Abstract
Both magnetic collection and “race track” purification techniques were highly effective for selective enrichment of magnetotactic bacteria (MTB) from complex communities, as suggested by amplified ribosomal DNA restriction analysis and denaturing gradient gel electrophoresis combined with sequence analysis of 16S rRNA genes. Using these purification methods, the occurrence and diversity of MTB in microcosms from various marine and freshwater environments were assayed by using a combined microscopic, molecular, and cultivation approach. Most microcosms were dominated by magnetotactic cocci. Consistently, the majority of retrieved 16S RNA sequences were affiliated with a distinct cluster in the Alphaproteobacteria. Within this lineage the levels of sequence divergence were <1 to 11%, indicating genus-level diversity between magnetotactic cocci from various microcosms, as well as between MTB from different stages of succession of the same microcosms. The community composition in microscosms underwent drastic succession during incubation, and significant heterogeneities were observed between microcosms from the same environmental sources. A novel magnetotactic rod (MHB-1) was detected in a sediment sample from a lake in northern Germany by fluorescence in situ hybridization. MHB-1 falls into the Nitrospira phylum, displaying 91% 16S rRNA sequence similarity to “Magnetobacterium bavaricum.” In extensive cultivation attempts, we failed to isolate MHB-1, as well as most other MTB present in our samples. However, although magnetotactic spirilla were not frequently observed in the enrichments, 10 novel isolates of the genus Magnetospirillum which had not routinely been isolated in pure culture before were obtained.
Magnetotactic bacteria (MTB) are a heterogeneous group of aquatic microorganisms which share the ability to orient themselves along magnetic field lines. Magnetic orientation is due to the presence of magnetosomes, which are intracellular membrane-bound crystals of magnetic iron mineral which consist of magnetite or greigite (3, 28, 29). Diverse MTB, including cocci, spirilla, rods, vibrios, and multicellular aggregates, have been found in different aquatic habitats (5, 7, 17). Because of their high abundance and their remarkable potential to accumulate and precipitate iron minerals, MTB are assumed to have great impact in the biogeochemical cycling in natural sediments, which, however, has remained poorly understood. Most cultivated and uncultivated MTB have been affiliated with the Alphaproteobacteria. Moreover, the magnetotactic sulfate-reducing bacterium Desulfovibrio magneticus (14) and a magnetotactic, many-celled prokaryote (9) belong to the Deltaproteobacteria, while a giant magnetotactic rod, tentatively named “Magnetobacterium bavaricum,” is affiliated with the Nitrospira phylum (34). Recently, a greigite-producing rod belonging to the γ-Proteobacteria was identified (32). The phylogenetic and morphological diversity of MTB is matched by notable variation in the shapes, organizations, and numbers of magnetosome crystals found in diverse MTB (28). While reasonable progress was recently made in the genetic and biochemical analysis of magnetosome formation in a limited number of cultivated strains (for a review see reference 29), the intriguing diversity found in magnetosome biomineralization has remained almost entirely unexplored at the metabolic and genetic levels. This is due to the fact that most MTB have proven to be recalcitrant to isolation, and only a very few strains are available in pure cultures. Because of their unknown growth requirements and presumptive metabolic diversity, there is no general strategy known for conventional enrichment of MTB based on metabolic selection. Therefore, there is an urgent need for innovative methods to explore the vast diversity of uncultivated MTB.
However, in contrast to most other uncultivated bacteria, MTB can be physically enriched by taking advantage of their directed swimming behavior in magnetic field lines, which has been successfully used in a number of previous attempts to isolate several strains of MTB (4, 13, 15, 16, 30). In addition, specific enrichment of MTB is a prerequisite for metagenomic analysis, which is a highly promising approach for analysis of uncultivated MTB from natural environments. However, it is unclear how selective these enrichment techniques were with respect to the recovered diversity of MTB. In addition, as previous detection and isolation of MTB were from laboratory enrichments, how prolonged incubation of environmental samples affected the community structure and diversity of MTB was not addressed.
Therefore, the aim of this study was to thoroughly evaluate different magnetic enrichment methods for selectivity and efficiency in collection and purification of uncultivated MTB from environmental samples, which was done by combined molecular and microscopic characterization. In addition, we investigated the diversity and succession of MTB populations in various microcosms from different aquatic habitats by using a combination of a culture-independent 16S rRNA gene-based approach with extensive cultivation experiments.
MATERIALS AND METHODS
Sampling and setup of microcosms.
During March 2000 and April 2003 sediment samples from the upper sediment layer and surface water were taken from more than 50 different sites in Germany and Sweden. The samples were transferred to aquaria or glass or plastic bottles (0.1 to 2 liters), covered loosely, and incubated in these microcosms under low-light conditions or in the dark for several months. The occurrence of MTB was studied in various microcosms from freshwater, brackish, and marine habitats. From 12 microcosms (microcosms A to L) from nine different habitats MTB were collected for subsequent molecular analysis. Typically, the maximum numbers of MTB in the upper sediment layers of microcosms were between 9.7 × 105/cm3 and 1.5 × 107/cm3 (10). Four of these microcosms from different marine and freshwater sites in Germany were selected for more detailed investigation because of their abundant and diverse MTB populations (microcosm A from the Wadden Sea near Sahlenburg in the German Bight; microcosm B from a public swimming area in a lake in Bremen in northern Germany [Waller See]; microcosm C from a drainage ditch in Bremen; microcosm D from a freshwater pond near Staßfurt, Sachsen-Anhalt, Germany).
