Abstract
The effects of sealing infected carious dentine below dental restorations on the phenotypic and genotypic diversity of the surviving microbiota was investigated. It was hypothesized that the microbiota would be subject to nutrient limitation or nutrient simplification, as it would no longer have access to dietary components or salivary secretion for growth. The available nutrients would be limited primarily to serum proteins passing from the pulp through the patent dentinal tubules to the infected dentine. Ten lesions were treated, and infected dentine was sealed below dental restorations for approximately 5 months. Duplicate standardized samples of infected dentine were taken at baseline and after the removal of the restorations. The baseline microbiota were composed primarily of Lactobacillus spp., Streptococcus mutans, Streptococcus parasanguinis, Actinomyces israelii, and Actinomyces gerencseriae. None of these taxa were isolated among the microbiota of the dentine samples taken after 5 months, which consisted of only Actinomyces naeslundii, Streptococcus oralis, Streptococcus intermedius, and Streptococcus mitis. The microbiota of the final sample exhibited a significantly (P < 0.001) increased ability to produce glycosidic enzymes (sialidase, β-N-acetylglucosaminidase, and β-galactosidase), which liberate sugars from glycoproteins. The genotypic diversity of S. oralis and A. naeslundii was significantly (P = 0.002 and P = 0.001, respectively) reduced in the final samples. There was significantly (P < 0.001) greater genotypic diversity within these taxa between the pairs of dentine samples taken at baseline than was found in the 5-month samples, indicating that the dentine was more homogenous than it was at baseline. We propose that during the interval between placement of the restorations and their removal, the available nutrient, primarily serum proteins, or the relative simplicity and homogeneity of the nutrient supply significantly affected the surviving microbiota. The surviving microbiota was less complex, based on compositional, phenotypic, and genotypic analyses, than that isolated from carious lesions which were also exposed to salivary secretions and pH perturbations.
The survival of bacteria in the mouth and in the oral biofilm, dental plaque, in particular, depends on the ability of the adherent biota obtaining nutrients from their immediate environment and being resistant to fluctuating environmental acid and nutrient stresses (3, 31). Dental plaque rapidly ferments dietary carbohydrates to acids, reversibly demineralizing the underlying enamel, which may ultimately develop into a carious lesion. The fluctuating nature of these acid exposures has significant effects on the microbiota of the oral biofilm (25). Perhaps the best documented are the increased representation of yeasts, lactobacilli, and mutans streptococci in dental plaque and saliva, especially of individuals with high caries scores and xerostomia (2, 20, 21). Other investigators have studied the effects of acids on the oral flora and demonstrated that other members of the oral biofilm are also affected. The nonmutans streptococci from the oral biofilm of individuals with caries produce more acid than similar strains from caries-free subjects, indicating that these species are heterogeneous with respect to this phenotype (32, 34). We have also reported the presence of distinct genotypes of Streptococcus oralis capable of growth at low pH (1) and have demonstrated that S. oralis strains from the plaque of caries-active subjects were more diverse than strains from caries-free subjects but Actinomyces naeslundii isolates were no more diverse (28). While the effect of acid stress has been well studied, the effect of nutrient limitation on the composition of dental plaque has received less attention. Previously, we investigated the effects of fasting on the microbiota of macaque monkeys and found that the only taxon to alter was “S. mitior” (now S. oralis), which increased in proportion sevenfold (3). When similar studies were undertaken in bone marrow transplant patients and liver transplant patients, we found that the proportions and numbers of S. oralis and Streptococcus mitis, two closely related species, increased significantly in the postoperative period when the patients took little food by mouth (23, 33). The ability of S. oralis to proliferate under such circumstances may be due to the particular ability of this species to deglycosylate human salivary and serum glycoproteins and its ability to utilize the liberated sugars for growth (12), although many other species, including other oral streptococci and Actinomyces spp., have a limited ability to degrade oligosaccharides from N- or O-linked glycoproteins (13, 14). The utilization of glycoprotein-derived sugars for growth is not restricted to bacteria producing the relevant glycosidic enzyme activities. Thus, Streptococcus sanguinis does not produce a sialidase activity but is able to utilize sialic acid for growth (11).
The effects of the withdrawal of the host's diet on the oral flora over a long period have not been investigated due to the complexities of establishing a model system in which to test these effects. In this study, we have devised a model system in which to test the long-term withdrawal of the host's diet on the phenotypic and genotypic diversity of the microbiota of infected carious dentine. In this model, infected carious dentine was sealed beneath dental restorations and the surviving flora recovered at a later date. In this model, the diet of the host was excluded and the bacteria had access to a limited supply of nutrients, primarily the serum proteins and glycoproteins, which pass from the pulp through the dentinal tubules of the uninfected dentine to the infected dentine (19). It has been proposed that applying a stress to the oral biofilm, in this case, limiting the nutrients available for growth, may reduce the complexity of the microbiota and also reduce the phenotypic and genotypic diversity of the surviving taxa (8). Those taxa that survive will be those best suited to survival and proliferation in this new environment.
