Abstract
Systemic lupus erythematosus (SLE) is associated with a high risk of venous and arterial thrombosis, not necessarily associated with prothrombotic antiphospholipid antibodies (Abs). Alternatively, thrombosis may be due to an increased titer of anti-dsDNA Abs that presumably promote thrombosis via direct platelet activation. Here, we investigated effects of purified anti-dsDNA Abs from the blood of SLE patients, alone or in a complex with dsDNA, on isolated normal human platelets. We showed that anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes induced strong platelet activation assessed by enhanced P-selectin expression and dramatic morphological and ultrastructural changes. Electron microscopy revealed a significantly higher percentage of platelets that lost their discoid shape, formed multiple filopodia and had a shrunken body when treated with anti-dsDNA Abs or anti-dsDNA Ab/dsDNA complexes compared with control samples. In addition, these platelets activated with anti-dsDNA Ab/dsDNA complexes typically contained a reduced number of secretory α-granules that grouped in the middle and often merged into a solid electron dense area. Many activated platelets released plasma membrane-derived microvesicles and/or fell apart into subcellular cytoplasmic fragments. Confocal microscopy revealed that platelets treated with anti-dsDNA Ab/dsDNA complex had a heterogeneous distribution of septin2 compared with the homogeneous distribution in control platelets. Structural perturbations were concomitant with mitochondrial depolarization and a decreased content of platelet ATP, indicating energetic exhaustion. Most of the biochemical and morphological changes in platelets induced by anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes were prevented by pre-treatment with a monoclonal mAb against FcγRIIA. The aggregate of data indicates that anti-dsDNA Abs alone or in a complex with dsDNA strongly affect platelets via the FcγRIIA receptor. The immune activation of platelets with antinuclear Abs may comprise a prothrombotic mechanism underlying a high risk of thrombotic complications in patients with SLE.
Keywords: systemic lupus erythematosus, thrombosis, anti-dsDNA-antibodies, platelet
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease associated with immune inflammation of various organs and tissues (joints, kidney, skin, central nervous system, etc.). In SLE the immune system works against various autologous cellular antigens, including cytoplasmic proteins, cell surface, and nuclear components such as DNA and histones[1,2].
Elevated levels of anti-dsDNA antibodies (Abs) in the blood is a characteristic sign of SLE, but the titer of anti-dsDNA Abs can vary during the disease depending on the disease activity[3]. Anti-dsDNA Abs form complexes with DNA that stimulate the peripheral blood mononuclear cells through toll-like receptors 9 followed by production of interferon-α, a strong pro-inflammatory factor[4–6]. Platelets also can be potentially activated by DNA-containing immune complexes through the FcγRIIA receptor[7] or by free anti-DNA Abs through the β3 subunit of the integrins αIIbβ3 and αvβ3[8].
SLE patients have an increased risk of venous and arterial thrombosis[9], which is one of the major causes of mortality in SLE. Cerebrovascular accidents, coronary occlusions, and pulmonary embolisms are the most common thrombotic events during SLE[10]. It has been shown that thrombosis during SLE is strongly associated with the presence of antiphospholipid Abs in the blood[11]. However, about 20% of SLE patients with thrombosis are negative for antiphospholipid Abs[12], suggesting that antiphospholipid Abs are not the only pathogenic factor responsible for the thrombosis during SLE. It is likely that the predisposition to thrombosis in SLE can be due to chronic platelet activation in the blood stream[13] followed by expression of procoagulant phosphatidylserine[14] and secretion of prothrombotic compounds assessed by increased expression of P-selectin[13]. Platelet activation in SLE is associated with complement deposition on cells that can activate platelets in combination with inflammatory cytokines[7,14]. Activated platelets can be a source of the soluble CD40 ligand, which can secondarily promote activation of antigen-presenting cells, including monocytes and plasmacytoid dendritic cells. These cellular reactions are critically important for the pathogenesis of SLE because they lead to the production of autoantibodies, including anti-DNA Abs that can aggravate the disease[7]. Importantly, platelet-mediated activation of plasmacytoid dendritic cells promotes the secretion of interferon-α[7] that may influence megakaryocytes, so that platelets derived from these megakaryocytes are pre-activated[15]. In the blood of SLE patients, platelet-monocyte complexes are formed[16], suggesting that activated platelets bearing the CD40 ligand (CD154) can trigger monocytes through CD154-CD40 interaction. Moreover, activated platelets can cross-activate neutrophils that form prothrombotic neutrophil extracellular traps (NETs)[17,18]. Despite strong evidence for involvement of platelets and anti-DNA Abs in the pathogenesis of SLE, many aspects of platelet activation and its relation to DNA-containing immune complexes remain unknown. Very little is known about ultrastructural changes of platelets in SLE and whether anti-dsDNA Abs or their complexes with dsDNA can induce these changes.
Here we studied functional and structural alterations of normal isolated human platelets induced by purified SLE-related anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes to investigate mechanisms of immune platelet activation, including a role of the FcγRIIA receptor. The results show that anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes affect platelet morphology, ultrastructure, septin 2 intracellular distribution, and lead to the α-granule secretion, indicating strong platelet activation followed by their energetic exhaustion. These changes are lessened substantially in the presence of an anti-FcγRIIA Ab, implying that the FcγRIIA receptors mediate the pathogenically important pro-thrombotic interactions between platelets and anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes.