Magnetic collection of MTB.
Collection of MTB was performed essentially as described previously (17). Cells were enriched by attaching the south pole of a permanent magnet outside a jar 1 cm above the sediment surface. After 2 to 4.5 h 200 to 400 μl of the water near the south pole of the magnet was collected with a pipette.
“Race track” purification of MTB.
MTB were purified by the capillary “race track” (RT) method (41), which was slightly modified (30). Briefly, the narrow tip of a Pasteur pipette was sealed in a gas flame, and the capillary (length, 1 to 9 cm) was filled with sterilized habitat water using a long hypodermic needle. Sample material (sediment or magnetically collected cells) was placed on top of a sterile, wetted cotton plug in the wide-mouth end of the pipette, which served as a reservoir. The filled pipette was exposed to a magnetic field produced by a permanent magnet along the capillary. MTB migrated through the cotton plug toward the end of the capillary. After 30 to 165 min the tip containing the accumulated MTB was broken off. Using a sterile hypodermic needle, the MTB were removed and transferred into sterilized habitat water.
Phase-contrast and electron microscopy.
The swimming behavior and cell morphology of MTB were investigated by the “hanging drop” method (27) using a phase-contrast microscope (Zeiss). The arrangement and morphology of the magnetosomes were analyzed by transmission electron microscopy. For transmission electron microscopy, cells were adsorbed onto 300-mesh Formvar-coated copper grids (Plano) and analyzed without staining. Alternatively, cells were negatively stained using 4% uranyl acetate. The samples were examined with an EM 301 transmission electron microscope (Philips) at 80 kV.
DNA extraction.
The DNA from sediment samples was extracted by the method of Zhou et al. (43), as modified by Sievert et al. (31). The DNA was dissolved in 100 μl PCR water.
PCR amplification.
For denaturing gradient gel electrophoresis (DGGE) the bacterial 16S rRNA genes were amplified with primers GM5F with a GC clamp and 907R (18) by using the MasterTaq system from Eppendorf or the RedTaq system from Sigma. The PCR mixtures were prepared according to the manufacturer's instructions with bovine serum albumin (final concentration, 0.3 mg/ml; Fluka) using the MasterTaq system and 4% (vol/vol) enhancer. Isolated DNA or magnetically collected cells were used as the template. The touchdown PCR was initiated by a heating step at 94°C. When the RedTaq system was used, the PCR mixture was cooled to 80°C, and Taq was added. In the following cycles, the temperature was decreased continuously at 1°C after two cycles from 65°C to 56°C. The PCR was finished with 19 cycles at 55°C. For cloning, nearly complete 16S rRNA genes were amplified using the RedTaq PCR system with the universal bacterial primers GM3F and GM4R (18). PCR was performed by using 33 cycles at an annealing temperature of 42°C. The inserts from clones were amplified using MasterMix from Promega and vector-specific primers. The PCR was performed for 35 cycles at an annealing temperature of 60°C. All PCR products were checked by electrophoresis in an agarose gel and by ethidium bromide staining.
DGGE.
DGGE was performed using the D-Gene system (Bio-Rad Laboratories), and the DNA fragments were separated in a 1-mm-thick polyacrylamide gel (6%, wt/vol) with a 20 to 70% denaturant gradient and 1× TAE electrophoresis buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA; pH 8.3) at 60°C. After 16 h of electrophoresis at a constant voltage of 100 V, the gel was stained with ethidium bromide. DNA bands were excised with a sterile scalpel and eluted in 100 μl PCR water overnight at 4°C. Aliquots of the eluates were reamplified with GM5F and 907R and were purified using an agarose gel and a Quiaquick gel extraction kit (QIAGEN). The purified DNA was used for sequencing.
Amplified ribosomal DNA restriction analysis (ARDRA).
The amplified DNA was cloned into a pCR2.1-TOPO or pCR4-TOPO vector and transformed into competent Escherichia coli cells (TOP10 One Shot [Invitrogen], DH5alpha [GIBCO BRL], or XL-10 Gold [Stratagene]). The clones were analyzed by PCR (see above). The PCR products from more than 60 positive clones were restricted with HindIII and RsaI (final activity, 0.5 U/μl) for 3 h at 37°C and were separated in a 3.5% agarose gel. Plasmids from clones with various band patterns were isolated with a Quiaprep Spin miniprep kit from QIAGEN and sequenced.
Sequencing.
The DNA was sequenced with the primers described above or vector-specific primers and GM1 (5′-CCAGCAGCCGCGGTAAT-3′) with a capillary sequencer (Applied Biosystems/Hitachi 3100 genetic analyzer).
Phylogenetic analysis.
