursts, yielding fraction 2 (disrupted cells). Sonicated cell suspensions were centrifuged (13,500 × g; 5 min; 4°C) to separate the supernatant of the crude cell extract (fraction 3; whole-cell extract) from the pellet (fraction 4), consisting primarily of cell debris and undisrupted whole cells. For experiments involving two-dimensional (2D) gel electrophoresis, whole-cell extracts were divided into two equal volumes (sample splits) to allow additional analysis by MALDI-TOF MS; reported CFU in the sample are corrected for the loss of biomass resulting from splitting of the samples.
In silico digestion.
Peptides resulting from tryptic digestion of the alpha and beta subunits of the dioxin dioxygenase were predicted from sequences deposited in the NCBI database (http://www.ncbi.nih.gov/) using MS-Digest (http://prospector.ucsf.edu/). The in silico digests were performed using trypsin and disallowing missed cleavages or posttranslational modifications, based upon screening of combinations of multiple search parameters. Cysteines were presumed to be unmodified, as were the N and C termini of the peptides. The mass range was specified as 500 to 5,000 Da; multiply charged ions were not considered.
MALDI-TOF MS analysis.
Samples (25 μl) were digested with 200 ng trypsin in 50 mM NH4HCO3 at 37°C overnight, vacuum dried in a Savant SVC100 Speed Vac (GMI, Albertville, MN), desalted using C18 Omix microextraction column tips (Varian, Palo Alto, CA), and mixed with matrix solution (∼1.5 μl) consisting of 10 mg ml−1 of alpha-cyano-4-hydroxy-cinnamic acid (CHCA) in 50% acetonitrile and 0.1% trifluoroacetic acid. A stainless steel 96-well MALDI target plate (Applied Biosystems, Foster City, CA) was spotted with approximately 1 μl of the sample-matrix solution, which was then air dried. Spectra were acquired using a Voyager DE-STR MALDI-TOF MS (Applied Biosystems, Foster City, CA) in positive reflector mode (m/z 500 to 5,000; 50 laser shots per spectrum). Initial external calibration was performed using a standard peptide mixture (human bradykinin fragments 1 to 7, 757.3997 Da; human adrenocorticotropic hormone fragments 18 to 39, 2,465.1989 Da; bovine insulin chain B, oxidized, 3,494.6513 Da) purchased from Sigma (St. Louis, MO). Additional internal calibration was carried out as described below.
Mass spectral data analysis.
Mass spectral data were analyzed and manipulated using Data Explorer software (Applied Biosystems, Foster City, CA). Spectra were deisotoped using the manufacturer's settings. Internal calibration was carried out using trypsin autolysis peaks. Acquired data were analyzed by comparison to in silico information contained in the NCBI databases (http://www.ncbi.nih.gov) using PMF. The 300 most intense peaks were searched against the NCBI taxonomy subset “All Bacteria” (>753,000 sequences) at a mass tolerance of 50 to 100 ppm using Mascot (http://www.matrixscience.com). Additional search parameters included disallowing missed cleavages and either fixed or variable posttranslational modifications. Probability scores for positive identification were determined using the statistical algorithm described by Pappin et al. (34).
Peptide sequencing.
Protein identifications obtained by PMF were confirmed in selected samples via sequencing of the target mass at m/z 3,036.3 using an ion trap mass spectrometer (LCQ Deca XP; Thermo Electron Corporation, MA) in conjunction with an atmospheric pressure MALDI source (Mass Tech Inc., MD). Presence of the alpha subunit of the dioxin dioxygenase was confirmed by submission of detected fragment ions to the National Center for Biotechnology Information database.
Two-dimensional gel analysis.
Cultures were dried by vacuum centrifugation and assayed for protein content by the bicinchoninic acid method using a commercial protein analysis kit according to the manufacturer's instructions (Pierce Biotech, Rockford, IL). Fifty micrograms of protein was reconstituted using 8 Murea-CHAPS (cholamidopropyl-dimethylammonio-1-propane-sulfonate)-45mM Tris-HCl. Samples for two-dimensional (2D) gel electrophoresis were separated in the first dimension on immobilized pH gradient (IPG) strips at 50 V for 10 h in an IPGphor isoelectric focusing system (Amersham Biosciences, Sunnyvale, CA). The IPG strips were equilibrated, and proteins were separated by molecular mass in the second dimension by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (4 to 12% NuPAGE TrisBis; Invitrogen, Carlsbad, CA). The resultant two-dimensional gel was silver stained using standard protocols and analyzed using Melanie 4.0 viewer software (http://us.expasy.org/melanie/) to obtain an unbiased estimate of the number of spots present on the gel. Since the visual dynamic range of gels is limited to about 2 orders of magnitude in concentration, obtained estimates of the total number of proteins were considered very conservative.
RESULTS
In silico analyses.
Theoretical (in silico) digestions were performed to construct peptide maps of the large (alpha) and small (beta) subunits of the dioxin dioxygenase (Tables 1 and 2, respectively). The porcine protease used in this study, trypsin (EC 3.4.21.4), cleaves proteins after the amino acids lysine and arginine, unless these are followed by a proline. Digestion yielded individual amino acids and peptides, the latter ranging in length from 2 to 35 amino acids. Summarized in Tables 1 and 2 are protein fragments of the alpha and beta subunits of the dioxin dioxygenase, the corresponding ions predicted to form during MALDI, and their individual contributions to the relative coverage of the two proteins, reported in percents. For the alpha subunit, there were 31 predicted potential MS targets in the experimentally defined detection range (mass-to-charge ratios of m/z 500 to 5,000), covering 94% of the amino acids of the total protein. The remaining 6% of the protein mass was composed of peptides situated outside of the detectable range (m/z <500). The beta subunit was calculated to yield a maximum of 15 detectable tryptic peptides, suggesting a maximum theoretical protein coverage of 84% (Table 2).
TABLE 1.