2. Materials and Methods
2.1. Patients, donors, blood collection and processing
The study was approved by the Ethical Committee of Kazan State Medical University (Kazan, Russia) and performed in accordance with the Declaration of Helsinki. With informed written consent, blood was withdrawn from 20 healthy donors and 37 SLE patients of whom 4 (11%) were men and 33 (89%) women at an average age of 37±10 years. Citrated platelet-rich plasma of healthy donors was used for platelet isolation and blood serum of SLE patients was used as a source of purified anti-dsDNA antibodies. SLE was documented using criteria of the American College of Rheumatology. The patients were selected based on the disease activity (an average SELENA-SLEDAI index was 12±5) and an abnormally high titer of anti-dsDNA Abs in the blood (11 patients had more than 200 IU/ml, 7 patients had 100200 IU/ml; the rest 19 patients had a titer of anti-dsDNA Abs <100 IU/ml but higher than the normal range 0–25 IU/ml). Antiphospholipid syndrome (APS) was diagnosed in 5 (13%) patients based on a history of deep vein thrombosis (3 patients) and recurrent miscarriage (2) combined with positive blood tests for lupus anticoagulant and anticardiolipin Abs. 3 SLE patients developed thrombosis and they all had elevated levels of anti-dsDNA Abs of 200, 63, and 30 IU/ml, while the reference range is <10 IU/ml. All the patients were on corticosteroids. Venous blood was drawn in vacuum containers containing 3.8% sodium citrate (Vacuette, Austria) (9:1, vol/vol) and centrifuged at 200g for 10 min at room temperature to obtain platelet-rich plasma (PRP) for platelets isolation. For serum, the blood was mixed with Z Serum Clot Activator (Vacuette, Austria) and allowed to clot and contract for 30 min at room temperature, followed by centrifugation at 800g for 10 min. Individual serum was collected and spun at 10,000g for 10 min, then frozen and stored at −72°C until antibodies purification. Blood for other biochemical, hematological, immunological, and hemostasis tests was processed accordingly.
2.2. Purity of the anti-dsDNA Abs
The purity of isolated anti-dsDNA Abs used in this study was confirmed by SDS-PAGE using 10 % resolving gel and 4% stacking gel. For non-reducing conditions, 1 μg protein per lane was applied after incubation in a sample buffer (62.5 mM Tris, 2% sodium dodecyl sulfate, 10 v/v% glycerol, 0.02% bromophenol blue) for 30 min at room temperature. For reducing conditions, 1 μg protein per lane was applied after incubation in a reducing sample buffer (62.5 mM Tris, 2% sodium dodecyl sulfate, 50 mM dithiothreitol, 10 v/v% glycerol, 0.02% bromophenol blue) for 10 min at 100°C in a water bath followed by cooling in ice. The gels were stained with Coomassie Brilliant Blue R-250 (BioRad, USA). Both the anti-dsDNA Abs and non-dsDNA binding IgG had ~95% purity according to the SDS-PAGE electrophoresis (Fig. S1A). The dsDNA-binding activity of anti-dsDNA Abs in concentration 50 μg/ml was 208.4 IU/ml and dsDNA-binding activity of non-DNA binding IgG in concentration 50 μg/ml was 5.9 IU/ml (Fig. S1E). The absence of contaminating DNA in the anti-dsDNA Ab preparations was confirmed by agarose gel (0.7%) electrophoresis with a GelRed nucleic acid gel stain (Biotium, USA). 10 μg of protein per lane was loaded and the results were analyzed using ChemiDoc™ XRS + System (Fig. S1B).
2.3. dsDNA-binding activity of the anti-dsDNA Abs
The specific dsDNA-binding activity of isolated Abs was confirmed by ELISA. Calf thymus dsDNA (Serva, Germany) in citrate buffer (0.1 M H2PO4, 0.05 M citric acid) (10 μg/ml; 100 μl/well) was adsorbed overnight at room temperature on a 96-well plate pre-activated with UV for 2 hrs. The plate was washed 3 times with 10 mM NaH2PO4/Na2HPO4, 100 mM NaCl, containing 0.2 v/v% Tween 20, pH 7.4 (PBST). The washed plate was coated with anti-dsDNA Abs or control IgG (80 μg/ml, 100 μl/well) diluted in PBST and incubated at 37°C for 1 h. The plate was washed 3 times with PBST. The washed plate was coated with 100 μl/well horseradish peroxidase-conjugated secondary mouse anti-human IgG diluted 1:10,0000 (“Sorbent” LLC, Russia) and incubated at 37°C for 1.5 hrs. After washing the plate 3 times with PBST and 2 times with milliQ water 100 μl/well of 3,3′,5,5′-tetramethylbenzidine (TMB) (Life Technologies, USA) was added. In 30 min the reaction was stopped by the addition of 100 μl/well of 1 M H2SO4. Absorbance at 450 nm was measured in a Microplate Reader Stat Fax 2100 (Awareness Technology, USA) (Fig. S1C). To calibrate the specific anti-ds-DNA-binding activity of isolated Abs in IU/ml, we used a Vecto-dsDNA-IgG KIT (Vector-BEST, Russia) that contained IgG standards with known activity in IU/ml (Fig. S1E).
2.4. Isolation of IgG using affinity chromatography on Protein G-Sepharose
HiTrap Protein G HP 1 ml column (GE Healthcare, Sweden) was equilibrated with PBS. 1 ml of individual or pooled blood serum from SLE patients diluted 7 times with PBS was passed through the column at a flow rate of 1 ml/min at room temperature. The column was washed with 5 column volumes of PBS and the adsorbed IgG was eluted with 100 mM glycine-HCl buffer, pH 2.3. The eluted IgG was immediately neutralized with 1 M Tris-NaOH, pH 9.8, at a 20:1 volume ratio. IgG preparations from at least 6 ml SLE serums were pooled and used for purification of anti-dsDNA Abs. Altogether, 9 batches of IgG from the blood of 37 SLE patients were purified and used in this study.
2.5. Synthesis of dsDNA-cellulose, a sorbent for purification of anti-dsDNA Abs
dsDNA-cellulose was synthesized as described[21]. Briefly, acid-washed microcristalline cellulose (Serva, Germany) was mixed with calf thymus dsDNA (Serva, Germany) dissolved in 10 mM Tris/10 mM NaCl, pH 7.4 (16 mg of DNA per 1 g of cellulose). The components were kneaded to achieve complete mixing, then spread as a thin layer over a glass surface and air-dried. The powder was suspended in 100 v/v% ethanol in the glass plate which was placed on ice in a shaker under UV illumination until complete drying due to ethanol evaporation. The dsDNA-cellulose was washed thoroughly with PBS using a Buchner funnel and stored at 4°C in PBS with 0.02% NaN3.