The partial and full-length 16S rRNA gene sequences retrieved in this study were added to an internal ARB data set containing approximately 31,500 partial and full-length small-subunit rRNA sequences, based on the database of the Technical University of Munich (June 2002 release). The tool ARB EDIT was used for automatic sequence alignment. The alignment was checked by eye and corrected manually. The phylogenetic tree presented below was reconstructed based on maximum-likelihood analysis of selected full-length sequences affiliated with known magnetotactic bacteria, the full-length 16S rRNA sequences generated in this study, and all sequences within the tree “tree-demo” of the public ARB data set from June 2002. The tree topology was evaluated by maximum-parsimony and neighbor-joining analyses of the full data set in combination with different filters excluding highly variable positions. A consensus tree was constructed by taking into consideration the results obtained by applying the various tree reconstruction methods. Discrepancies in the different reconstruction methods are indicated by multifurcations. Partial 16S rRNA gene sequences obtained by DGGE analysis were added to the tree using the “Parsimony quick add” tool of ARB.
Fluorescence in situ hybridization.
Cells of a magnetotactic rod (MHB-1) were magnetically collected and fixed with paraformaldehyde as described by Pernthaler et al. (21). Cells were then embedded in agarose (final concentration, 0.02%) and fixed on glass slides (Amann type; Paul Marienfelder KG). A second fixation (dehydration) was performed by applying increasing concentrations of ethanol (50, 80, and 96%). The dried samples were hybridized with a specific Cy3-labeled probe for “M. bavaricum” (34) for 90 min at 46°C without any formamide. The fluorescein-labeled probe EUB338 (2) and the non-EUB probe (38) were used as controls. The samples were washed without EDTA for 20 min at 48°C, rinsed with water (Millipore), and stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma). The samples were washed with H2O and 70% ethanol. After drying they were embedded in Citifluor (Citifluor Products) and investigated with a Zeiss fluorescence microscope.
Cultivation experiments.
Various minimal and complex media with 0 to 0.8% agar were used for cultivation experiments. The medium constituents used as potential electron donors included reduced sulfur compounds and a variety of organic compounds, such as glucose, amino acids, lactate, malate, succinate, alpha-ketogluterate, citrate, pyruvate, acetate, formate, propionate, butyrate, isobutyrate, 2-methylbutyrate, valerate, and capronate, which were added either individually or as mixtures as sole carbon or energy sources. Different potential electron acceptors, including oxygen, nitrate, N2O, and sulfate, were added in different combinations and at different concentrations. In addition, several media were used in combination with oxygen-sulfide gradients (19, 30). Ferric quinate and ferric citrate were added as iron sources. Sulfide and cysteine were used as reducing agents, and resazurine was used as a redox indicator in most media. Most media were supplemented with various vitamin cocktails, yeast extracts, peptones, or sediment extract. The pH was between 6.8 and 7.2. Twenty-milliliter Hungate tubes were filled with 10 ml media and closed with screw caps. Anaerobic media were prepared by the method of Widdel and Bak (40). For anaerobic or microaerobic growth the gas phase of the tubes was replaced by different gas mixtures having various concentrations of O2, N2, N2O, and CO2, and the tubes were sealed with butyl stoppers. Inocula of purified MTB were applied to the media in dilutions down to 10−6. All tubes were incubated at room temperature under low-light conditions. Growth was analyzed by visual screening (colony or band formation) and microscopy. For further cultivation of isolated magnetic spirilla the medium contained (per liter) 0.068 g KH2PO4, 0.108 g MgSO4 · 7H2O, 0.1 g NH4Cl, 0.166 g sodium succinate, 2 ml ferric citrate (10 mM), 5 ml mineral solution (42), and 0.2% Noble agar (Difco). The medium was adjusted to pH 7.0 and autoclaved. Further details regarding the culture conditions will be provided on request.
Nucleotide sequence accession numbers.
The nucleotide sequences of partial 16S rRNA genes of MTB have been deposited in the GenBank, EMBL, and DDJB libraries under accession numbers AJ863135 to AJ863158.
RESULTS
Magnetic collection and enrichment of MTB.
In order to find optimal conditions for collection of MTB, we first evaluated two different magnetic enrichment techniques with respect to recovered cell numbers and selectivity. Magnetic collection from the water columns of several microcosms yielded a visible pellet of accumulated MTB close to the south pole of the magnet, which was equivalent to 107 to 108 magnetotactic cells. Typically, up to 100 ng genomic DNA could be obtained from this amount of cells. Small magnetotactic crustaceans (Ostracoda) apparently grazing on MTB were frequently observed among collected cells. Several magnetic morphotypes, which could be observed in samples collected directly from the sediment, were reluctant to swim out of the sediment, even if the water column was rendered anoxic by sparging with nitrogen. Repeated collections (approximately 5 to 10) within several days resulted in increasing depletion of magnetotactic cells. In the example shown in Fig. 1, a DGGE profile of magnetically collected cells (lane M) revealed two distinct bands (bands a and b), which were consistently obtained and yielded sequences with high similarity to sequences of uncultivated magnetic cocci belonging to the Alphaproteobacteria from the database. The sequence of band b matched the sequence of a weak band in the complex DGGE patterns obtained from DNA extracted from the upper sediment layer (lane S) (depth, approximately 0 to 5 mm), indicating that the collected MTB were among the detectable species in this layer. DGGE and ARDRA of magnetically collected MTB resulted in identical results (not shown).