MATERIALS AND METHODS
Subjects and sampling sites.
The local ethics committee approved this study, and all subjects gave informed consent to the procedures. A total of 10 teeth, in six patients, with discrete carious lesions through the enamel with infection of the underlying dentine were studied. Radiographs were used to classify dentinal lesion size, and all pulps were vital, indicating that sound, uninfected dentine was present between the infected dentine and the pulp of the tooth. Preliminary sample size calculations indicated that 8 to 10 teeth were required to demonstrate statistically significant (P < 0.05) differences between the initial and final dentine samples with respect to the predicted changes in the number of bacteria and genotypes recovered and the changes in the frequency of production of glycosidic enzyme activities. Ten lesions were therefore studied.
Treatment procedures and microbiological sampling.
Local anesthesia was given and a rubber dam applied to isolate the tooth and prevent contamination of the sites by saliva. Access to the infected dentine was gained using a sterile Jet 330 burr in the air rotor to remove the enamel over the lesion. The consistency (soft or hard), wetness (dry or wet), and color (dark brown, midbrown, or pale) clinical status of the dentine at each excavation was recorded using the previously reported criteria (17).
A sterile, round steel number 3 burr was used to clear all infected dentine from the enamel-dentine junction, leaving the infected dentine over the pulp. Two replicate, standardized dentine samples (initial or baseline samples) were taken for microbial analysis using a sterile number 3 round burr (17) dampened in PBSTC (1.58 g of K2HPO4 · 3H2O, 0.34 g of KH2PO4, 8 g of NaCl, 1.0 g of sodium thioglycolate, 0.001 g of cetyltrimethylammonium bromide per liter of distilled water). The cetyltrimethylammonium bromide at this low concentration assists in the dispersing of the bacterial aggregates, facilitating the enumeration of the microbiota in the samples. The standardization of the samples of infected dentine involves using the burr at a slow speed, which is used such that the head of the burr is pushed into the dentine and the adherent dentine constitutes the sample. The position of the samples was marked on the chart of the cavity to ensure that subsequent samples were not removed from sites that had already been sampled. The burr was placed in 2 ml of sterile PBSTC and stored on ice until processed. To prepare the cavities for sealing, they were etched for 30 s using 30% phosphoric acid (Kerr, United Kingdom), thoroughly washed with water for 20 s, and dried using a high-volume aspirator. Another dentine sample (postetch) was taken from a different area of each carious lesion and the position recorded. The teeth were restored with a light-cured dentine bonding agent (Optibond Solo; Kerr) and a light-cured composite (Herculite XR; Kerr).
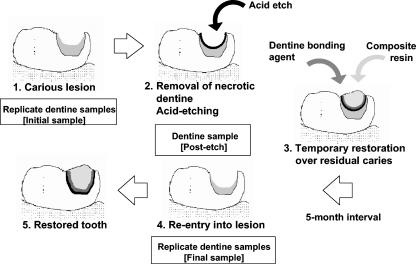
After approximately 5 months, the restorations were removed under a rubber dam to prevent contamination of the site by saliva. A Jet 330 burr in the air rotor was used to remove the restoration from the periphery of the cavity, creating a trough between the filling and the cavity wall. The restoration was lifted off the dentine with an excavator placed in the prepared trough between the cavity wall and the restoration. Two replicate samples of dentine (final samples) were taken as described above from discrete previously unsampled areas. This is shown diagrammatically in Fig. 1.
FIG. 1.
Design of study. 1. Carious lesion identified and replicate samples of infected dentine taken. 2. Dentine acid etched and postetching sample taken. 3. Carious lesion restored; restoration placed over infected dentine. 4. After 5 months, restoration removed, lesion reentered, and replicate dentine samples taken. 5. Tooth restored.
Microbial analysis of samples.
The standardized dentine samples were dispersed by vortexing with sterile 3.5- to 4.5-mm-diameter glass beads (BDH; Lutterworth, Leicester, United Kingdom) for 30 s and decimally diluted in PBSTC. Conventional plating methods were used to determine the number of bacteria (aerobic and anaerobic counts), streptococci, Actinomyces spp., lactobacilli, and yeasts (5, 6, 9, 10). Candida spp. were identified and enumerated on Chromagar (M-Tech Diagnostics Ltd, Cheshire, United Kingdom) and Veillonella spp. on Veillonella agar (Becton-Dickinson, Oxford, United Kingdom) (24). The streptococci and Actinomyces spp. were identified to the species level on the basis of biochemical and physiological characteristics (10, 35). Actinomyces naeslundii isolates were further examined using specific antiserum and classified as A. naeslundii genospecies 1 or A. naeslundii genospecies 2 using whole-cell agglutination (30). The detection limit for individual species was 10 CFU per sample.