2.6. Affinity chromatography of anti-dsDNA Abs on dsDNA-cellulose
dsDNA-cellulose was packed into a Tricorn 10/50 column (GE Healthcare, Sweden) and equilibrated with a binding buffer (20 mM Tris, 50 mM NaCl, 2 mM EDTA, pH 7.4). IgG obtained from the serum of SLE patients (1.7–2.3 mg/ml in 12–20 ml) was pre-heated on a water bath at 56°C for 45 min to destroy DNases[22] and passed through the column at a flow rate of 0.05 ml/min at room temperature. The column was washed with the binding buffer and the absorbed IgGs comprising anti-dsDNA Abs were eluted with a high ionic strength elution buffer with (20 mM Tris, 1 M NaCl, 2 mM EDTA, pH 7.4). The protein-containing fractions were collected, concentrated and transferred into PBS using Amicon Ultra Centrifugal Filters with NMWL of 50, 000 Da (Merc Millipore, USA). The portion of IgG not absorbed on the dsDNA-cellulose column was collected and used as a negative control for anti-dsDNA Abs. It is noteworthy that the non-dsDNA binding IgG used as a non-immune control IgG was isolated exclusively from patient serum not containing antiphospholipid Abs. As an alternative source of control IgG we used commercially available native human IgG protein (Abcam, USA) that had no activating effect on platelets, either in the presence or absence of DNA. To remove the possible aggregates the anti-dsDNA Abs were filtered through Amicon Ultra Centrifugal Filters with NMWL of 300,000 Da (Merc Millipore, USA).
2.7. Platelet isolation and treatment
Platelets were isolated from PRP of healthy donors (1.0–1.5 ml) by gel-filtration on a 2 cm·8 cm column filled with Sepharose 2B (GE Healthcare, Sweden) and equilibrated with Tyrode’s buffer (4 mM HEPES, 135 mM NaCl, 2.7 mM KCl, 2.4 mM MgCl2, 5.6 mM D-glucose, 3.3 mM NaH2PO4, 0.35 mg/ml bovine serum albumin, pH 7.4). About 2 ml of isolated platelets were collected in the void volume at a concentration of 80,000 to 240,000 platelets per 1 μl (counted in a hemocytometer at 400). Isolated platelets were untreated (negative control) or treated with anti-dsDNA Abs (50 μg/ml) in the absence or presence of dsDNA (50 μg/ml) (Sigma-Aldrich, USA) at 37°C for 15 min in Tyrode’s buffer containing 2 mM CaCl2. The 50 μg/ml concentration of anti-dsDNA Abs was determined by titration (Fig. S1D) and corresponded to ~200 IU/ml based on ELISA (Fig. S1E). A non-DNA-binding IgG either purified from SLE serum (50 μg/ml with specific anti-dsDNA activity <10 IU/ml) or native human IgG purchased from Abcam was used as an additional negative control for anti-dsDNA Abs and dsDNA alone (50 μg/ml) was used as a negative control for dsDNA-containing immune complexes. In some experiments, 40 μg/ml of a monoclonal anti-FcγRIIA antibody (mAb, clone IV.3) against the FcγRIIA receptor purified from supernatants of hybridoma cells (HB-217, American Type Culture Collection) as described previously[19] was added to platelets 15 min prior to the incubation with anti-dsDNA Abs alone anti-dsDNA Abs/dsDNA complexes. In some experiments platelets were pre-treated with 20 U/ml DNase I (Thermo Scientific, USA) for 30 min at 37°C in Tyrode’s buffer in the presence of 0.1 mM CaCl2.
2.8. Flow cytometry of platelets
Functionality of isolated platelets and degree of activation were assessed by the mitochondrial membrane potential (ΔΨm) and expression of P-selectin using flow cytometry. Platelets untreated or treated with various stimulants were mixed with a ΔΨm-sensitive dye, Mito Tracker Deep Red FM (300 nM final concentration) (Invitrogen, US) or with 1.5 μl of anti-human CD62P phycoerythrin-labeled murine antibodies (BD Biosciences, USA). The samples labeled for P-selectin (50 μl final volume) and with Mito Tracker Deep Red FM (150 μl final volume) were incubated in Tyrode’s buffer containing 2 mM CaCl2 for 15 or 30 min at 37°C in the dark before the measurements. The samples were analyzed using a FacsCalibur flow cytometer equipped with BD CellQuest™ software (BD Biosciences, USA). Platelets were gated based on the size and granularity using Forward Light Scatter (LFS) and Side Scatter (LSS) channels and 5,000 cells were counted in each sample. FlowJo X software was used for data analysis.
2.9. Measurement of ATP content in platelets
Isolated platelets (0.1 ml at 107 cells/ml) in Tyrode’s buffer were incubated for 15 and 30 min with the anti-dsDNA Ab/dsDNA complexes or non-DNA-binding IgG (purified from SLE serum) mixed with dsDNA in the presence of 2 mM CaCl2. After the incubation, platelets were spun down for 5 min at 2,000g and lysed with 0.2 v/v% Triton X-100 in Tyrode’s buffer for 20 min at 4°C with continuous shaking. Debris was removed from the lysates by centrifugation at 8,000g for 10 min. ATP concentration in the supernatant of platelet lysates was measured in a plate reader Infinite 200 PRO (Tecan, Switzerland) using an ATP Bioluminescent Assay Kit (Sigma-Aldrich, USA) according to the manufacturer’s instructions. The ATP concentration was normalized by the protein content in the platelet lysates determined with a Pierce BCA Protein Assay Kit (ThermoFisher Scientific, USA).
2.10. Scanning electron microscopy of platelets
Isolated platelets untreated or treated with various stimulants were fixed in 2% glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.5), containing 150 mM NaCl, for 90 min at room temperature. The fixed gel-filtered platelets were layered on a carbon filter (0.4-μm-pore size) and centrifuged at 150g for 5 min. The samples were rinsed three times with the cacodylate buffer for 5 min, dehydrated in ascending concentrations of ethanol, and dried overnight with hexamethyldisilazane. A thin film of gold-palladium was layered on the samples using a sputter coater Quorom Q 150T ES (Quorum, UK). Micrographs were taken with a scanning electron microscope Merlin (Carl Zeiss, Germany).