Protein coverage, amino acid sequences, and corresponding theoretical masses of peptides of the dioxin dioxygenase alpha subunita
| Theoretical mass (M + H) | No. of amino acids | % Coverage of total protein | Amino acid sequence |
|---|---|---|---|
| 506.3 | 4 | 0.92 | IFAR |
| 534.3 | 4 | 0.92 | DIMR |
| 559.3 | 5 | 1.15 | DAVVR |
| 573.3 | 5 | 1.15 | LGDIR |
| 586.3 | 5 | 1.15 | WGAPR |
| 632.4 | 6 | 1.38 | DTGVLK |
| 639.3 | 6 | 1.38 | VETYK |
| 685.4 | 5 | 1.15 | VWQPR |
| 730.4 | 6 | 1.38 | LCLADR |
| 761.4 | 6 | 1.38 | AAHYLR |
| 845.4 | 7 | 1.61 | NMPQEVK |
| 919.5 | 8 | 1.84 | NISSANWK |
| 951.5 | 9 | 2.07 | GVSEGYIAR |
| 962.5 | 8 | 1.84 | GLIFGNWR |
| 1,050.5 | 10 | 2.30 | LGHASSGFFK |
| 1,234.6 | 11 | 2.53 | NAVDVADLFDR |
| 1,282.7 | 12 | 2.76 | QTHLNMALGLGK |
| 1,393.7 | 11 | 2.53 | TEVWNYVIVDR |
| 1,541.8 | 14 | 3.22 | QGDGSFAAFLNQCR |
| 1,584.7 | 13 | 2.99 | SWLFLGHESQIPK |
| 1,740.7 | 15 | 3.45 | AFYSHWQDMLAGDEA |
| 1,847.8 | 16 | 3.68 | ASAYSDQAVYDLEMER |
| 2,005.0 | 18 | 4.14 | FGDYITTTMGEDSVILSR |
| 2,180.1 | 19 | 4.37 | TDHMGTLVMTVFPNFSLNR |
| 2,194.0 | 21 | 4.83 | GGPNPDYPGTINDVYSEEGGP |
| 2,222.1 | 19 | 4.37 | SYEHIFHPGEQGHQFALPK |
| 2,798.3 | 25 | 5.75 | GCTLAFNASGLLEQDDAENVAMCQR |
| 3,036.3b | 27 | 6.21 | CSYHGWVFNNAGGLVSMPHEANYTIDK |
| 3,081.4 | 27 | 6.21 | NMNDGDAAMLQQFPPHPAPEYYYGPGR |
| 3,450.7 | 32 | 7.36 | WQAEQHATDHLHVAVSHFSGFAALAPEGSPPR |
| 3,703.8 | 35 | 8.05 | ADAPDLLSSLGEATWYLDAFLDANEGGVELIGPQR |
| 49,389.3 | 435d (197c) | 94d (45c) |
Obtained by in silico tryptic digestion of the protein (gi|3426122) from Sphingomonas wittichii strain RW1. Peptide masses outside of the monitoring range (m/z 500) were omitted. Peptides detected experimentally by MALDI-TOF MS are shown in boldface.
The mass at m/z 3,036.3 served as the target of confirmatory analysis by peptide sequencing.
Empirical protein coverage observed in all experiments combined.
Theoretical maximum protein coverage in the range of m/z 500 to 5,000.
TABLE 2.
Protein coverage, amino acid sequences, and corresponding theoretical masses of peptides of the dioxin dioxygenase beta subunita
| Theoretical mass (M + H) | No. of amino acids | % Coverage of total protein | Amino acid sequence |
|---|---|---|---|
| 563.3 | 5 | 2.79 | SLGMR |
| 607.3 | 5 | 2.79 | TTDDR |
| 627.3 | 5 | 2.79 | LSDHR |
| 639.4 | 5 | 2.79 | HDLVR |
| 679.3 | 6 | 3.35 | MSSQVK |
| 760.4 | 7 | 3.91 | TAVTNVR |
| 832.5 | 7 | 3.91 | QFLPLSK |
| 848.4 | 8 | 4.47 | SDGPLGFR |
| 1,077.6 | 9 | 5.03 | VYPPLIGYR |
| 1,080.5 | 9 | 5.03 | GAHFEDNYK |
| 1,420.7 | 11 | 6.15 | TWVENPPMYQR |
| 1,890.0 | 18 | 10.06 | TVYLDHAVLPGSGISTFL |
| 2,035.9 | 18 | 10.06 | ETDVAGEYEAYSNIAFTR |
| 2,279.1 | 19 | 10.61 | FEDWYALIAEDIHYAVPAR |
| 2,337.2 | 19 | 10.61 | IQWEVEQFLYEEAALLAER |
| 21,361.28 | 179 (50b) | 84c (33b) |
Obtained by in silico tryptic digestion of the protein (gi|3426123) from Sphingomonas wittichii strain RW1. Peptide masses outside of the monitoring range (m/z <500) were omitted. Peptides detected experimentally by MALDI-TOF MS are shown in boldface.
Empirical protein coverage observed in all experiments combined.
Theoretical maximum protein coverage in the range of m/z 500 to 5,000.
Screening of various fractions of RW1 cells for the dioxin dioxygenase.
Initial experiments concentrated on the feasibility of detecting the dioxin dioxygenase in four different fractions of processed cell cultures (Fig. 1). Proteins contained in the various cell fractions were digested, purified, and desalted via passage through a pipette tip functioning as a C18 microextraction column. Purified digests were mixed with matrix and analyzed by PMF using MALDI-TOF MS, as shown in the schematic (Fig. 1). The investigated cell fractions included undisrupted cells (fraction 1), cells disrupted by sonication (fraction 2), whole-cell extracts representing the supernatant of disrupted, centrifuged cells (fraction 3), and the corresponding pellet consisting of cell debris and residual whole cells (fraction 4). Since the optimal amount of biomass for the assay was not known a priori, experiments were performed using a range of initial cell quantities (105 to 1010 cells).
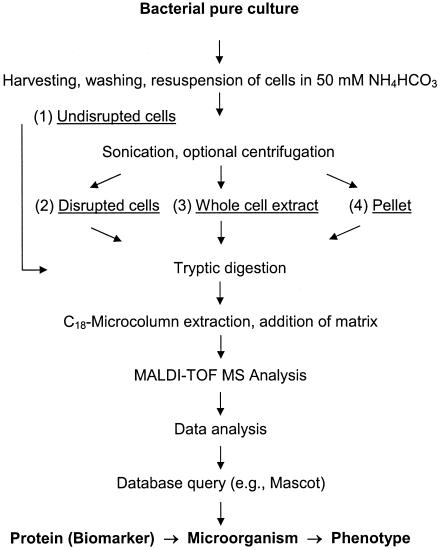
FIG. 1.
Experimental approach used for screening of cell preparations for the dioxin dioxygenase, an enzyme that served as a proteinaceous biomarker of dioxin degradation-enabled phenotypes of Sphingomonas wittichii strain RW1. Four different preparations of cells (105 to 1010 CFU) were obtained and tryptically digested for analysis by peptide mass fingerprinting using vacuum MALDI-TOF MS: (1) undisrupted cells, (2) disrupted cells, and (3) whole-cell extracts, as well as (4) pellets obtained by centrifugation of disrupted cells.
(i) Fraction 1.