FIG. 1.
DGGE profiles of amplified 16S rRNA gene fragments from microcosm D. Lane S, isolated DNA from the upper sediment layer; lane M, cells obtained by magnetic collection; lane RT, cells obtained by magnetic collection followed by RT purification. Bands that were excised for reamplification and sequencing are indicated (bands a and b). All attempts to analyze additional weak bands, which were observed only sporadically after magnetic collection and RT purification, resulted in a failure to reamplify or yielded only poor sequences.
Using the RT method, MTB could be enriched to virtual homogeneity, as shown by microscopy, and were visible as a pellet at the tip of the capillary. Nonmagnetic cells were present only occasionally and in very low numbers (estimated concentration, <0.01%), as revealed by microscopy. ARDRA and DGGE combined with 16S rRNA gene sequence analysis of RT-purified MTB confirmed their identity with the MTB obtained by magnetic collection (Fig. 1 and 2). Approximately 105 to 107 cells could be obtained per run. All attempts to scale up this method did not result in significantly increased cell numbers. Only addition of a reductant (200 μM sodium cysteine) to the water within the capillary seemed to increase the yield of MTB. With a running distance of 9 cm, visible amounts of MTB reached the tip of the capillary after 10 min. This is equivalent to a migration speed of about 150 μm/s, and in most cases the major fraction of MTB arrived at the tip after 30 min. The variations in running times, conditions, and distances had no marked effect on the selectivity of magnetic collection and RT purification, as indicated by identical DGGE band patterns (Fig. 2).
FIG. 2.
DGGE band patterns for amplified 16S rRNA gene fragments of microcosm C obtained after repeated collection under various enrichment conditions. Unless indicated otherwise, the running length of the capillary was 1 cm for RT purification. Lane 1, 2 h of magnetic collection; lane 2, 2 h of magnetic collection plus 0.5 h of RT purification; lane 3, 2 h of magnetic collection plus 3 h of RT purification; lane 4, 0.5 h of RT purification from sediment sample; lane 5, 2 h of magnetic collection under anoxic conditions (i.e., the water column of the microcosm was stripped of oxygen by sparging with N2 prior to and during magnetic collection); lane 6, 2 h of magnetic collection plus 0.5 h of RT purification in the presence of 0.2 mM cysteine; lane 7, 2 h of magnetic collection plus 0.5 h of RT purification (capillary length, 9 cm).
Diversity and succession of MTB in microcosm experiments.
In a survey of more than 150 samples from marine coastal and freshwater habitats, MTB were detected by microscopy in the majority of samples. We failed to detect MTB in wet soil samples and in sediments from highly eutrophic habitats, including swine waste pits and sewage sludges. Conspicuous magnetotactic multicellular aggregates were observed in several marine samples from the Wadden Sea (Sahlenburg) and sediment cores from the Baltic Sea (Eckernförder Bight), and these aggregates strongly resembled those that were previously described as magnetotactic many-celled prokaryotes by Rodgers et al. (24). Most samples from different sites were dominated in terms of numbers by various morphotypes of MTB, such as cocci, rods, vibrios, and spirilla (Fig. 3).
FIG. 3.
Selected electron micrographs of different MTB collected from various habitats. The morphotypes observed include various cocci (A to C), spirilla (D), and vibroid to rod-shaped cells (E to H). The black arrows indicate magnetosomes; the arrowheads indicate insertion of flagellum bundles. Bars = 0.5 μm. A conspicuous intracellular structure (grey arrow) was occasionally observed in a large magnetotactic vibrio (G); this structure consisted of two electron-dense ovoid spheres at the cell poles that were connected by a spine-like structure.
Most of the sequences obtained from magnetically collected cells were closely related to previously identified uncultivated magnetotactic cocci in the Alphaproteobacteria (Fig. 4, cluster I), with levels of sequence divergence between 0.1 and 11%. Sequences unrelated to known MTB from the database were occasionally obtained from magnetic collections and often corresponded to irreproducible bands in DGGE gels. Interestingly, several of these sequences were affiliated with the Rhodospirillaceae but were not closely related to Magnetospirillum species. Although we cannot fully exclude the possibility that some of them potentially represent unknown phylogenetic lineages of MTB, they were attributed to nonmagnetic bacteria present in these samples.
FIG. 4.
Phylogenetic tree for magnetotactic bacteria based on comparative sequence analysis of 16S rRNA genes. Bar = 10% estimated sequence divergence. If identical 16S rRNA gene sequences from both DGGE and ARDRA from the same microcosms were available, only the clone sequences were included. Sequence differences that were less than 0.3% for isolates from the same habitat were not itemized. The sequences determined in this study are indicated by boldface type, and the date of magnetic collection is indicated. The shaded branches indicate multiple sequences that were retrieved from a single sample (microcosm D) after collection at different times. The asterisks indicate sequences obtained from marine habitats.