2.11. Transmission electron microscopy of platelets
Isolated platelets incubated under various experimental conditions were fixed in 2% glutaraldehyde solution in phosphate-buffered saline (PBS) for 90 min at room temperature. The fixed platelets were centrifuged at 1,000g for 5 min. The pellet was washed with PBS and the samples were post-fixed with 1% osmium tetroxide in the same buffer with addition of sucrose (25 mg/ml) for 2 hrs. The samples were dehydrated in ascending concentrations of ethanol, then in acetone and propylene oxide. Epon 812 was used as the embedding resin. Samples were polymerized for 3 days under increasing temperature from 37°C to 60°C. Sections were obtained on an LKB-III ultramicrotome (LKB, Sweden). The sections were contrasted with saturated aqueous solution of uranyl acetate for 10 min at 60°C and then with an aqueous solution of lead citrate for 10 min. The preparations were examined using a JEM1200EX electron microscope (JEOL, Japan).
2.12. Immunofluorescent staining of isolated platelets
Isolated treated or untreated platelets (1.2×106 platelets in 100 μl of Tyrode’s buffer) were fixed in suspension by 4% paraformaldehyde for 90 min at room temperature. Fixed platelets were attached to polylysine-coated glass cover slips by centrifugation (800g, 5 min). Immunofluorescent staining of the fixed platelets was performed as described previously[20] with modifications. Briefly, after three consecutive washes in phosphate-buffered saline (PBS), 5 min each, cells were permeabilized by incubation with 0.1% Triton X-100 for 5 min. Then cells were incubated with normal goat serum (Vector Laboratories, USA) in the presence 0.1% Triton X-100 and 5% bovine serum albumin for 30 min at room temperature. Immunostaining of septin 2 was performed by 1-hour incubation with polyclonal anti-septin 2 antibodies (Sigma Prestige, 1:50), followed by 1-hour incubation with Alexa Fluor 488-conjugated anti-rabbit secondary antibodies (Invitrogen, 1:200). Staining was detected by confocal microscopy using a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss Microscopy GmbH, Germany) and analyzed using ImageJ software.
2.13. Determination of the working concentration of isolated anti-dsDNA Abs by the ability to induce expression of P-selectin on platelets
To determine the working concentration of anti-dsDNA Abs that effectively activates platelets, platelets were labeled with 1.5 μl of anti-human CD62P phycoerythrin-labeled murine antibodies (BD Biosciences, USA) and treated with anti-dsDNA Abs at various concentrations (1, 5, 10, 20, 30, 50, 60, 70, 80 and 90 μg/ml). After incubation for 15 min at 37°C in Tyrode’s buffer (150 μl final volume) containing 2 mM CaCl2 the level of P-selectin expression was measured with flow cytometry. The samples were analyzed using a FacsCalibur flow cytometer equipped with BD CellQuest™ software (BD Biosciences, USA). Platelets were gated based on the size and granularity using in Forward Light Scatter (LFS) and Side Scatter (LSS) channels and 5,000 cells were counted in each sample. FlowJo X software was used for data analysis (Fig. S1D). Based on the results of these titration experiments (Fig. S1D), the working concentration of anti-dsDNA Abs used in this study was determined as 50 μg/ml, which corresponds to a high titer (~200 IU/ml) of anti-dsDNA Abs in the blood of SLE patients.
2.14. Statistical analyses
Statistical analyses were performed using a GraphPad Prism 5.0 package (GraphPad Software, La Jolla, CA, USA). After assessing normality with the Shapiro-Wilk and D’Agostino-Pearson criteria, samples were analyzed for statistical significance using a two-tailed Student’s t-tests, a Mann-Whitney U-test, a Kruskal-Wallis test or a chi-square test (for categorical values) with a 95% level of significance (alpha=0.05). Significance is represented as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
3. Results
3.1. Anti-dsDNA Abs induce FcγRIIA-mediated expression of P-selectin in platelets
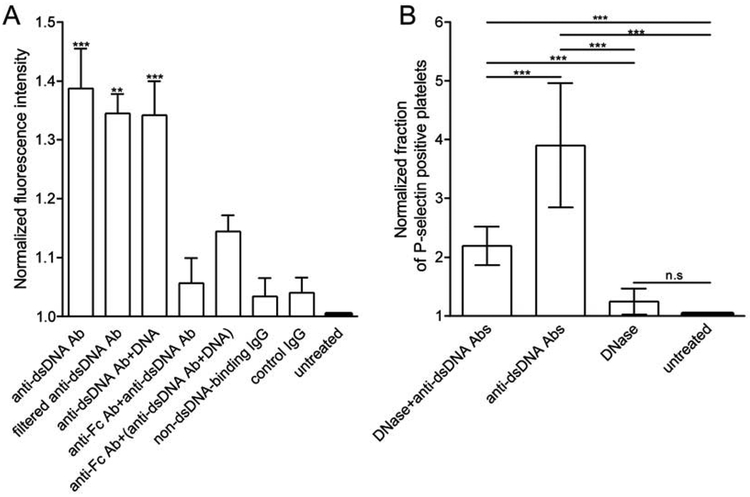
Flow cytometry showed that normal platelets isolated from the blood of healthy donors had a significantly increased level of P-selectin expression after incubation with anti-dsDNA Abs alone or mixed with dsDNA compared to control untreated platelets (Fig. 1A). Importantly, the activating effect of anti-dsDNA Abs was observed at a concentration and activity that corresponded to the elevated titer of Abs observed in the blood of SLE patients (~200 IU/ml). Platelets incubated with non-DNA-binding IgG, either purified from SLE serum or purchased from Abcam, had no significant difference from untreated platelets, implying that the increased P-selectin expression was due to anti-dsDNA Abs alone or in a complex with dsDNA. To exclude the possibility that the activating effect of anti-dsDNA Abs in the absence of dsDNA was due to non-specific aggregation of Abs that could mimic formation of immune complexes, we filtered Abs to remove possible aggregates. The filtered monomeric anti-dsDNA Abs maintained their ability to stimulate platelets without addition of the antigen (Fig. 1A). To see if the activating effect of isolated anti-dsDNA Abs was mediated by surface-bound DNA, platelets were pre-treated with DNase I before the addition of the Abs. Flow cytometry showed that DNase I-treated platelets incubated with anti-dsDNA Abs had a significantly lower percentage of P-selectin positive platelets compared with platelets untreated with DNase (Fig. 1B). Importantly, pre-treatment of platelets with an FcγRIIA blocking antibody (mAb IV.3) reduced the level of P-selectin expression down to the baseline, indistinguishable from the background control values in unstimulated platelets, indicating that platelet activation with anti-dsDNA Abs, alone or in a complex with dsDNA, was mediated by the FcγRIIA receptor (Fig. 1A).