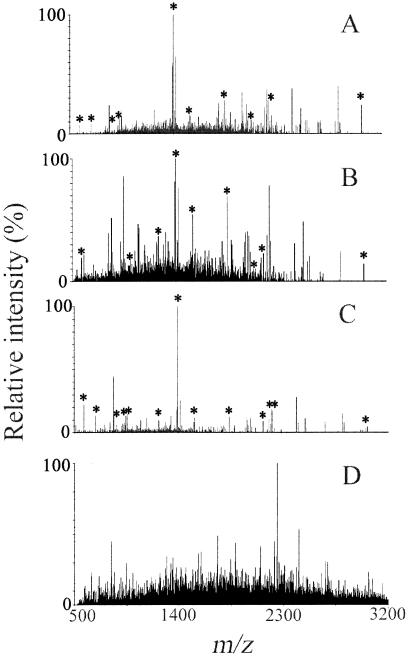
Analysis by peptide mass fingerprinting of a digest of 108 undisrupted cells of RW1 yielded the deisotoped mass spectrum shown in Fig. 2A. Ten target peaks rose above the baseline noise: m/z 685.4, 951.5, 1,234.6, 1,393.7, 1,541.8, 1,847.8, 2,005.0, 2,194.0, 2,222.1, and 3,036.3. A list of up to 300 ions having the greatest signal intensities was generated and submitted to online protein databases representing the kingdom or domain of Bacteria. The data query returned the alpha subunit of the dioxin dioxygenase as the best fit among >753,000 proteins. The resultant Mascot score of 52 indicated that the search result was not statistically significant (P > 0.1), however. Overall, the 10 target peptides provided 31% protein coverage (Table 3).
FIG. 2.
Deisotoped MALDI mass spectra obtained from various fractions of digested cells of RW1: (A) undisrupted cells; (B) disrupted cells; (C) whole-cell extract; and (D) pellet of disrupted cells. Peaks labeled with an asterisk correspond to peptides of the alpha subunit of the dioxin dioxygenase. Simple physical fractionation of cell components significantly improved the identification of the protein, with whole-cell extract producing the best results.
TABLE 3.
Experimental results obtained during screening of various bacterial cell fractions for the alpha subunit of the dioxin dioxygenase by MALDI-TOF MS peptide mass fingerprintinga
| Digested sample fractionb | log CFUc | No. of target ions observedd | % Protein coverage | Mascot score | P value |
|---|---|---|---|---|---|
| 1 (undisrupted cells) | 8 | 10 | 31 | 52 | >0.1 |
| 2 (disrupted cells) | 8 | 6 | 30 | 77 | <0.01 |
| 7 | 9 | 32 | 69 | <0.05 | |
| 5-6 | 0 | ||||
| 3 (whole-cell extract) | 7 | 13 | 34 | 105 | <0.00001 |
| 4 (pellet of disrupted cells) | 5-8 | 0 |
Cells of Sphingomonas wittichii strain RW1 were grown in liquid phosphate-buffered minimal medium containing dibenzofuran as the sole carbon and energy source. Mass lists were generated from spectra using Data Explorer software and entered into the Mascot search engine. Statistically significant observations (P < 0.05) scoring >68 are shown in boldface.
Refer to Materials and Methods and Fig. 1 for details of sample preparation.
CFU were determined by plate count and estimated photospectrometrically from absorbance at λ560 nm.
Ions matching the theoretical masses of peptides of the alpha subunit of the dioxin dioxygenase, as determined by Mascot.
(ii) Fraction 2.
Analysis of a digest of 107 disrupted cells of RW1 produced the spectrum shown in Fig. 2B. Nine target peptides of the alpha subunit of the dioxin dioxygenase were detected. Compared to fraction 1, the mass at m/z 951.5 was missing, and an increase in the level of noise was observed in the range from m/z 1,000 to 3,200. Again, database searching returned the alpha subunit of the dioxin dioxygenase as the best match, with a statistically significant (P < 0.05) Mascot score of 69. Protein coverage was 32%.
(iii) Fraction 3.
Analysis of supernatant obtained by centrifugation of 107 disrupted cells of RW1 yielded the best result. A typical mass spectrum recorded for the experimentally defined whole-cell extract is shown in Fig. 2C. The spectrum had a very low level of noise across the entire m/z range of interest. Major detectable ions were clustered between m/z 500 and 3,200. In the spectrum shown, four of the eight most intense peaks—detected at m/z 1,393.7 (100% relative intensity), 586.3 (22%), 2,222.1 (17%), and 962.5 (15%)—matched in silico values calculated for peptides of the alpha subunit of the dioxin dioxygenase (Table 1); the second intense ion, at m/z 842.5, corresponded to a trypsin autolysis product that was used as an internal standard for mass calibration. A total of 13 target peaks were detected, resulting in confident protein identification (P < 0.00001) by Mascot searching, with a score of 105 and a protein coverage of 34%. Target ions detected at lesser intensities included m/z 685.4 (13% relative intensity), 919.4 (7%), 951.5 (13%), 1,234.6 (9%), 1,541.8 (8%), 2,005.0 (10%), 2,194.0 (11%), and 3,036.3 (4%). The ease of detection of the alpha subunit in whole-cell extract is consistent with previous reports that localized dioxin dioxygenase activity to the soluble proteome of extracts from cells grown on DF (1, 5, 13).
(iv) Fraction 4.
Analysis of digested pellets obtained by centrifugation of disrupted cells yielded noisy mass spectra (e.g., Fig. 2D) that did not show any target m/z regardless of the amount of biomass processed (Table 3). This finding was consistent with reports in the literature indicating cell pellets to be depleted in dioxin dioxygenase activity relative to whole-cell extracts of RW1 (fraction 3) (1, 5, 13). Overall, the results demonstrated that the dioxin dioxygenase is most easily detectable by PMF in digested whole-cell extract. Therefore, the sensitivity of PMF analysis was further investigated in that matrix.
Sensitivity analyses and robustness of the assay.