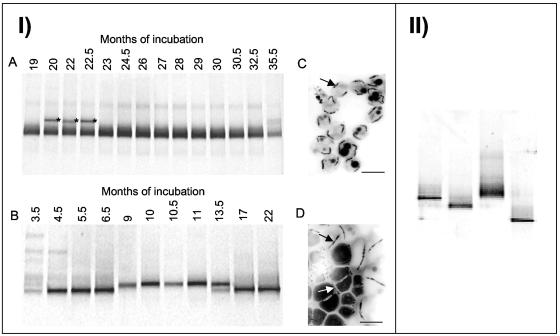
Generally, freshwater microcosms underwent a characteristic succession upon prolonged incubation. We often noticed an increase in the total numbers of MTB cells and a rapid change in morphological diversity, as revealed by microscopy. Therefore, we were interested in whether the compositions of the populations of MTB in our aged microcosms remained stable after the mass development of magnetotactic cocci, which occurred in most microcosms after several days to months. Figure 5IA and B shows the DGGE profiles of magnetically collected cells from two microcosms over several months of incubation. In microcosm C, the same phylo- and morphotypes (Fig.5IC) were persistently detected for 16 months. In contrast, a shift in the detectable MTB species was indicated by divergent community profiles in DGGE profiles from microcosm D, while the collected magnetotactic cells displayed virtually the same morphology (cocci) over many months, as revealed by light microscopy. Nevertheless, detailed analysis by electron microscopy showed the presence of two different morphotypes of magnetotactic cocci in a sample (Fig.5ID). Sequences obtained from different times displayed up to 7% divergence.
FIG. 5.
(I) DGGE community profiles of amplified 16S rRNA gene fragments from microcosms C (A) and D (B) collected at different times during incubation. The asterisks indicate bands which yielded sequences unrelated to known MTB. (C and D) Electron micrographs of magnetotactic cocci from microcosms C (C) and D (D). The arrows indicate magnetosomes. Bars = 1 μm. (II) DGGE profiles of amplified 16S rRNA gene fragments obtained from magnetically collected cells from four microcosm samples, which were simultaneously taken from a freshwater pond (Staßfurt, Germany) and incubated in the laboratory for 22 months under identical conditions.
We also noticed that stored samples from the same location sometimes contained different morphotypes of MTB. Therefore, four aliquots of the same sample taken from a single site were incubated for 22 months under identical conditions in the lab. All four microcosms developed populations of abundant magnetic cocci, which displayed virtually the same morphology. However, DGGE analysis clearly revealed the presence of different MTB phylotypes in every microcosm (Fig.5II), indicating that the development of an MTB population was not reproducible and apparently depended on very subtle differences in physicochemical conditions between the microcosms.
MHB-1 is a novel magnetotactic rod affiliated with the Nitrospira phylum.
In one microcosm (microcosm B), the MTB population was found to be dominated (estimate, >50% of the total MTB number) after 19 months of incubation by a small (length, 2 to 3 μm; width, 1 μm), slowly moving magnetotactic rod. Sequence analysis of a 16S rRNA gene clone obtained from magnetically collected cells revealed high similarity (91%) to “M. bavaricum,” which is affiliated with the Nitrospira phylum (34) (Fig. 4, cluster III). In order to verify the identity of the clone, we used the “M. bavaricum” probe previously described by Spring et al. (34), which perfectly matched the target sequence of the clone. In fluorescence in situ hybridization experiments, both the EUB338 probe (eubacteria) and the “M. bavaricum” probe recognized cells apparently identical to slowly moving magnetotactic rods, which were designated MHB-1 (Fig. 6A to C). Electron micrographs of MHB-1 cells revealed magnetosome crystals, which were aligned in multiple chains and had the same bullet-shaped morphology as those from “M. bavaricum” (Fig. 6D). However, unlike the several hundred magnetosome particles organized in three to five bundles of chains which were present in the giant cells of “M. bavaricum” (11), MHB-1 cells contained only 30 to 60 magnetosomes in a single bundle.
FIG. 6.
(A to C) Cells of MHB-1 that were magnetically collected from microcosm B (Waller See, Bremen, Germany): DAPI-stained cells (A) and cells hybridized with bacterial probe EUB338 (B) and a probe for “M. bavaricum” (C). (D) Electron micrograph of MHB-1. Bars = 1 μm. The arrow indicates magnetosomes.
Cultivation experiments.
In extensive cultivation experiments, a multitude of different growth media were used in an attempt to isolate heterotrophic, autotrophic, or mixotrophic MTB. Typically, 105 to 107 viable (motile) MTB obtained by magnetic collection or RT purification were used as the inoculum in an experiment and were added either directly or in 10-fold dilution steps. If inocula were prepared by magnetic collection, growth of nonmagnetic contaminants was observed occasionally in dilutions up to 10−4. In contrast, RT-purified samples only very infrequently gave rise to contaminant growth, indicating that this purification method was strictly selective for MTB.