The predominant aciduric microorganisms were isolated and enumerated by a most probable number (MPN) method (1). A maximum of 20 microorganisms capable of growth at pH 4.8, 5.2, and 7.0 were subcultured onto Columbia blood agar base (Oxoid, Basingstoke, United Kingdom) supplemented with 5% (vol/vol) horse blood. Isolates were identified to the species level using the procedures described above.
Screening of bacterial isolates for enzyme activities.
Representative isolates recovered from all dentine samples were screened for sialidase, β-galactosidase, and β-N-acetylglucosaminidase activity using fluorogenic substrates (4). These activities sequentially remove sugars from glycoproteins (12). The isolates tested were from the dentine samples collected at each sampling of all 10 teeth and consisted of all the streptococcal isolates and 50% of the lactobacilli and Veillonella spp. isolated from the respective selective medium and 50% of the Actinomyces spp. from the nonselective medium. The lactobacilli, Veillonella spp., and Actinomyces spp. were selected at random.
Genotyping of A. naeslundii genospecies 2 and S. oralis.
Up to 20 randomly chosen A. naeslundii genospecies 2 and S. oralis isolates, recovered from the nonselective medium and Mitis-Salivarius agar, respectively, from the initial and final dentine samples in five patients, were genotyped using repetitive extragenic palindromic-PCRs (REP-PCRs). DNA was extracted from A. naeslundii using microLYSIS (Microzone Ltd., Sussex, United Kingdom). The REP-PCR patterns for A. naeslundii strains were obtained as previously described (10). S. oralis DNA was extracted, and REP-PCR patterns were obtained as described previously (1). The REP-PCR patterns were compared (Applied Maths, Belgium), and patterns ≥95% similar were considered identical (1, 28).
Statistical analysis.
The number of microorganisms, expressed as log10(CFU + 1), and the proportion of each taxon per sample were calculated from the colony counts of the appropriate media. The MPN of organisms was calculated for each sample (http://members.ync.net/mcuriale/mpn/). The bacteria recovered from the MPN aciduric media (pH 4.8 and 5.2) were expressed as a percentage of the MPN count at pH 7.0. Data were compared using the Wilcoxon signed-ranks test, and Spearman correlation coefficients were calculated. Changes in proportions of enzyme-producing bacteria, distribution of genotypes, and changes in the clinical scores were compared using the χ2 test. The genotypic diversity of A. naeslundii and S. oralis was compared using a Student t test (SPSSPC, Chicago, Ill.).
RESULTS
Changes in the composition of the dentinal microbiota.
The reproducibility of the standardized sampling procedure was examined by comparing the total anaerobic colony counts from the two samples taken at the initial and final samplings. The mean (± standard error [SE]) of the counts [log10(CFU + 1) per sample] for the initial samples were 4.55 ± 0.13, with a median of 4.65, and 4.63 ± 0.29, with a median of 4.74 (P = 0.879), and for the final samples, 2.17 ± 0.04, with a median of 2.16, and 2.24 ± 0.08, with a median of 2.22 (P = 0.271). The total counts were significantly correlated, being 0.68 (P = 0.041) and 0.84 (P = 0.012) for the initial and final pairs of samples, respectively. On the basis of these analyses, the microbiological data from the pairs of samples at each of the two sampling times were combined by calculating the arithmetic mean of the untransformed counts for individual taxa prior to the log10 transformation.
The dentine of the initial samples was soft and wet; however, on reentry, it appeared harder (χ2 = 5.00; P = 0.025) and dryer (χ2 = 8.57; P = 0.003), but the color of individual lesions had not changed significantly (P > 0.05).
There were significant differences between the composition of the microbiota of the initial and final samples determined using conventional isolation and enumeration methods (Table 1). The total numbers of organisms per standardized sample decreased significantly (P < 0.05), while the proportion of streptococci recovered in the final sample was significantly (P < 0.01) higher. The number of streptococcal species recovered was reduced from eight in the baseline samples to three in the final samples. Only S. mitis (isolated from two subjects), Streptococcus intermedius and S. oralis were recovered in the final samples. The proportion of gram-positive pleomorphic rods in the samples decreased significantly (P < 0.05) but the proportion of A. naeslundii increased significantly (P < 0.05). Actinomyces israelii and Actinomyces gerencseriae were isolated from the initial samples but neither was recovered from the final samples, while A. naeslundii and S. oralis were recovered from all final samples. The taxa isolated from the dentine after 5 months being sealed beneath the restorations was limited to only those taxa reported in Table 1.
TABLE 1.