To determine the biomass range suitable for positive identification of strain RW1 via PMF of the alpha subunit of the dioxin dioxygenase, whole-cell extracts of 105 to 1010 DF-grown CFU were analyzed following digestion with a standard amount of 200 ng of trypsin. Positive protein identification with significant probability-based Mascot scores of >68 (P < 0.05) were obtained consistently when >106 cells were processed and analyzed. Analysis of extracts obtained from 107 and 108 DF-grown cells yielded Mascot scores ranging from 73 to 105 (P < 0.01 to 0.00001) and 84 to 111 (P < 0.001 to 0.00001), respectively (Table 4); in these experiments, the number of matched peptide masses ranged from 10 to 13 and 12 to 14, respectively, with protein coverages for the alpha subunit of the dioxin dioxygenase ranging from 31 to 34% (107 CFU) and from 37 to 43% (108 CFU). Analysis of ≤106 CFU yielded no target ions and no significant matches for either the two target proteins or any of the more than 753,000 proteins contained in the nonredundant NCBI database at the time of data analysis. Similarly, no database matches were found in experiments using ≥109 CFU (Table 4). A total of 15 different peptide masses, corresponding to the alpha subunit of the dioxin dioxygenase, were detected in more than 100 experiments conducted with biomass harvested in the early, mid-, and late exponential growth phase (total protein coverage, 45%) (Table 1). In contrast, none of these target peptides were found and no positive identifications of the dioxin dioxygenase were obtained during analysis of the more than 20 negative control strains that represented a broad spectrum of microorganisms capable of catabolizing dioxin-related aromatic compounds.
TABLE 4.
Results of database searches for ions corresponding to the alpha subunit of the dioxin dioxygenase detected in tryptically digested extracts of Sphingomonas wittichii strain RW1 grown on various substratesa
| Growth substrate | log CFU | No. of target ions observed | % Protein coverage | Mascot score | P value |
|---|---|---|---|---|---|
| DF | 10 | 0 | |||
| 9 | 10 | 31 | 59 | >0.10 | |
| 8 | 14 | 43 | 111 | <0.00001 | |
| 8b | 12 | 37 | 84 | <0.001 | |
| 7 | 13 | 34 | 105 | <0.00001 | |
| 7b | 10 | 31 | 73 | <0.01 | |
| 5-6 | 0 | ||||
| 6b | 0 | ||||
| Glucose + DF | 9 | 0 | |||
| 8 | 10 | 31 | 61 | >0.05 | |
| 7 | 9 | 22 | 59 | >0.10 | |
| 6 | 0 | ||||
| Glucose | 6-9 | 0 | |||
| LB broth | 6-9 | 0 |
Unless otherwise stated, cells were harvested in mid-exponential growth phase. Statistically significant observations (P < 0.05) scoring >68 are shown in boldface.
Cells harvested in the deceleration growth phase. True exponential growth was not observed with dibenzofuran due to the limited solubility of the compound.
The results of repeatedly performed experiments were very consistent. The following variables had no detectable effect on the outcome of the experiment (data not shown): substituting CHCA for 3,5-dihydroxybenzoic acid as the ionization matrix, type of C18 microextraction column used (n = 2), and identity of the operator (n = 3). However, when cells were harvested late into the exponential growth phase (deceleration phase), a slight drop in Mascot scores was observed (Table 4).
Interestingly, the beta subunit of the dioxin dioxygenase was never identified by database searching in any of these experiments. This is surprising, because the observed removal of DF during the growth of RW1 cultures indicated the presence of this essential protein at quantities equimolar to those of the alpha subunit (1). Although some target ions of the beta subunit were present, as determined by manual identification, the signal intensities of these peptide masses at m/z 563.4 (1% relative intensity), 607.3 (2%), 693.3 (4%), 832.5 (8%), 848.5 (10%), and 1,077.6 (6%) typically were at or near the baseline noise level. Following spectral processing and data reduction using a peak threshold of approximately 5 to 10% relative intensity, these ions were mostly rejected and did not enter the online database query; this effectively prevented a potential identification of the beta subunit when the online search algorithm was used.
Effect of growth substrate on strain identification.
Cultures of RW1, grown in phosphate-buffered mineral salt solution supplemented with the growth substrates (i) DF, (ii) DF plus glucose, and (iii) glucose only were processed and analyzed by MALDI-TOF MS and 2D gel electrophoresis. The alpha subunit of the dioxin dioxygenase—i.e., the previously established biomarker of dioxin degradation-enabled cells of RW1—was identified readily in the digested soluble proteome of DF-grown cells with scores as high as 111, indicating a very low probability of false-positive misidentification (P < 0.00001) (Table 4). Detection of up to 14 target peptides in whole-cell extracts of 108 CFU resulted in protein coverage of 43%, the best result achieved (Table 4). Again, the ions corresponding to peptides of the alpha subunit were among the most prominent in the mass spectra (e.g., Fig. 3A).
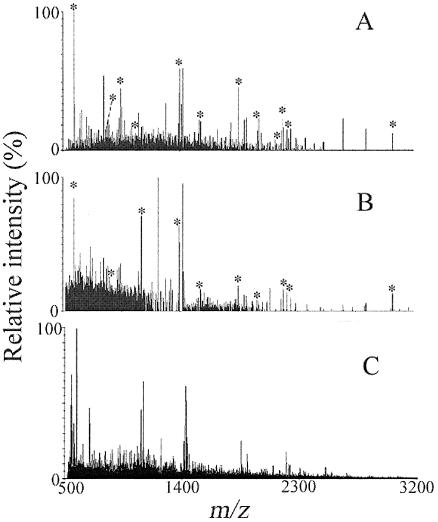
FIG. 3.
Deisotoped MALDI mass spectra of digested whole-cell extracts from 108 cells of Sphingomonas wittichii strain RW1 grown under the following conditions: phosphate-buffered minimal medium supplemented with (A) dibenzofuran, (B) dibenzofuran plus glucose (50 mM), and (C) glucose. Peaks labeled with an asterisk correspond to peptides of the dioxin dioxygenase. Submission of mass lists generated from spectra A and B to the Mascot search engine returned the alpha subunit of the dioxin dioxygenase as the best possible match (P < 0.00001 and P > 0.05, respectively).
The alpha subunit of the dioxin dioxygenase also was returned as the best database match when analyzing glucose-grown cells of RW1 that were coexposed to DF for enhanced expression of the dioxin dioxygenase (Fig. 3B); however, the corresponding score was not significant (P > 0.05), necessitating peptide sequencing for unambiguous protein identification. Compared to DF-grown cells (Fig. 3A), the signal intensities of target peaks were lower in glucose-grown biomass coexposed to DF (Fig. 3B). No target peaks were detected when analyzing biomass grown on glucose in the absence of DF, and no significant matches were found for any of the >753,000 proteins contained in the nonredundant NCBI database (Fig. 3C and Table 4). Analysis of cells grown using nonselective complex media, e.g., Luria-Bertani broth (35), also revealed no ions of interest in the mass spectra recorded (Table 4). Lack of detection of the alpha subunit in LB-grown cells was consistent with information in the literature indicating repressed dioxin dioxygenase expression during growth of RW1 on complex media (1).
2D gel electrophoresis.