In numerous repeated experiments, we failed to detect growth of magnetotactic cocci, which were very abundant in the inoculum. Neither the addition of complex supplements or redox-active compounds nor incubation with different oxygen levels or in oxygen-sulfide gradients stimulated the growth of these MTB. However, in slush agar cultures, which were inoculated with MTB from three different sampling sites (Ihle, Staßfurt, and Bremen), growth of magnetotactic bacteria was repeatedly detected by establishment of a distinct microaerophilic plate of bacteria after several days of incubation. Microscopic examinations revealed the presence of magnetic spirilla, which initially displayed a polar north-seeking motility that was lost upon repeated transfer. While several of these cultures contained mixed populations with nonmagnetic contaminants and could not be purified further, 10 isolates were obtained in pure cultures from various complex and minimal media containing a mixture of different organic substrates. Most strains were obtained from media with oxygen-sulfide gradients (30). All isolates were microaerophiles which grew heterotrophically on minimal media on succinate and had a morphology very similar to that of previously isolated magnetotactic spirilla (Fig. 7). Phylogenetic analysis resulted in affiliation of all of them with the genus Magnetospirillum, with the greatest similarity to either MSM-6 (30) or Magnetospirillum gryphiswaldense (25) (Fig. 4, cluster II). Levels of sequence divergence of up to 2.4% were found between isolates.
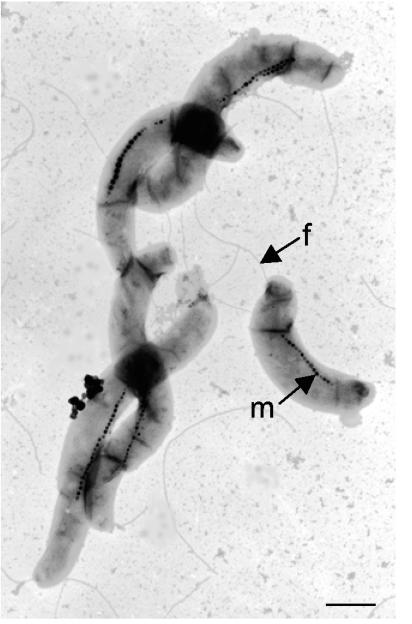
FIG. 7.
Electron micrograph of a representative of the isolated Magnetospirillum strains. Cells were negatively stained with uranyl acetate. Other strains isolated in this study had identical morphology. M, magnetosomes; f, flagellum. Bar = 0.5 μm.
DISCUSSION
The active migration of MTB along magnetic field lines provides a unique tool for detection, manipulation, and enrichment of these organisms from environmental sources. Our study demonstrated by molecular analysis that uncultivated MTB can be selectively separated from sediment particles and nonmagnetic bacteria by magnetic collection and RT purification. While only 105 to 107 cells could be recovered by RT purification in each run, significantly larger amounts (up to 108 cells) could be obtained by magnetic collection from the water columns of several microcosms. The total yield of harvested MTB could be increased by repeated collection but hardly reached 109 MTB due to increasing depletion of the microcosms. Although the collected cells still contained rare contaminants, they were sufficiently enriched for isolation of high-molecular-weight genomic DNA (data not shown). However, both methods were biased for fast-swimming MTB and did not fully retrieve the diversity of MTB observed by direct microscopy of sediment samples. Instead, both ARDRA and DGGE analysis revealed low diversity within the MTB population collected. Both methods can be influenced by differential amplification of the 16S rRNA genes during the PCR and give no reliable data concerning the abundance of the organism(s) detected in the sample (23, 35).
Ubiquitous occurrence and great diversity of MTB, including previously unknown morpho- and phylotypes, were found in a survey of numerous aquatic habitats. 16S rRNA gene analysis showed that all MTB from this study were affiliated with known MTB belonging to either two phylogenetic clusters within the Alphaproteobacteria (clusters I and II) or the Nitrospira phylum (cluster III). Nevertheless, we cannot exclude the possibility that some of the sequences, which do not correspond to known MTB, represent unknown phylogenetic lineages of MTB, which requires further investigation. The general dominance of magnetotactic cocci in most of our samples may have been a consequence of the selective enrichment conditions in our microcosms, which seemed to favor the mass development of single or a few magnetotactic species at the expense of diversity. The occurrence of closely related phylotypes of magnetotactic cocci in single samples may represent either microdiversity of MTB or the presence of different 16S rRNA gene operons of a single species, which has been demonstrated for many other bacteria (1, 20, 22, 37). Nevertheless, the high levels of sequence divergence (up to 11%) indicate that MTB with this morphotype seem to fall into different genera. On the other hand, samples from different sites contained nearly identical phylotypes, while sequences retrieved from a single sample often displayed remarkable heterogeneity. This indicates that the correlation between the phylogenetic diversity and the geographical and geochemical heterogeneities of microcosms is not obvious.