Comparison of the microbiota recovered from demineralized dentine sampled from enamel caries lesions (initial sample) and 5 months later following removal of the restoration (final sample)a
| Parameter | Initial sample (n = 10)
|
Final sample (n = 10)
|
||
|---|---|---|---|---|
| Mean ± SE | Median | Mean ± SE | Median | |
| Total microbial counts | ||||
| Anaerobic count | 4.6 ± 0.2b | 4.7 | 2.2 ± 0.1b | 2.2 |
| Aerobic count | 4.6 ± 0.2 | 4.6 | 2.2 ± 0.1 | 2.2 |
| Count (pH 4.8) MPN | 2.9 ± 0.3 | 3.1 | 0.6 ± 0.1 | 0.7 |
| Count (pH 5.2) MPN | 3.1 ± 0.3 | 3.3 | 0.9 ± 0.1 | 0.9 |
| Count (pH 7.0) MPN | 4.5 ± 0.2 | 4.4 | 2.1 ± 0.1 | 2.1 |
| Proportion of total anaerobic count | ||||
| Lactobacilli | 14.8 ± 5.5 (8)c | 9.7 | NDd | ND |
| Veillonella spp. | 0.2 ± 0.2 (5) | <0.01 | ND | ND |
| Total streptococci | 4.4 ± 1.1 (10) | 3.8 | 39.0 ± 2.1 (10)c | 37.8 |
| S. parasanguinis | 1.9 ± 0.6 (8) | 1.61 | ND | ND |
| S. gordonii | 0.2 ± 0.01 (4) | <0.01 | ND | ND |
| S. oralis | 0.5 ± 0.2 (7) | 1.7 | 37.9 ± 1.8 (10) | 37.7 |
| S. mitis | 0.2 ± 0.01 (6) | <0.01 | 0.6 ± 0.4 (2) | <0.01 |
| S. intermedius | 0.3 ± 0.1 (6) | 0.2 | 0.5 ± 0.5 (1) | <0.01 |
| S. anginosus | 0.7 ± 0.3 (8) | 0.2 | ND | ND |
| S. mutans | 1.4 ± 0.4 (9) | 1.1 | ND | ND |
| S. sobrinus | 0.01 ± 0.01 (3) | <0.01 | ND | ND |
| Gram-positive pleomorphic rods | 80.7 ± 5.4 (10) | 87.1 | 61.0 ± 2.1 (10) | 62.2 |
| A. naeslundii | 44.4 ± 2.8 (10) | 46.6 | 61.0 ± 2.1 (10) | 62.2 |
| A. israelii | 13.0 ± 4.0 (10) | 9.5 | ND | ND |
| A. gerencseriae | 23.4 ± 3.3 (10) | 22.9 | ND | ND |
| Proportion of MPN count at pH 7.0 | ||||
| Total count (pH 4.8) MPN | 3.5 ± 0.8 | 3.1 | 2.7 ± 0.3 | 2.8 |
| Total count (pH 5.2) MPN | 6.2 ± 1.7 | 4.9 | 5.3 ± 0.5 | 5.8 |
Numbers in boldface indicate significant differences between initial and final samples (P < 0.05).
Log10 (CFU per sample + 1) ± SE.
Proportion (number of samples positive for the microorganism).
ND, not detected.
Significantly more bacteria were recovered at each pH using the MPN method from the initial sampling than from the final sampling (P < 0.05), significantly more were recovered at pH 7.0 than at pH 5.2, and fewer were recovered at pH 4.8 (P < 0.05).
Acid etching significantly (P < 0.05) reduced the number of microorganisms recovered but did not significantly (P > 0.05) reduce the number of species recovered (data not shown). The mean (± SE) of the count for S. oralis was 0.59 ± 0.24 following acid etching, but this had increased significantly to 1.79 ± 0.09 after the removal of the filling material, while the number of A. naeslundii did not change significantly. Further, the proportion of S. oralis increased significantly from 3.6 ± 2.0 percent of the flora to 37.9 ± 1.8 percent of the flora while the proportion of A. naeslundii did not change significantly. In contrast, the proportions of lactobacilli and A. gerencseriae were 28.3 ± 10.4 and 5.8 ± 2.4 percent of the flora after etching, but both were present at below the detection level following removal of the restoration.
Identification of the predominant aciduric microbiota.
The predominant aciduric microbiota is shown in Table 2. Of 339 isolates identified from pH 4.8 at initial sampling, the majority were lactobacilli. However, no lactobacilli were recovered in the final sample. The recovery of A. naeslundii, all identified as genospecies 2, was pH dependent; significantly more were recovered at pH 7.0 than at 5.2 or 4.8. Conversely, A. israelii was isolated in greater numbers at pH 4.8 and 5.2 than at pH 7.0. The recovery of Streptococcus parasanguinis increased with pH, while S. oralis was optimally recovered at pH 5.2. The aciduric microbiota recovered in the final sample was less diverse as A. naeslundii predominated at pH 7.0, S. oralis at pH 5.2, and S. intermedius at pH 4.8.
TABLE 2.