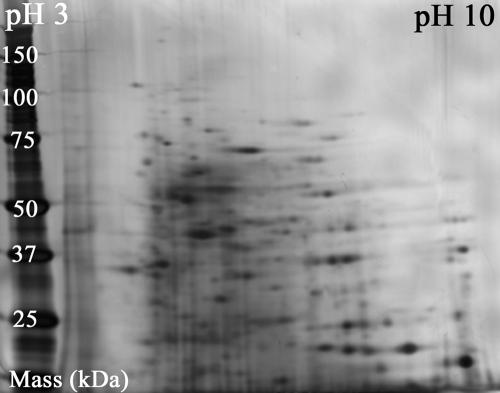
To characterize the level of matrix complexity in whole-cell extracts, the soluble proteome of RW1 was separated by two-dimensional gel electrophoresis using isoelectric focusing in the first dimension and size-dependent electrophoretic mobility in the second dimension (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Figure 4 shows a silver-stained 2D gel of an extract of RW1 cells grown on DF. Digital image analysis of the gel revealed the presence of a minimum of 350 different proteins. Analysis of 2D gels of extracts from cells grown on glucose and glucose supplemented with DF yielded similar results (data not shown). It is important to note that the 2D sample separation was done exclusively for the purpose of investigating the complexity of the sample matrix. More commonly, this technique is employed as a cleanup procedure in preparation for protein analysis by PMF (24). However, this labor-intensive separation of proteins followed by in-gel digestion of selected protein spots was unnecessary in this study because the alpha subunit of the dioxin dioxygenase was detected successfully with great confidence (P < 0.00001 to 0.01) in whole-cell extracts of DF-grown cells by PMF—a result that was successfully confirmed by an additional assay involving sequencing of the target peptide at m/z 3,036.3 using an ion trap mass spectrometer equipped with an atmospheric pressure MALDI source.
FIG. 4.
Scanned image of a two-dimensional gel used to separate soluble proteins in whole-cell extracts of Sphingomonas wittichii strain RW1 grown on dibenzofuran. Digital image analysis of the silver-stained gel demonstrated the presence of at least 350 proteins in the complex sample matrix.
DISCUSSION
Mass spectrometric identification of microorganisms.
Mass spectrometry has been used extensively in the past for the identification of microbial pure cultures at the genus, species, and strain levels (reviewed in references 11, 28, and 39). The most common approach is the MALDI MS analysis of matrix-embedded intact or disrupted vegetative cells, spores, or cysts in linear detector mode in the range of m/z 5,000 to 30,000, yielding mass spectral “barcodes” for the microorganisms of interest. This technique can serve to identify microbial species and strains with the important prerequisite that, in order to interpret the data, standard spectra are available which were obtained from authentic cultures grown, harvested, and processed under highly standardized conditions identical to those used for unknown samples (11). The technique is ultrafast (<5 min) but limited in its informational value because the chemistry and function of the ion-producing molecules remain unknown throughout the process. Successful applications of mass spectral fingerprinting by MALDI-TOF MS include the automated bacterial identification of various Firmicutes and Proteobacteria (23), rapid characterization of spores of the Bacillus cereus group (36), and differentiation of oocysts of Cryptosporidium parvum and Cryptosporidium muris (30).
To overcome some of the limitations associated with traditional mass spectral fingerprinting of whole cells and cell lysate, additional research has concentrated on the targeted detection of strain-specific cell components whose corresponding ions are predictable from DNA sequence information. One strategy is the analysis of cell preparations by MALDI-TOF MS in linear detector mode to scan for ions of intact proteins; good targets are, for example, ribosomal proteins, because they are abundant in vegetative cells regardless of culturing conditions (∼20% of protein content) and sufficiently unique to allow confident identification (2). This technique represents a significant improvement over conventional microbial fingerprinting, but it is not without limitations. Since MALDI is a soft ionization technique, it leaves target molecules unfragmented and produces predominantly singly charged (MH+) ions (19, 25). Detection of an intact protein in linear mode typically yields a single ion on which the identification has to be based. In the mass range of m/z <30,000, experimentally determined molecular masses have a mass accuracy of ±1 Da or better. Since proteins possessing (nearly) identical molecular masses can differ dramatically in structure and function, protein identifications obtained based on a single ion are often only tentative. To achieve statistically significant results, mass spectral analysis of target proteins in linear mode necessitates the use of customized databases containing a limited number of proteins having distinct masses (9). For environmental applications in which the identities of bacterial isolates are completely unknown, searching against small databases can be challenging, although some successes have been reported (8).
An alternative strategy for the targeted analysis of proteinaceous biomarkers is the use of PMF, as demonstrated in the present study and elsewhere (e.g., references 18 and 29). It involves the digestion of partially purified cell components followed by mass spectrometric analysis in positive (or negative) reflector mode, typically in the mass range of m/z 500 to 5,000. In contrast to mass spectral microbial fingerprinting, PMF is more powerful because specific target proteins can be selected a priori and their corresponding ions (peptide masses) can be predicted in silico, as shown by the theoretical mass lists presented in Tables 1 and 2. Identification is based on the detection of multiple fragments of a given protein rather than on a single molecular ion. Therefore, protein matches by PMF have a quantifiable confidence level and often are statistically highly significant even when searching nonrestricted, complex databases containing hundreds of thousands of proteins (Table 4). The identities of detected proteins can be ascertained without having to obtain and analyze authentic protein standards, an important advantage when attempting to identify environmental isolates whose proteins have never been purified. Since the function of the detected biomarker either is known or can be inferred, PMF of microbial cells can reveal critical information on biomass physiology that otherwise would be difficult or impossible to obtain (32).
The few studies performed to date suggest that successful use of PMF for bacterial identification requires extensive sample preparation steps to separate the proteins and peptides of interest from nontarget interferences prior to mass spectrometric analysis (7, 31, 33). Commonly applied tools used for this purpose include one- or two-dimensional gel electrophoresis (31, 33), one- or two-dimensional chromatography (21), affinity chromatography (10), and retentate chromatography using protein chips in conjunction with surface-enhanced laser desorption ionization-time of flight MS (3), to name just a few. Even when performed on a routine basis in high-throughput mode, these more sophisticated sample preparation steps can be time-consuming, labor-intensive, and expensive. While these techniques are indispensable for the analysis of low-abundance proteins, less sophisticated sample preparation techniques may suffice.
Due to these real or perceived constraints, PMF currently is not considered a suitable and cost-effective tool for the identification and characterization of environmental isolates. The present study challenges this view by employing PMF on minimally processed microbial cells. Experimental results of the present study reveal the value and power of this nontraditional usage of PMF by MALDI-TOF MS for the identification and phenotypic characterization of certain environmental microorganisms, such as strain RW1.