In light of the considerable diversity observed in different environmental samples, an important question is, which environmental factors control the occurrence and development of different MTB under natural conditions and in the microcosms stored under laboratory conditions? Consistent with earlier reports (17), neither of our attempts to amend the microcosms with various electron donors and acceptors, such as iron [Fe0 or Fe(III)], sulfate, or nitrate, under aerobic or anaerobic growth conditions resulted in increased numbers or increased diversity of MTB based on metabolic selection (data not shown). We found that smaller microcosms were less likely to develop stable MTB populations than larger microcosms. As we have previously shown in a different study that the occurrence of MTB is closely associated with various vertical geochemical gradients (10), this is probably because of the instability of these gradients within the small microcosm systems. Parallel samples originating from the same site frequently developed populations of MTB which were dominated by different phylotypes. Likewise, aged microcosms that had developed apparently stable populations still exhibited long-term dynamics characterized by either sporadic collapses and subsequent recovery of the whole MTB population or temporary shifts towards different, yet morphologically similar species. This suggests that MTB are strictly adapted to particular microniches and even subtle change in the environmental conditions may affect the community structure of MTB.
Only one uncultivated magnetotactic member of the Nitrospira phylum was previously identified and was named “M. bavaricum.” This large magnetotactic rod was detected in the sediments of various lakes in Upper Bavaria (Germany) (34). Our observation of MHB-1 in a sediment from a lake in northern Germany indicates that the occurrence of MTB belonging to this lineage is not restricted to oligotrophic Bavarian lakes. Although the presence of bullet-shaped magnetosomes seems to be a common trait of MTB belonging to the Thermodesulfovibrio-“Magnetobacterium” lineage of the Nitrospira phylum (8), there is significant morphological and phylogenetic diversity within this magnetotactic group. Whereas isolation and cultivation-independent studies have identified Nitrospira-like bacteria as nitrite oxidizers (8, 26, 39), cultivation experiments with Thermodesulfovibrio yellowstonii and Thermodesulfovibrio islandicus, which represent the closest cultured relatives of MHB-1, have demonstrated their ability to reduce oxidized sulfur compounds (12, 33). In addition, in “M. bavaricum” and a closely related nonmagnetic bacterium from deep-sea hydrothermal systems the oxidation of sulfide as energy source was implicated (34, 36). Further studies are required to analyze if these metabolic traits are present in uncultivated MTB belonging to the Nitrospira phylum.
Although magnetic cocci were highly abundant both in the sediment of microcosms and in our purified magnetic enrichments, they failed to grow with a multitude of the cultivation media and conditions tried. Although magnetotactic spirilla were not abundant and could not be detected by DGGE, they were repeatedly isolated on simple media preferentially containing opposing gradients of oxygen and sulfide (30). However, the cultivated diversity of magnetotactic spirilla seems to be confined to species of the genus Magnetospirillum, which in physiological and morphological traits strongly resemble previously isolated species (6, 13, 25, 30).
In summary, our study demonstrated that despite extensive attempts only a small fraction of MTB is amenable to cultivation by conventional methods. Beside detailed geochemical analysis of microhabitats, which may lead to improved isolation strategies, innovative approaches are required, which should aim to reconstruct the complex chemical gradients found in stratified sediments. On the other hand, our data demonstrate that several MTB can be propagated in lab-scale microcosms, from which they can be readily collected in high numbers and enriched to obtain virtually monospecific populations. In addition to the PCR-based RNA approach, genomic DNA could be isolated from those collected cells in amounts sufficient for the construction of large-insert genomic libraries (A. Meyerdierks, personal communication). Thus, cloning and analysis of the genome of single magnetotactic species, or ultimately the “magnetotactic metagenome,” seem to be one of the most powerful approaches for obtaining further insight into the genetic diversity of magnetosome biomineralization within uncultivated MTB.
Acknowledgments
We are grateful to Frank Oliver Glöckkner and Rudolf Amann, MPI Bremen, for help with the phylogenetic analysis and valuable discussions. We thank Frank Mayer and Michael Hoppert, University Göttingen, for providing access to the electron microscope.
This work was supported by the BMBF and the Max Planck Society.