Fig. 1. Biochemical signs of platelet activation induced by anti-dsDNA Abs and dsDNA-containing immune complexes.
(A) Average P-selectin (CD62P) expression levels on isolated normal human platelets under various experimental conditions normalized by control untreated platelets. Bars from left to right represent the following samples: platelets treated with anti-dsDNA Abs unfiltered (n=8) or filtered to remove aggregates (n=3); with anti-dsDNA Ab/dsDNA complexes (n=7); with anti-DNA Abs after pre-treatment with anti-FcγRIIA mAb (n=5); with anti-dsDNA Ab/dsDNA complex after pre-treatment with anti-FcγRIIA mAb (n=4); with non-DNA-binding IgG purified from SLE serum (n=7); with control IgG (Abcam) (n=3) or incubated without treatment (untreated, n=8). n is the number of experiments with platelets from different donors. A Mann-Whitney U-test was used to compare with the negative control. (B) Percent of P-selectin- positive isolated normal human platelets under various experimental conditions normalized by control untreated platelets. Bars from left to right represent the following samples: platelets pre-treated with DNase I followed by incubation with anti-dsDNA Abs (DNase+anti-dsDNA Abs) (n=3); treated with anti-dsDNA Abs only (anti-dsDNA Abs) (n=3); pre-treated with DNase I without further adding anti-dsDNA Abs (DNase) (n=3); untreated platelets (n=3). “n” is the number of experiments with platelets from different donors. A chi-square test was used for statistical analysis. ***p<0.001.
3.2. Anti-dsDNA Abs induce platelet shape change
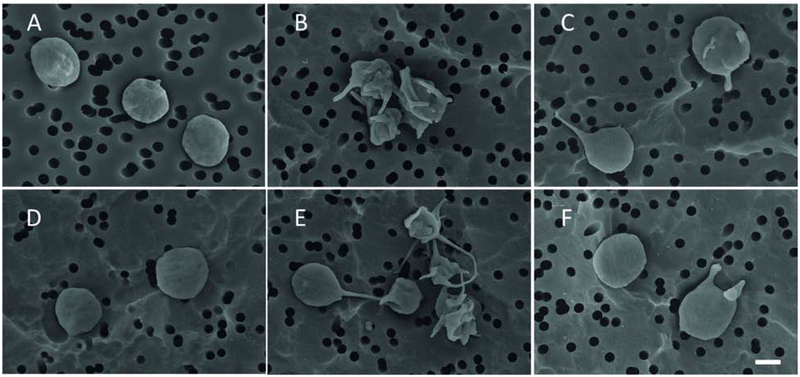
To confirm the direct activating effects of anti-dsDNA Abs on platelets, we used scanning electron microscopy to follow morphological changes of isolated normal platelets under various experimental conditions. The extent of platelet activation was quantified as a fraction of morphologically altered cells after segregation of individual platelets into activated versus non-activated cells based on their distinct morphology. Quiescent or resting platelets had a discoid shape without or with a few (1–2) thin filopodia. Activated platelets lost their discoid form, and formed multiple membrane protrusions, such as thinner or thicker filopodia. To assess the extent of platelet activation based on the morphological signs, we counted the number of filopodia per one platelet (Table 1). The mean number of filopodia per one platelet was significantly higher in the samples treated with anti-dsDNA Abs alone (3.8±0.1) and the anti-dsDNA Ab/dsDNA complexes (4.0±0.1) compared with untreated samples (1.4±0.1) (p<0.0001, one-way ANOVA test) (Fig. 2). As in the flow cytometry experiments, preincubation of platelets with an anti-FcγRIIA mAb significantly decreased the mean number of filopodia per one platelet to 1.5±0.1 when they were treated with anti-dsDNA Abs alone and to 1.7±0.1 when platelets were treated with the anti-dsDNA Ab/dsDNA complexes (Fig. 2C,F). Platelets treated with control non-DNA-binding IgG (Fig. 2D) were minimally activated at the same level as control untreated platelets (Table 1).
Table 1.
Morphological sign of platelet activation induced by anti-dsDNA Abs and dsDNA-containing immune complexes (from scanning electron microscopy).
| Anti-DNA Abs | Anti-DNA Abs+DNA | mAb IV.3 +Anti-DNA Abs | mAb IV.3 + Anti-DNA Abs+DNA | Non-DNA binding control IgG | Untreated platelets | |
|---|---|---|---|---|---|---|
| Mean number of filopodia per 1 platelet | 3.8±0.1**** | 4.0±0.1**** | 1.5±0.1 | 1.7±0.1 | 1.3±0.1 | 1.4±0.1 |
p<0.0001 compared with untreated platelets. One-way ANOVA test was used. N>200 platelets per sample, data presented as mean±SEM.
Fig. 2. Representative scanning electron micrographs of isolated normal human platelets under various experimental conditions (see Methods for details).
(A) An untreated platelet (control); (B-F) platelets incubated with: anti-dsDNA Ab/dsDNA complexes (B), anti-dsDNA Ab/dsDNA complexes after pre-treatment with anti-FcγRIIA mAb (C), non-immune IgG (D), anti-dsDNA Abs (E), and anti-dsDNA Abs after pre-treatment with anti-FcγRIIA mAb (F). In B and E platelets have morphological signs of activation: characteristic shape changes, formation of filopodia and aggregates. Scale bar = 1 μm.