The alpha subunit as a biomarker for dioxin degradation-enabled phenotypes of RW1.
The dioxin dioxygenase is the preferred biomarker for dioxin degradation-enabled phenotypes of RW1 because the protein complex is essential for turnover of dioxin-related substrates (1, 5). Fortunately, the dioxygenase fulfilled the basic requirements of PMF: its corresponding DNA sequence information was known and contained in searchable on-line databases. Localization of the relevant genes on a single open reading frame, situated within the bacterial chromosome (1), provided an additional incentive to target this catabolic enzyme, since chromosomal genes generally are more stably expressed and less frequently transferred between species and genera than are plasmid-encoded genes (17).
For PMF of complex protein mixtures to be successful, the target protein must be present in detectable quantities, and multiple peptides must have ionization behaviors superior to those of competing peptides that are present in the sample at similar or greater concentrations. Our experimental results demonstrate that only one of the two subunits of the dioxin dioxygenase fulfills these requirements. Theoretically, both subunits of the protein complex represent viable targets for mass spectrometry by offering 15 or more peptides, each within the experimental m/z range (Tables 1 and 2). However, recorded mass spectra were dominated by peptide ions of the alpha subunit (Fig. 2 and 3), whereas those of the beta subunit were weak or not detected at all.
The dichotomy of these results illustrates a yet-unsolved challenge in MALDI-TOF MS analysis: the difficulty of predicting with confidence the ionization behavior of peptides (12). As stated previously, the genes dxnA1 and dxnA2 encoding the alpha and beta subunits of the dioxin dioxygenase protein complex, respectively, are contained on a single transcriptional unit (1). Consequently, both proteins were present in approximately equimolar quantities in whole cells. The sample preparation strategy used is known to concentrate both components of the dioxin dioxygenase complex in whole-cell extracts (1, 5, 13). Experimental results of MALDI-TOF MS analysis showed that peptides of the alpha subunit (Mr, ∼49 kDa; pI, ∼5.8) have much more favorable ionization properties than those of the beta subunit (Mr, ∼22 kDa; pI, ∼7.9). In addition, these peptides also dominated over anticipated ones corresponding to the 29 mostly hypothetical proteins that are listed for RW1 in the nonredundant NCBI database (none of these were found by online database searches in any of the experiments). Furthermore, the thousands of tryptic peptides anticipated to result from digestion of the >350 unidentified proteins contained in whole-cell extracts (Fig. 4) also could have interfered with the ionization and detection of the alpha subunit. This was not the case, however. It is remarkable that the alpha subunit could be identified by database searching in minimally processed samples of such great complexity without the need for peptide sequencing.
The favorable ionization behavior of the alpha subunit was not predictable in silico. Arginine and a number of hydrophobic residues are known to increase the ionization of peptides, whereas lysine suppresses it (4, 12, 27). The frequency of these signal-modulating residues in peptides of the alpha and beta subunits was analyzed (data not shown) but could not fully explain experimental findings. This underscores the observation by others (4, 12, 27) that the prediction of peptide ionization is a challenging task, with currently available models delivering only rudimentary and imprecise results. Since the ionization behavior of peptides from pure proteins already is challenging, peptide ionization in complex protein mixtures is unpredictable and requires an empirical approach.
Cell fractions suitable for biomarker detection.
The alpha subunit of the dioxygenase was one of the most prominent components detectable by PMF within the entire bacterial proteome: many target peaks were significantly more intense than observed nontarget peaks (Fig. 2). However, the noticeable increase in noise in whole-cell preparations reduced the confidence of protein identification (Fig. 2A and B; Table 3). In MALDI-TOF MS, only a finite number of molecules are actually analyzed at the detector, so the greater the interference, the less chance there is for detecting a peak of interest (26). Therefore, sample preparation is an important mechanism for increasing the probability of successful protein identification by concentrating the targets into a more manageable chemical matrix. The mass spectra and Mascot search results presented in Fig. 2 and Table 3 illustrate that the three simple and inexpensive sample preparation techniques employed—sonication of cells, followed by centrifugation and C18 microextraction cleanup of digested soluble proteins—were highly effective in reducing baseline noise and improving the overall results of PMF analysis of microorganisms by MALDI-TOF MS.
Effects of culture conditions on microbial identification.
In addition to sample-processing techniques, microbial culture conditions were identified as important determinants influencing the success and significance of positive protein identification. The age of the culture did not affect the overall result as much as the growth medium itself. This is consistent with results obtained from the proteomic analysis of Helicobacter pylori, grown in media of various pHs (24). Statistically significant identification of RW1 via detection of the alpha subunit of the dioxin dioxygenase was dependent on the use of the selective growth substrate DF (Fig. 3 and Table 4). Cells grown on DF and harvested just prior to entering the stationary phase were positively identified (Table 4). Although a drop in significance levels resulted when analyzing these maturing cultures, the qualitative result of positive identification did not change (Table 3). These observations were consistent with other studies exploring the effects of various sample-processing parameters on the quality and reproducibility of MALDI mass spectra (40).
The various culture media caused significant differences in the corresponding mass spectra (Fig. 3). This was expected and exploited in the experimental design. Cells of RW1 grown on complex media that are known to suppress dioxin dioxygenase expression (5) were used as negative controls for the biocatalyst (LB-grown RW1). Growth on glucose also represses dioxygenase expression. The detection of multiple target peaks in cells grown on glucose and coexposed to dibenzofuran (Fig. 3B) hints at an interesting application in bioremediation; dioxin degradation-enabled biomass may be produced economically and rapidly by using inexpensive growth substrate of high biomass yield in conjunction with inducers of enzyme expression. Identification of the dioxygenase under such culture conditions by PMF was only tentative, however (Table 4). To achieve positive identification of the target protein by PMF, these cell fractions would have required additional sample purification. Sequencing of the few detected target masses by MS/MS analysis represents an alternative means of confirming protein identifications. This approach was used successfully in this study on the mass at m/z 3,036.3. Other examples of peptide sequencing for positive identification of microorganisms are the detection of Norovirus particles in minimally processed clinical stool samples (14) and the detection of purified Sindbis virus AR 339 (45) and enterobacteriophage MS2 (44).
Sensitivity, ease of use, and robustness of PMF analysis.