REFERENCES
- 1.Amann, G., K. O. Stetter, E. Llobet-Brossa, R. Amann, and J. Anton. 2000. Direct proof for the presence and expression of two 5% different 16S rRNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373-376. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., R. B. Frankel, and H. W. Jannasch. 1988. Anaerobic magnetite production by a marine magnetotactic bacterium. Nature 334:518-519. [Google Scholar]
- 5.Blakemore, R. P. 1975. Magnetotactic bacteria. Science 190:377-379. [DOI] [PubMed] [Google Scholar]
- 6.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, L., R. Popa, D. Bazylinsky, B. Lanoil, S. Douglas, A. Belz, D. Engler, and K. H. Nealson. 2002. Organization and elemental analysis of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiol. J. 19:387-406. [Google Scholar]
- 8.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLong, E. F., R. B. Frankel, and D. A. Bazylinski. 1993. Multiple evolutionary origins of magnetotaxis in bacteria. Science 259:803-806. [DOI] [PubMed] [Google Scholar]
- 10.Flies, C., H. Jonkers, D. deBeer, K. Bosselmann, M. Böttcher, and D. Schüler. 2005. Diversity and vertical distribution of magnetotactic bacteria along chemical gradients in freshwater microcosms. FEMS Microbiol. Ecol. 52:185-195. [DOI] [PubMed] [Google Scholar]
- 11.Hanzlik, M., M. Winklhofer, and N. Petersen. 2002. Pulsed-field-remanence measurements on individual magnetotactic bacteria. J. Magn. Magn. Mater. 248:258-267. [Google Scholar]
- 12.Henry, E. A., R. Devereux, J. S. Maki, C. C. Gilmour, C. R. Woese, L. Mandelco, R. Schauder, C. C. Remsen, and R. Mitchell. 1994. Characterization of a new thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol. 161:62-69. [PubMed] [Google Scholar]
- 13.Kawaguchi, R., J. G. Burgess, and T. Matsunaga. 1992. Phylogeny and 16S rRNA sequence of Magnetospirillum sp. AMB-1, an aerobic magnetic bacterium. Nucleic Acids Res. 20:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi, R., J. G. Burgess, T. Sakaguchi, H. Takeyama, R. H. Thornhill, and T. Matsunaga. 1995. Phylogenetic analysis of a novel sulfate-reducing magnetic bacterium, RS-1, demonstrates its membership of the delta-Proteobacteria. FEMS Microbiol. Lett. 126:277-282. [DOI] [PubMed] [Google Scholar]
- 15.Meldrum, F. C., S. Mann, B. R. Heywood, R. B. Frankel, and D. A. Bazylinski. 1993. Electron microscopy study of magnetosomes in a cultured coccoid magnetotactic bacterium. Proc. R. Soc. Lond. Ser. B Biol. Sci. 251:231-236. [Google Scholar]
- 16.Meldrum, F. C., S. Mann, B. R. Heywood, R. B. Frankel, and D. A. Bazylinski. 1993. Electron microscopy study of magnetosomes in two cultured vibrioid magnetotactic bacteria. Proc. R. Soc. Lond. Ser. B Biol. Sci. 251:237-242. [Google Scholar]
- 17.Moench, T. T., and W. A. Konetzka. 1978. A novel method for the isolation and study of a magnetotactic bacterium. Arch. Microbiol. 119:203-212. [DOI] [PubMed] [Google Scholar]
- 18.Muyzer, G., A. Teske, and C. O. Wirsen. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 19.Nelson, D. C., and H. W. Jannasch. 1983. Chemoautotrophic growth of a marine Beggiatoa in sulfide-gradients cultures. Arch. Microbiol. 136:262-269. [Google Scholar]
- 20.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 22.Rainey, F. A., N. L. Ward-Rainey, P. H. Janssen, H. Hippe, and E. Stackebrandt. 1996. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology 142:2087-2095. [DOI] [PubMed] [Google Scholar]
- 23.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodgers, F. G., R. P. Blakemore, N. A. Blakemore, R. B. Frankel, D. A. Bazylinski, D. Maratea, and C. Rodgers. 1990. Intercellular structure in a many-celled magnetotactic prokaryote. Arch. Microbiol. 154:18-22. [Google Scholar]
- 25.Schleifer, K. H., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst.Appl. Microbiol. 14:379-385. [Google Scholar]
- 26.Schramm, A., D. De Beer, J. C. Van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 28.Schüler, D. 1999. Formation of magnetosomes in magnetotactic bacteria. J. Mol. Microbiol. Biotechnol. 1:79-86. [PubMed] [Google Scholar]
- 29.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1-7. [DOI] [PubMed] [Google Scholar]
- 30.Schüler, D., S. Spring, and D. A. Bazylinski. 1999. Improved technique for the isolation of magnetotactic spirilla from a freshwater sediment and their phylogenetic characterization. Syst. Appl. Microbiol. 22:466-471. [DOI] [PubMed] [Google Scholar]
- 31.Sievert, S. M., T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmons, S. L., S. M. Sievert, R. B. Frankel, D. A. Bazylinski, and K. J. Edwards. 2004. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 70:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonne-Hansen, J., and B. K. Ahring. 1999. Thermodesulfobacterium hveragerdense sp. nov., and Thermodesulfovibrio islandicus sp. nov., two thermophilic sulfate reducing bacteria isolated from a Icelandic hot spring. Syst. Appl. Microbiol. 22:559-564. [DOI] [PubMed] [Google Scholar]
- 34.Spring, S., R. Amann, W. Ludwig, K.-H. Schleifer, H. Van Gemerden, and N. Petersen. 1993. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, Y., F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2004. Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb. Ecol. 47:186-196. [DOI] [PubMed] [Google Scholar]
- 37.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindstrom, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of Rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 39.Watson, S. W., E. Bock, F. W. Valois, J. B. Waterbury, and U. Schlosser. 1986. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol. 144:1-7. [Google Scholar]
- 40.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, W. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, Berlin Germany.
- 41.Wolfe, R. S., R. K. Thauer, and N. Pfennig. 1987. A capillary racetrack method for isolation of magnetotactic bacteria. FEMS Microbiol. Ecol. 45:31-36. [Google Scholar]
- 42.Wolin, E. A., M. J. Wolin, and R. S. Wolfe. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882-2886. [PubMed] [Google Scholar]
- 43.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]