3.3. Anti-dsDNA Abs alter platelet ultrastructure
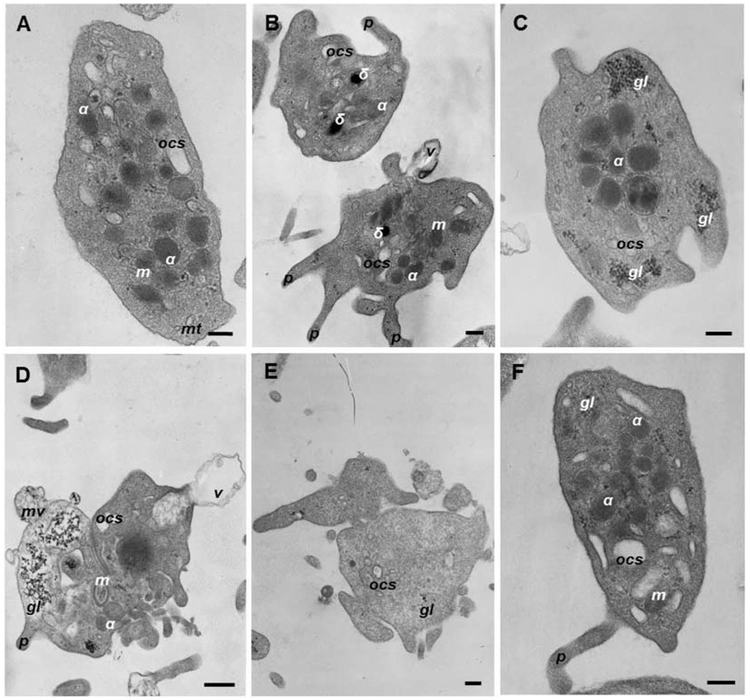
To reveal ultrastructural changes in platelets induced by anti-dsDNA Abs, we used transmission electron microscopy. Segregation of non-activated versus activated platelets from the electron micrographs was based on the following criteria. Resting platelets had discoid or round shape depending on the projection; they also contained round-shaped α-granules that were usually located randomly in the platelet body; all other organelles were clearly visualized, such as dense bodies (δ-granules), glycogen granules, open canalicular system (OCS), and mitochondria (Fig. 3A). The resting platelets might form 1 or 2 short filopodia. Unlike resting platelets that were largely homogenous, activated platelets had diverse and greatly altered ultrastructural features (Fig. 3). After treatment with isolated anti-dsDNA Abs (Fig. S2E) 87% platelets had pronounced signs of activation, including a loss of the discoid or round shape, formation of multiple filopodia (Fig. 3B), a substantial loss of α-granules (Fig. S3A) and vesiculation of the plasma membrane (Fig. 3E). A considerable fraction of the anti-dsDNA-Ab-activated platelets fell apart into cellular fragments (Fig. 3D) or had α-granules grouped in the middle of the platelet (Fig. 3C), sometimes even merging into a solid electron dense area (Fig. 3D, Table 2). After treatment with anti-dsDNA Abs/dsDNA complexes (Fig. S2B), 98% of platelets had the same ultrastructural signs of activation, but there were 2 times more platelets with α-granules merged into a solid electron dense area rather than with separate α-granules centralized in the middle of the platelet (Table 2). Also, we noticed that platelets treated with anti-dsDNA Abs/dsDNA complexes contained ~10% of membrane-surrounded “empty” platelet bodies or platelet “ghosts” that had a uniformly low electron density and no organelles inside (Fig. 3E). Platelets pre-treated with an anti-FcγRIIA mAb (Fig. S2C,F) still had moderate signs of activation, like a loss of the discoid or round shape, formation of filopodia and centralization of α-granules in the middle of platelet body. However, the overall changes were much less pronounced than in the corresponding platelet samples not pre-treated with the anti-FcγRIIA mAb. In platelets incubated in the presence of non-DNA-binding IgG (Fig. S2D) used as a negative control, the resting non-activated platelets prevailed with a relatively small fraction of platelets (27%) that displayed moderate ultrastructural signs of activation (Figs. 3F, S3; Table 2). Untreated platelets also had 16% of platelets with moderate ultrastructural signs of activation (Figs. 3A; S3; Table 2).
Fig. 3. Representative transmission electron micrographs of human platelets without or with characteristic ultrastructural alterations induced by anti-dsDNA Abs or anti-dsDNA Abs/dsDNA complexes.
Ultrastructure of resting (A, F) and activated (B, C, D, E) platelets: (B) shape change, shrinkage and formation of multiple filopodia; (C) shape change, α-granules grouped in the middle of the platelet body; (D) dramatic shape change, partial fragmentation, a solid electron dense area in the center, vesiculation of the plasma membrane; (E) a “grey” platelet without α-granules. Designations: α, α-granules; δ, δ-granules, gl, glycogen granules; m, mitochondria; mt, microtubules; OCS, open canalicular system; f, filopodia; v, microvesicle with a single membrane; mv, a multivesicular particle. Scale bars are 0.2 μm (A, B, D, E) and 0.5 μm (C).
Table 2.