Identification of cultures of RW1 via detection of the alpha subunit of the dioxin dioxygenase was successful in whole-cell extracts prepared from >106 to <109 CFU (Table 4). The practical lower detection limit likely was dictated by both the diminished mass of target protein in the sample and the complexity of the sample matrix, which is known to obscure signals and interfere with the ionization and detection of target peptides (28). The observed detection limit was consistent with results from a study of Bacillus subtilis in which 2.2 × 107 CFU were detected using MALDI-TOF MS (18). Since the oxygenase complex constitutes about 4% of the total soluble proteome of DF-grown cells of RW1 (5), the lower detection limit for the 49-kDa alpha subunit of the dioxygenase in whole-cell extracts of 107 CFU was calculated to equal about 500 femtomoles. Thus, the sensitivity of the assay was excellent, particularly considering the complexity of the sample matrix. The observed upper limit of detection (Table 4) likely resulted from a combination of incomplete digestion due to saturation of trypsin and saturation of the microextraction column resin during sample cleanup and incomplete lysis of cells by sonication.
The developed assay for the identification of RW1 by PMF of minimally processed whole cells is easy to use; does not require expensive sample preparation materials, e.g., protein chips (3); and is robust, as demonstrated by the negative results obtained for all control microorganisms and for LB-grown cells of RW1 devoid of dioxin dioxygenase. Qualitative results of analyses were relatively insensitive to culture age (cells harvested in the early, mid-, or late logarithmic phase) and independent of sample cleanup materials (C18 microextraction columns), sample matrix used (3,5-dihydroxybenzoic acid or CHCA), and individuals performing the experiment. All sample manipulation steps can easily be automated to allow unattended high-throughput analysis, an important goal for the analysis of both environmental and clinical microorganisms (6).
Applying PMF of whole-cell extracts in bioremediation.
The analysis strategy and methodology presented here are attractive for application in the field of bioremediation for several reasons. The principal advantage of the assay is its ability to identify cells of RW1 and simultaneously yield information on their most critical phenotypic characteristic that drives the removal of dioxins from contaminated environments during bioaugmentation: the expression of appreciable quantities of the dioxin dioxygenase.
Analysis of whole-cell extracts by PMF can inform on the extent to which vegetative cells of RW1 are charged with this enzyme. Since the assay is performed on a nonpurified bacterial proteome fraction, only cells containing appreciable quantities of the dioxin dioxygenase are detectable by PMF. Cells from at least two origins could be assayed during field-scale bioremediation: biomass grown in the laboratory for introduction into contaminated target environments and environmental cultures isolated from field samples taken at the bioaugmented site. The experiments conducted in this study inform on assay performance under both scenarios.
Compared to molecular methods for the detection of microorganisms, PMF by MALDI-TOF MS has a much lower sensitivity, which is not necessarily undesirable. At a bioremediation site where substantial amounts of biomass were introduced to accelerate degradation of pollutants, the use of extremely sensitive methods, such as PCR, may be of limited value. Although a few cells may trigger a positive result, their sparse presence will be inconsequential for the fate of pollutants at a contaminated site. The real question is whether there is enough of the enzyme to cause detectable biodegradation. To determine this, PMF of whole-cell preparations represents an attractive strategy. This was clearly demonstrated for cells maximally induced and moderately induced and for those repressed in dioxin dixoygenase expression (cells grown on DF, glucose plus DF, and LB broth, respectively). Additional benefits of the assay are its reproducibility and robustness and the potential for unattended high-throughput analysis during routine screening of environmental isolates.
The methodology demonstrated here for a dioxin-degrading bacterium may be extended to other microorganisms containing large quantities of characteristic proteins. Enzymes expressed in moderate quantities also may be suitable targets, as long as their corresponding peptides ionize favorably, similar to those of the alpha subunit of the dioxin dioxygenase. In summary, this study of strain RW1 suggests that PMF of whole-cell extracts may be a promising technique for the identification and phenotypic characterization of certain microbial pure cultures offering high-abundance proteins composed of peptides with favorable ionization behavior. Since the technique is inexpensive and may be automated, it could prove valuable in bioremediation and other areas of applied and environmental microbiology.
Acknowledgments
This project was supported by a seed grant from the Johns Hopkins Bloomberg School of Public Health Technology Transfer Committee. Support for E.S.W. was provided by NIH training grant T32ES07141. Training of D.R.C. in the use of MALDI mass spectrometers was made possible by a predoctoral fellowship from the Johns Hopkins Center for a Livable Future.
We thank Robert Cole and the friendly staff of the JHMI AB mass spectrometry facility for their technical assistance. We thank Peter Scholl for critiquing an early draft of the manuscript.
The technology described in this paper has pending patents and may be licensed for use.
REFERENCES
- 1.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, R. J., and J. P. Reilly. 1999. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 269:105-112. [DOI] [PubMed] [Google Scholar]
- 3.Barzaghi, D., J. D. Isbister, K. P. Lauer, and T. L. Born. 2004. Use of surface-enhanced laser desorption/ionization-time of flight to explore bacterial proteomes. Proteomics 4:2624-2628. [DOI] [PubMed] [Google Scholar]
- 4.Baumgart, S., Y. Lindner, R. Kuhne, A. Oberemm, H. Wenschuh, and E. Krause. 2004. The contributions of specific amino acid side chains to signal intensities of peptides in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 18:863-868. [DOI] [PubMed] [Google Scholar]
- 5.Bünz, P. V., and A. M. Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J. Bacteriol. 175:6467-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalcoli, J. D., R. A. VanBogelen, P. C. Andrews, and B. Moldover. 1997. Unique identification of proteins from small genome organisms: theoretical feasibility of high throughput proteome analysis. Electrophoresis 18:2703-2708. [DOI] [PubMed] [Google Scholar]
- 7.Cordwell, S. J., and I. Humphery-Smith. 1997. Evaluation of algorithms used for cross-species proteome characterisation. Electrophoresis 18:1410-1417. [DOI] [PubMed] [Google Scholar]
- 8.Demirev, P. A., Y. P. Ho, V. Ryzhov, and C. Fenselau. 1999. Microorganism identification by mass spectrometry and protein database searches. Anal. Chem. 71:2732-2738. [DOI] [PubMed] [Google Scholar]
- 9.Demirev, P. A., J. S. Lin, F. J. Pineda, and C. Fenselau. 2001. Bioinformatics and mass spectrometry for microorganism identification: proteome-wide post-translational modifications and database search algorithms for characterization of intact H. pylori. Anal. Chem. 73:4566-4573. [DOI] [PubMed] [Google Scholar]
- 10.Di Napoli, A., E. Maltese, M. Bucci, P. Pagnotti, J. Seipelt, S. Duquerroy, and R. Perez Bercoff. 2004. Molecular cloning, expression and purification of protein 2A of hepatitis A virus. New Microbiol. 27:105-112. [PubMed] [Google Scholar]
- 11.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 12.Gay, S., P. A. Binz, D. F. Hochstrasser, and R. D. Appel. 2002. Peptide mass fingerprinting peak intensity prediction: extracting knowledge from spectra. Proteomics 2:1374-1391. [DOI] [PubMed] [Google Scholar]
- 13.Halden, R. U. 1997. Engineered in situ biodegradation of dioxins and related compounds. Ph.D. thesis. University of Minnesota, Minneapolis.