Comparison of the microbiota recovered from demineralized dentine sampled from enamel caries lesions (initial sample) and 5 months later (final sample) in media adjusted to pH 4.8, 5.2, and 7.0
| Microbial taxon | Initial sample (n = 10)
|
Final sample (n = 10)
|
||||
|---|---|---|---|---|---|---|
| pH 4.8 | pH 5.2 | pH 7.0 | pH 4.8 | pH 5.2 | pH 7.0 | |
| Lactobacilli | 62.0 (7)a | 22.9 (7) | 1.7 (5) | NDb | ND | ND |
| Streptococci | 12.1 (7) | 21.7 (8) | 25.8 (9) | 100 (10) | 98.3 (10) | 12.0 (8) |
| S. parasanguinis | 0.6 (2) | 4.8 (3) | 19.8 (8) | ND | ND | ND |
| S. gordonii | 1.5 (2) | 1.7 (2) | ND | ND | ND | ND |
| S. oralis | 6.8 (4) | 15.0 (7) | 6.0 (4) | 2.7 (1) | 84.2 (10) | 7.3 (7) |
| S. mitis | ND | ND | ND | ND | 1.3 (2) | ND |
| S. intermedius | 3.3 (3) | 0.3 (1) | ND | 97.3 (10) | 12.8 (8) | 4.7 (5) |
| Gram-positive pleomorphic rods | 26.0 (8) | 55.34 (10) | 72.5 (10) | ND | 1.7 (2) | 88.0 (10) |
| A. naeslundii | 0.3 (1) | 3.4 (5) | 44.1 (10) | ND | 1.7 (2) | 88.0 (10) |
| A. israelii | 20.7 (4) | 17.0 (3) | 2.9 (4) | ND | ND | ND |
| A. gerencseriae | 5.0 (4) | 35.03 (8) | 25.5 (10) | ND | ND | ND |
| No. identified/no. subcultured | 339/339 | 354/354 | 349/356 | 150/150 | 234/234 | 317/317 |
Percentage of isolates (number of samples positive).
ND, not detected.
Production of sialidase, β-galactosidase, and N-acetyl-β-glucosaminidase activities.
The proportion of the microbiota producing sialidase (χ2 = 138.98; P < 0.001), β-galactosidase (χ2 = 46.91; P < 0.001), and N-acetyl-β-glucosaminidase (χ2 = 78.11; P < 0.001) activity significantly increased in the final sample compared to the initial sample (Table 3). In the final sample, all isolates were positive for sialidase and β-galactosidase and all, except for S. mitis, produced N-acetyl-β-glucosaminidase. All S. oralis isolates were positive for the three activities, while the A. naeslundii isolates exhibited variable production in the initial sample, but in the final sample, all isolates were positive for all activities.
TABLE 3.
Comparison of the production of sialidase, β-galactosidase, and N-acetyl-β-glucosaminidase by the total microbiota and A. naeslundii recovered from demineralized dentine sampled from enamel caries lesions (initial sample) and following removal of the restoration (final sample)
| Enzyme activity | % of flora positive
|
% of A. naeslundii isolates positive
|
||
|---|---|---|---|---|
| Initial sample (n = 4,148) | Final sample (n = 518) | Initial sample (n = 360) | Final sample (n = 377) | |
| Sialidase | 18.0 | 100 | 59.0 | 100 |
| β-Galactosidase | 62.3 | 100 | 6.2 | 100 |
| N-Acetyl-β-glucos- aminidase | 42.2 | 98.8 | 6.2 | 100 |
Genotypic comparison of A. naeslundii genospecies 2 and S. oralis strains.
To investigate changes in genotypic diversity, subjects from whom 20 A. naeslundii and 20 S. oralis isolates were recovered from both dentine samples were identified. Only five subjects fulfilled these criteria. The other patients did not yield sufficient isolates of both species and so were excluded from this aspect of the analysis. To examine the homogeneity of the dentine sampled at baseline and after 5 months, the similarity of the genotypes in the duplicate samples taken at both times was considered in order to get an understanding of the homogeneity of the dentine at the two sampling times. We found that only 18% of the S. oralis and 18% of the A. naeslundii isolates genotyped at baseline were common between two sampling sites on the teeth, with the remaining isolates of both taxa being recovered from only one, but not both, of the sampling sites. This compares with 64% of S. oralis and 97% of A. naeslundii being recovered from both sampling sites in the 5-month dentine samples (χ2 = 45.6, P < 0.0001, and χ2 = 127.9, P < 0.0001, respectively), indicating that the two dentine sites in each cavity were more homogenous at the end of the study than when they were first sampled.
We also compared the number of different genotypes of the two species between the two sampling times. In the initial sample, the A. naeslundii isolates yielded an average of 13.4 (± 3.3) genotypes among the 20 strains examined per sample, while in the final sample, only 1.6 (± 1.0) genotypes were identified (P = 0.002). Similarly, 13.6 (± 2.8) S. oralis genotypes were identified among the 20 strains investigated from the initial samples compared to only 3.4 (± 0.8) genotypes in the final sample (P = 0.001).