Fractions of activated platelets with characteristic structural features under various experimental conditions (from transmission electron microscopy)
| Structural platelet variants | Platelets treated with: | Untreated platelets | ||||
|---|---|---|---|---|---|---|
| Total platelets taken as 100% (n*) |
100 (n=131) |
100 (n=164) |
100 (n=38) |
100 (n=107) |
100 (n=27) |
100 (n=262) |
| Activated platelets, % | 87 | 98 | 78 | 73 | 26 | 16 |
| centered α-granules, % | 7 | 4 | 14 | 7 | 0 | 0 |
| merged α- granules, % | 6 | 15 | 0 | 2 | 0 | 0 |
| no α-granules, % | 8 | 9 | 0 | 0 | 0 | 0 |
| Cytoplasmic fragmentation, % | 11 | 13 | 0 | 0 | 0 | 0 |
| Platelet “ghosts”, % | 0 | 10 | 0 | 0 | 0 | 0 |
n is the absolute number of individual platelets analyzed
3.4. Rearrangements of septin 2 in platelets induced by anti-dsDNA Abs and their immune complexes with dsDNA
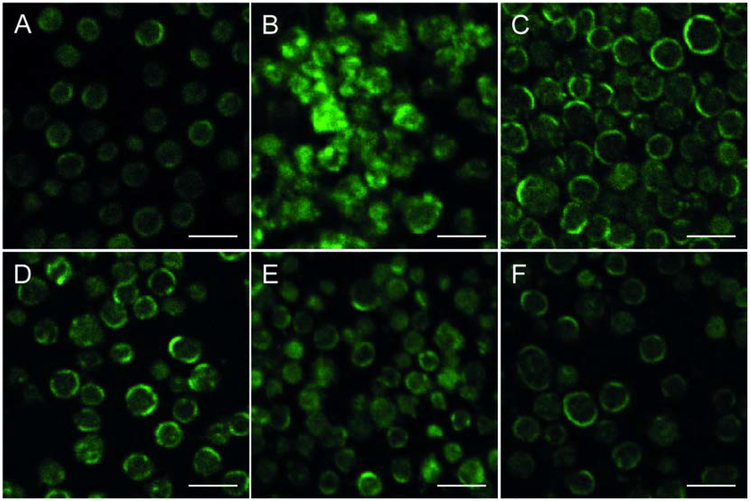
To analyze intra-platelet localization of immunostained cytoskeletal protein septin 2 under various experimental conditions, we used confocal microscopy. Control untreated (resting) platelets displayed a homogeneous distribution of septin 2 over platelet bodies with a prevalent concentration at the cell periphery, suggesting its association with the plasma membrane (Fig. 4A). Dramatic changes were observed in platelets treated with the complex of anti-dsDNA Abs and dsDNA (Fig. 4B), namely the distribution of septin 2 was highly heterogeneous and multiple fluorescent clusters were located mostly at the cell periphery, as well as in the middle of deformed platelet bodies. The observed changes of septin 2 distribution indicate dramatic alterations in platelet cytoskeleton upon activation induced by the anti-dsDNA Abs/ds-DNA complexes. In the samples treated with anti-dsDNA Abs alone, the distribution of septin 2 was much like in control untreated cells with the prevailing concentration at the cell periphery, albeit with clustering and moderate cytoplasmic spreading (Fig. 4C). Platelets treated with dsDNA alone showed a distribution of septin 2 similar to control untreated samples, but some platelets had clusters of septin 2 (Fig. 4D). Platelets preincubated with FcγRIIA mAb IV.3 before adding anti-dsDNA Abs (alone or in a complex with dsDNA), displayed a distribution of septin 2 similar to the control untreated platelets, with a homogeneous fluorescence intensity concentrated at the cell periphery (Fig. 4E,F).
Fig. 4. Representative images of septin 2 distribution in isolated normal human platelets under various experimental conditions as detected by immunofluorescence.
(A) Control (untreated) platelets. (B-F) Platelets incubated with: anti-dsDNA Ab/dsDNA complexes (B), anti-dsDNA Ab/dsDNA complexes after pre-treatment with anti-FcγRIIA mAb (C), dsDNA (D), anti-dsDNA Abs (E), and anti-dsDNA Abs after pre-treatment with anti-FcγRIIA mAb (F) In A, C, D, and F, platelets display homogeneous intracellular distribution of septin 2 with a prevalent concentration at the cell periphery. In B, septin 2 forms multiple fluorescent clusters mostly at the cell periphery as well as in the middle of platelet bodies. In E, platelets contain clusters of septin 2 with prevailing concentration at the cell periphery, albeit with moderate clustering and cytoplasmic spreading. All the images were acquired with the same microscope settings. Scale bar = 5 μm.
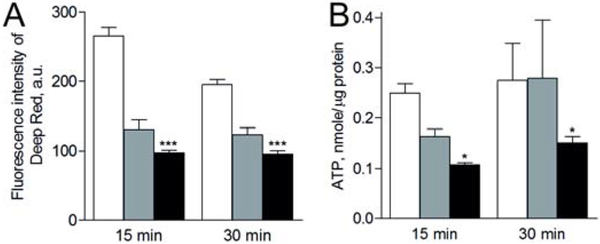
3.5. Effect of anti-dsDNA Abs on the energy metabolism in platelets
To evaluate changes in energy metabolism of platelets, we measured the mitochondrial membrane potential (Fig. 5A) and intracellular ATP content (Fig. 5B) after incubation of platelets for 15 and 30 min with either anti-dsDNA Ab/dsDNA complexes or a non-DNA-binding IgG/dsDNA mixture (used as an additional control for anti-dsDNA Abs). The average values of the mitochondrial membrane potential descended in the following order: untreated (resting) platelets > platelets treated with non-DNA-binding IgG mixed with dsDNA > platelets treated with anti-dsDNA Abs/dsDNA complex (Fig. 5A). The average ATP content decreased in the same order as the mitochondrial membrane potential (Fig. 5B). There was no time dependence for both parameters, indicating that the energetic exhaustion of platelets resulting in a decrease of the mitochondria membrane potential and ATP depletion was basically developed within 15 minutes of platelet activation with the anti-dsDNA Ab/dsDNA immune complexes.
Fig. 5. Mitochondrial membrane potential (A) and ATP content (B) in untreated platelets (white bars) and in platelets after incubation for 15 and 30 min with non-DNA-binding IgG/DNA complex (grey bars) or anti-dsDNA Ab/dsDNA complexes (black bars).
Final concentrations: 50 μg/ml anti-dsDNA Abs, 50 μg/ml non-DNA-binding IgG purified from SLE serum and 50 μg/ml dsDNA. Results of a representative experiment out of three independent but consistent experiments. A paired Student’s t-test was used for statistical analysis. *p<0.05 compared with non-DNA-binding IgG mixed with DNA.