- 14.Halden, R. U., R. N. Cole, C. Bradford, D. Chen, and K. J. Schwab. 2003. Presented at Exploring the Proteome II, Bethesda, Md., 1 and 2 May, 2003.
- 15.Halden, R. U., and D. F. Dwyer. 1997. Biodegradation of dioxin-related compounds: a review. Bioremed. J. 1:11-25. [Google Scholar]
- 16.Halden, R. U., B. G. Halden, and D. F. Dwyer. 1999. Removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soils inoculated with Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 65:2246-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halden, R. U., S. M. Tepp, B. G. Halden, and D. F. Dwyer. 1999. Degradation of 3-phenoxybenzoic acid in soil by Pseudomonas pseudoalcaligenes POB310(pPOB) and two modified Pseudomonas strains. Appl. Environ. Microbiol. 65:3354-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, W. A., and J. P. Reilly. 2002. On-probe digestion of bacterial proteins for MALDI-MS. Anal. Chem. 74:4410-4416. [DOI] [PubMed] [Google Scholar]
- 19.Hillenkamp, F., and M. Karas. 1990. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 193:280-295. [DOI] [PubMed] [Google Scholar]
- 20.Hong, H. B., Y. S. Chang, I. H. Nam, P. Fortnagel, and S. Schmidt. 2002. Biotransformation of 2,7-dichloro- and 1,2,3,4-tetrachlorodibenzo-p-dioxin by Sphingomonas wittichii RW1. Appl. Environ. Microbiol. 68:2584-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoving, S., M. Munchbach, H. Schmid, L. Signor, A. Lehmann, W. Staudenmann, M. Quadroni, and P. James. 2000. A method for the chemical generation of N-terminal peptide sequence tags for rapid protein identification. Anal. Chem. 72:1006-1014. [DOI] [PubMed] [Google Scholar]
- 22.James, P., M. Quadroni, E. Carafoli, and G. Gonnet. 1993. Protein identification by mass profile fingerprinting. Biochem. Biophys. Res. Commun. 195:58-64. [DOI] [PubMed] [Google Scholar]
- 23.Jarman, K. H., S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217-1223. [DOI] [PubMed] [Google Scholar]
- 24.Jungblut, P. R., D. Bumann, G. Haas, U. Zimny-Arndt, P. Holland, S. Lamer, F. Siejak, A. Aebischer, and T. F. Meyer. 2000. Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 36:710-725. [DOI] [PubMed] [Google Scholar]
- 25.Karas, M., and F. Hillenkamp. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60:2299-2301. [DOI] [PubMed] [Google Scholar]
- 26.Karty, J. A., M. M. Ireland, Y. V. Brun, and J. P. Reilly. 2002. Artifacts and unassigned masses encountered in peptide mass mapping. J. Chromatogr. B 782:363-383. [DOI] [PubMed] [Google Scholar]
- 27.Krause, E., H. Wenschuh, and P. R. Jungblut. 1999. The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal. Chem. 71:4160-4165. [DOI] [PubMed] [Google Scholar]
- 28.Lay, J. O., Jr. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 29.Loo, J. A., D. E. DeJohn, P. Du, T. I. Stevenson, and R. R. Ogorzalek Loo. 1999. Application of mass spectrometry for target identification and characterization. Med. Res. Rev. 19:307-319. [DOI] [PubMed] [Google Scholar]
- 30.Magnuson, M. L., J. H. Owens, and C. A. Kelty. 2000. Characterization of Cryptosporidium parvum by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 66:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marvin, L. F., M. A. Roberts, and L. B. Fay. 2003. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin. Chim. Acta 337:11-21. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, M., O. N. Jensen, E. Barofsky, D. F. Barofsky, and D. J. Reed. 1994. Thioredoxin alkylation by a dihaloethane-glutathione conjugate. Chem. Res. Toxicol. 7:659-665. [DOI] [PubMed] [Google Scholar]
- 33.Molloy, M. P., N. D. Phadke, J. R. Maddock, and P. C. Andrews. 2001. Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis 22:1686-1696. [DOI] [PubMed] [Google Scholar]
- 34.Pappin, D. J. C., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass finger printing. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 35.Powell, B. S., and D. L. Court. 1998. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J. Bacteriol. 180:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryzhov, V., Y. Hathout, and C. Fenselau. 2000. Rapid characterization of spores of Bacillus cereus group bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Appl. Environ. Microbiol. 66:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Thakur, I. S. 1996. Use of monoclonal antibodies against dibenzo-p-dioxin degrading Sphingomonas sp. strain RW1. Lett. Appl. Microbiol. 22:141-144. [DOI] [PubMed] [Google Scholar]
- 39.van Baar, B. L. 2000. Characterisation of bacteria by matrix-assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol. Rev. 24:193-219. [DOI] [PubMed] [Google Scholar]
- 40.Williams, T. L., D. Andrzejewski, J. O. Lay, and S. M. Musser. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 14:342-351. [DOI] [PubMed] [Google Scholar]
- 41.Wittich, R. M. 1998. Degradation of dioxin-like compounds by microorganisms. Appl. Microbiol. Biotechnol. 49:489-499. [DOI] [PubMed] [Google Scholar]
- 42.Wittich, R. M., H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, W. Z., K. W. Schramm, Y. Xu, and A. Kettrup. 2002. Contamination and distribution of polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/F) in agriculture fields in Ya-Er Lake area, China. Ecotoxicol. Environ. Saf. 53:141-147. [DOI] [PubMed] [Google Scholar]
- 44.Yao, Z. P., C. Afonso, and C. Fenselau. 2002. Rapid microorganism identification with on-slide proteolytic digestion followed by matrix-assisted laser desorption/ionization tandem mass spectrometry and database searching. Rapid Commun. Mass Spectrom. 16:1953-1956. [DOI] [PubMed] [Google Scholar]
- 45.Yao, Z. P., P. A. Demirev, and C. Fenselau. 2002. Mass spectrometry-based proteolytic mapping for rapid virus identification. Anal. Chem. 74:2529-2534. [DOI] [PubMed] [Google Scholar]