DISCUSSION
The results of this study indicate that the dentinal microbiota under the restorations were subject to significant environmental change. The changes in the microbiota were characterized by a reduction in the microbial load in the infected dentine, a reduction in the microbial diversity, and a reduction in the genotypic diversity of the surviving microbiota. Although the total number of bacteria recovered following acid etching was similar to that recovered at the end of the study, in the final samples, the dentine bonding system used here may also have had antimicrobial activity, which could also have reduced the number of surviving microbiota further. It cannot be known what the overall effect of the procedures used to restore the teeth had on the survival of the microbiota under the restoration. However, it should be noted that none of the genotypes of either S. oralis or A. naeslundii recovered in the final sample were detected among the strains genotyped in the initial samples. This suggests that the surviving genotypes did replicate during the period of the study. Further, the numbers of S. oralis cells in the final samples were significantly greater than those recovered following acid etching. Together, these data suggest that bacterial growth occurs beneath the restoration and that growth within both of the taxa studied here in some detail was restricted to those genotypes best able to exploit the environment beneath the restorations.
The most obvious environmental change for the members of the microbiota surviving the procedures required for the restoration of the teeth was the principal sources of nutrient available to the microbiota for growth. The microbiota of carious lesions are exposed to saliva, its components, and the diet of the host, while the flora beneath the restorations would be exposed primarily to serum constituents, including glycoproteins passing down the patent dentine tubules from the pulp to the infected dentine. It is also likely that components of bacteria, which did not survive sealing beneath the restoration, may have contributed to nutrients available to those bacteria which survived below the restorations. It is not known to what extent these potential nutrient sources supported bacterial growth, nor for how long. It is likely that the nutrients available to the microbiota in the 5-month samples were limited in both their amount and their complexity compared to those available to the microbiota of open carious lesions restored at the onset of the study. Additionally, in the carious lesion, the pH would be subject to large perturbations dependant upon the consumption of fermentable carbohydrates, especially di- and monosaccharides, which result in rapid acid production and consequent establishment of an acidic environment.
Serum glycoproteins are present in dentinal tubule fluid (19), which passes through the patent dentinal tubules into the infected dentine. The changes in composition of the microbiota of the infected dentine and in the production of glycosidic enzymes suggest that the surviving taxa were selected on the basis of their ability to derive nutrient, specifically carbohydrates, from serum glycoproteins. In this environment, the major sugars available for growth will be sialic acid, galactose, and N-acetylglucosamine. Previously, we have reported that parental feeding in bone marrow and liver transplant patients results in the development of an oral microbiota with an increased capacity to utilize host-derived glycoproteins (23, 33). The same effect is apparent in the microbiota sealed under the restoration. We found that the microbiota in the infected dentine sampled after sealing consisted entirely of bacterial taxa which produced glycosidic enzymes with the ability to liberate individual carbohydrates from the N-linked glycans of serum glycoproteins. The distribution of these activities was significantly increased among the surviving taxa. The ability of bacterial consortia to degrade oligosaccharides by glycosidic enzyme activity has been widely reported (8). S. oralis, S. mitis, and S. intermedius produce sialidase, which liberates N-acetylneuraminic acid (sialic acid) from glycans and may subsequently utilize this amino sugar as a carbon source (4, 11). These species usually produce N-acetylglucosaminidase and β-galactosidase activities, which also liberate sugars from N-linked glycans and are used by bacteria for growth (4, 13). A. naeslundii may produce sialidase (27) and liberates sialic acid from immunoglobulin A (14). The A. naeslundii strains also produced β-N-acetylglucosaminidase and β-galactosidase as reported previously (18, 29).
In this study, only bacteria capable of producing the enzymes required to cleave the terminal sugars from the glycoproteins were recovered from the dentine after cavity sealing. This supports the hypothesis that the microbiota were subjected to a significant nutrient stress with only bacteria able to utilize the limited nutrient source capable of proliferation and survival. The survival, however, may not be long-term since tubular sclerosis and reduced permeability of the dentine (22) will subsequently limit nutrient availability, resulting in further reductions in the numbers of bacteria surviving long-term under restorations (15, 16).
Here, we have also investigated the effects of placing the restoration on the genotypic diversity of the two prominent taxa surviving, S. oralis and A. naeslundii genospecies 2. It has previously been postulated that environmental stresses in the oral cavity should lead to a reduced number of genotypes best able to proliferate in a particular environment (8). In a previous study, we found that caries activity, indicative of a local acidic stress, was associated with the increased genotypic diversity of S. oralis but not A. naeslundii, which we postulated was due to an increase in niche diversity which could be exploited by S. oralis but not by A. naeslundii (28). In this study, we anticipated that sealing bacteria below a restoration would be more stressful and that it would result in a reduction in genotypic diversity; this was seen in both the A. naeslundii and S. oralis populations. The applied stress may therefore exert its effects by modulating the diversity of niches that can be populated by the resident microbiota: the more homogeneous the niches, the more pronounced the observed reduction in genotypic diversity. This was apparent in the analysis of the distribution of genotypes of both S. oralis and A. naeslundii between the duplicate samples of dentine taken at baseline and in the final samples. Other environmental pressures have also been shown to affect clonal populations: A. naeslundii from sound surfaces and carious lesions treated with chlorhexidine and fluoride exhibit reduced clonal diversity (8).
While it may be argued that the surviving A. naeslundii strains were selected for on the basis of their ability to produce all three relevant glycosidic enzymes, the same cannot be the case for S. oralis since this species produces all three of these activities. It may therefore be that survival was predicated by the production of the glycosidic enzymes and the possession of other uncharacterized traits that might include, for example, the possession of more efficient sugar transport systems. Nonetheless, the conditions under the restorations were less supportive of a genotypically heterogeneous population than were the conditions within the carious lesion.
The use of charts of the cavities enabled the distinct sampling sites (initial, postetch, and 5-month samples) to be selected and identified accurately without the possibility of resampling regions of dentine which had previously been sampled. Thus, differences in the number and proportions of bacteria in the dentine samples were not the result of the prior physical removal of demineralized infected dentine. Sealing carious dentine for 5 months below a restoration resulted in a significant reduction in the numbers of bacteria recovered per sample in accord with the previous observations (7, 26). The clinical appearance of the dentine under the restorations also underwent significant changes associated with caries arrest (17).
These results provide support for the previous suggestion that bacterial diversity within the oral biofilm is determined by the diversity of niches present within the biofilm. Thus, the apparently homogenous environment beneath dental restorations supported less-diverse microbiota than those recovered from infected dentine in the caries cavity.
Acknowledgments
We thank George Bowden for kindly providing the antiserum for Actinomyces identification.
This project was supported by the Joint Research Committee of the King's Health Care Trust and by The Wellcome Trust (grant number 063952).
REFERENCES
- 1.Alam, S., S. R. Brailsford, S. Adams, C. Allison, E. Sheehy, L. Zoitopoulos, E. A. Kidd, and D. Beighton. 2000. Genotypic heterogenity of Streptococcus oralis and distinct aciduric subpopulations in human dental plaque. Appl. Environ. Microbiol. 66:3330-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almstahl, A., M. Wikstrom, and U. Kroneld. 2001. Microflora in oral ecosystems in primary Sjogren's syndrome. J. Rheumatol. 28:1007-1013. [PubMed] [Google Scholar]
- 3.Beighton, D., and H. Hayday. 1986. The influence of diet on the growth of streptococcal bacteria on the molar teeth of monkeys (Macaca fascicularis). Arch. Oral Biol. 31:449-454. [DOI] [PubMed] [Google Scholar]
- 4.Beighton, D., J. M. Hardie, and R. A. Whiley. 1991. A scheme for the identification of viridans streptococci. J. Med. Microbiol. 35:367-372. [DOI] [PubMed] [Google Scholar]
- 5.Beighton, D., P. H. Hellyer, E. Lynch, and M. R. Heath. 1991. Salivary levels of mutans streptococci, lactobacilli, yeasts, and root caries prevalence in non-institutionalized elderly dental patients. Comm. Dent. Oral Epidemiol. 19:302-307. [DOI] [PubMed] [Google Scholar]
- 6.Beighton, D., E. Lynch, and M. R. Heath. 1993. A microbiological study of primary root-caries lesions with different treatment needs. J. Dent. Res. 72:623-629. [DOI] [PubMed] [Google Scholar]
- 7.BjØrndal, L., and T. Larsen. 2000. Changes in the cultivable flora in deep carious lesions following a stepwise excavation procedure. Caries Res. 34:502-508. [DOI] [PubMed] [Google Scholar]
- 8.Bowden, G. H. W., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 9.Brailsford, S. R., E. Lynch, and D. Beighton. 1998. The isolations of Actinomyces naeslundii from sound root surfaces and root carious lesions. Caries Res. 32:100-106. [DOI] [PubMed] [Google Scholar]
- 10.Brailsford, S. R., R. B. Tregaskis, H. S. Leftwich, and D. Beighton. 1999. The predominant Actinomyces spp. isolated from infected dentin of active root caries lesions. J. Dent. Res. 78:1525-1534. [DOI] [PubMed] [Google Scholar]
- 11.Byers, H. L., K. A. Homer, and D. Beighton. 1996. Utilization of sialic acid by viridans streptococci. J. Dent. Res. 75:1564-1571. [DOI] [PubMed] [Google Scholar]
- 12.Byers, H. L., E. Tarelli, K. A. Homer, and D. Beighton. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469-479. [DOI] [PubMed] [Google Scholar]
- 13.Byers, H. L., E. Tarelli, K. A. Homer, H. Hambley, and D. Beighton. 1999. Growth of viridans streptococci on human serum alpha1-acid glycoprotein. J. Dent. Res. 78:1370-1380. [DOI] [PubMed] [Google Scholar]
- 14.Frandsen, E. V. 1994. Carbohydrate depletion of immunoglobulin A1 by oral species of gram-positive rods. Oral Microbiol. Immunol. 9:352-358. [DOI] [PubMed] [Google Scholar]
- 15.Handelman, S. L., F. Washburn, and P. Wopperer. 1976. Two-year report of sealant effect on bacteria in dental caries. J. Am. Dent. Assoc. 93:967-970. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, O. E., and S. L. Handelman. 1980. Effect of autopolymerizing sealant on viability of microflora in occlusal dental caries. Scand. J. Dent. Res. 88:382-388. [DOI] [PubMed] [Google Scholar]
- 17.Kidd, E. A. M., S. Joyston-Bechal, and D. Beighton. 1993. Microbiological validation of assessment of caries activity during cavity preparation. Caries Res. 27:402-408. [DOI] [PubMed] [Google Scholar]
- 18.Kiel, R. A., J. M. Tanzer, and F. N. Woodiel. 1977. Identification, separation, and preliminary characterization of invertase and beta-galactosidase in Actinomyces viscosus. Infect. Immun. 16:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knutsson, G., M. Jontell, and G. Bergenholtz. 1994. Determination of plasma proteins in dentinal fluid from cavities prepared in healthy young human teeth. Arch. Oral Biol. 39:185-190. [DOI] [PubMed] [Google Scholar]
- 20.Kuramitsu, H. K. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14:331-1344. [DOI] [PubMed] [Google Scholar]
- 21.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love, R. M., and H. F. Jenkinson. 2002. Invasion of dentinal tubules by oral bacteria. Crit. Rev. Oral Biol. Med. 13:171-183. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, V. S., D. Beighton, G. J. Roberts, and S. J. Challacombe. 1997. Changes in the oral streptococcal flora of children undergoing allogeneic bone marrow transplantation. J. Infect. 35:135-141. [DOI] [PubMed] [Google Scholar]
- 24.Marchant, S., S. R. Brailsford, A. C. Twomey, G. J. Roberts, and D. Beighton. 2001. The predominant microflora of nursing caries lesions. Caries Res. 35:397-406. [DOI] [PubMed] [Google Scholar]
- 25.Marsh, P. D. 2004. Dental plaque as a microbial biofilm. Caries Res. 38:204-211. [DOI] [PubMed] [Google Scholar]
- 26.Mertz-Fairhurst, E. J., G. S. Schuster, and C. W. Fairhurst. 1986. Arresting caries by sealants: results of a clinical study. J. Am. Dent. Assoc. 112:194-197. [DOI] [PubMed] [Google Scholar]
- 27.Moncla, B. J., and P. Braham. 1989. Detection of sialidase (neuraminidase) activity in Actinomyces species by using 2′-(4-methylumbelliferyl)α-d-N-acetylneuraminic acid in a filter paper spot test. J. Clin. Microbiol. 27:182-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paddick, J. S., S. R. Brailsford, E. A. Kidd, S. C. Gilbert, D. T. Clark, S. Alam, Z. J. Killick, and D. Beighton. 2003. Effect of the environment on genotypic diversity of Actinomyces naeslundii and Streptococcus oralis in the oral biofilm. Appl. Environ. Microbiol. 69:6475-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirieva, D. A., A. G. Chigaleichik, N. A. Tiunova, and S. S. Rylkin. 1977. Physiological and biochemical properties of Actinomyces kurssanovii, active producter of chitinase. Mikrobiologiia 46:661-666. (In Russian.) [PubMed] [Google Scholar]
- 30.Putnins, E. E., and G. H. Bowden. 1993. Antigenic relationships among oral Actinomyces isolates, Actinomyces naeslundii genospecies 1 and 2, Actinomyces howellii, Actinomyces denticolens, and Actinomyces slackii. J. Dent. Res. 72:1374-1385. [DOI] [PubMed] [Google Scholar]
- 31.Rudney, J. D. 2000. Saliva and dental plaque. Adv. Dent. Res. 14:29-39. [DOI] [PubMed] [Google Scholar]
- 32.Sansone, C., J. Van Houte, K. Joshipura, R. Kent, and H. C. Margolis. 1993. The association of mutans streptococci and non-mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J. Dent. Res. 72:508-516. [DOI] [PubMed] [Google Scholar]
- 33.Sheehy, E. C., D. Beighton, and G. J. Roberts. 2000. The oral microbiota of children undergoing liver transplantation. Oral Microbiol. Immunol. 15:203-210. [DOI] [PubMed] [Google Scholar]
- 34.van Ruyven, F. O., P. Lingstrom, J. van Houte, and R. Kent. 2000. Relationship among mutans streptococci, “low-pH” bacteria, and lodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J. Dent. Res. 79:778-784. [DOI] [PubMed] [Google Scholar]
- 35.Whiley, R. A., and D. Beighton. 1998. Current classification of the oral streptococci. Oral Microbiol. Immunol. 13:195-216. [DOI] [PubMed] [Google Scholar]