4. Discussion
Thrombosis is a common complication associated with autoimmune disorders, such as SLE. Several potential mechanisms may lead to a pro-thrombotic status in patients with autoimmune disorders. Antiphospholipid Abs and their complexes often formed in SLE have been shown to promote thrombosis through multiple pathways, including binding to β2 glycoprotein, negatively charged phospholipids, and to the FcγRIIA receptors on platelets, thus triggering a release of platelet α-granules and formation of highly procoagulant extracellular microvesicles[23]. Abs against tissue factor pathway inhibitor (TFPI) co-exist with antiphospholipid Abs and block the activity of TFPI followed by a decrease of the inhibition of Factor Xa[11,24]. In addition, inflammation by itself may change the hemostatic balance toward the pro-thrombotic status via enhanced expression of tissue factor and adhesion molecules on the endothelial cells and leukocytes and suppression of thrombomodulin and heparin-like glycosaminoglycans[25,26]. Inflammation also increases production of “acute phase proteins”, including fibrinogen, which is a strong risk factor for venous thrombosis[26,27]. However, thrombosis in SLE patients often develops irrespective of formation of antiphospholipid Abs and hyperfibrinogenemia, suggesting the existence of alternative pro-thrombotic mechanisms. Importantly, a higher incidence of thrombosis in SLE patients has been associated with an elevated level of anti-DNA Abs[28] as well as with hyperproduction of other antinuclear Abs[29], supporting the possibility that the circulating immune complexes containing dsDNA play an important role in the pathogenesis of venous thromboembolism in SLE. We have recently found that anti-dsDNA Abs purified form the blood of SLE patients increase the rate and degree of platelet-driven clot contraction, suggesting that they activate platelets and promote platelet contractility[30]. Therefore, we hypothesized that anti-dsDNA Abs formed at a high titer in the blood of SLE patients could cause the pro-thrombotic state through direct platelet activation.
Here, using various independent methods, we have shown that platelets are strongly activated by anti-dsDNA Abs alone and in a complex with dsDNA. Evidence for platelet activation is based on the release of α-granules assessed by P-selectin expression (Fig. 1A), a decrease of the mitochondrial membrane potential and cellular ATP content (Fig. 5), as well as on dramatic changes in platelet morphology (Tables 1, 2, Figs. 2; 3; S2), and septin 2 redistribution (Fig. 4). In platelets treated with anti-dsDNA Abs alone or in a complex with dsDNA, we observed grouped α-granules in the middle of a body and even merged α-granules (Fig. 3C,D) that likely comprise two consecutive steps of granular secretion through the open canalicular system[31,32]. Septins are cytoskeletal proteins[33] with poorly investigated involvement into platelet activation. Changes in septin 2 were shown to be tightly connected with alterations in platelet functionality, including apoptosis[34–37]. Previously, it has been demonstrated that septin 2 is expressed in platelets and is co-localized with actin and transferrin receptor (endocytosis marker)[38]. Also, septin 2 is co-localized with actin filaments primarily at or near the cell membrane, which is in agreement with our finding that in resting platelets septin 2 is mostly concentrated near the platelet periphery (Fig. 4A). We determined that the intracellular distribution of septin 2 is changed under the influence of the anti-dsDNA Ab/dsDNA complex from homogeneous to heterogeneous (clusters) (Fig. 4B). Together with the results of transmission and scanning electron microscopy (Figs. 2B; 2S,B), these data indicate profound cytoskeletal changes in the platelets activated by DNA-containing immune complexes.
A decreased mitochondrial membrane potential and content of ATP in platelets treated with the anti-dsDNA Ab/dsDNA complex (Fig. 5) is a sign of energetic exhaustion that follows platelet activation[39]. It is noteworthy that alterations in the platelet shape, intracellular location of α-granules and destruction of organelles have been described earlier in relation to other immune or inflammatory platelet stimulations induced by aggregated IgGs, immune complexes containing platelet factor 4 and heparin, or bacteria[40]. Formation of extracellular microvesicles and platelet fragmentation induced by the immune complex of anti-dsDNA Abs with dsDNA and revealed in this study are similar to platelet disintegration observed after stimulation with thrombin[41].
It is noteworthy that most, if not all, of the revealed effects of anti-dsDNA Abs on platelets were mediated via the FcγRIIA receptor, because blockage of this receptor with a mAb abolishes many of the changes described and therefore protects platelets from the activation induced by anti-dsDNA Abs and anti-dsDNA Ab/dsDNA complexes (Figs. 1A,B; 2C,F; S2C,F; 3C,F; 4C,F; Tables 1,2). The central role of FcγRIIA in platelet activation was shown earlier for immune complexes of various types and composition[40,42]. For example, the immune complexes of IgG with platelet factor 4 and heparin, which are formed in heparin-induced thrombocytopenia, lead to thrombocytopenia and thrombotic disorders through the FcγRIIA receptor[40,43,44]. Furthermore, platelets can be activated through the FcγRIIA receptor by bacteria as a part of an immune complex with IgG or in an IgG-independent way[40].
What was somewhat unexpected is that platelets were activated not only by immune complexes composed of anti-dsDNA Abs and dsDNA, but also by anti-dsDNA Abs alone in the absence of the exogenously added antigen. An artifact due to aggregation of anti-dsDNA Abs has been excluded by removing the potential aggregates via filtration with a 300-kDa exclusion limit, after which monomeric anti-dsDNA Abs maintains the ability to activate platelets (Fig. 1A). The lack of platelet-activating effects of the non-DNA-binding IgG purified from the same source with a similar procedure excludes the possibility of activation via potential contamination of endotoxin in the anti-dsDNA Ab preparations. There are several possible explanations of the direct activating effect of anti-dsDNA Abs on platelets. First, DNA has been shown to reside on the platelet surface[45], such that anti-dsDNA Abs can interact with the endogenous antigen on the plasma membrane and form immune complexes in situ that could activate platelets through the FcγRIIA receptor (Fig. 1B). Our results with pre-treatment of platelet with DNase I (Fig. 1B) confirm this scenario. Second, there is a cross-reactivity of anti-dsDNA Abs with the integrin β3 subunit[8], suggesting that anti-dsDNA Abs can either bind and activate the platelet integrins αIIbβ3 and αvβ3 and/or induce bridging and clustering between FcγRIIA and the β3 integrins, leading to platelet activation. The interplay between FcγRIIA and the integrins on platelets has been proposed as a mechanism for platelet activation in eptifibatide-dependent thrombocytopenia[46].
Irrespective of the activating stimulus and underlying mechanisms, platelet activation has multiple pathogenic implications in SLE beyond the risk of thrombotic complications[47]. For example, activated platelets are involved in dysregulation of the immune system by promoting NETosis[17,48], by expressing a soluble and platelet-associated protein CD154 or CD40 ligand belonging to the TNFα family[49], via expression on the plasma membrane and release of S100A8/A9 (calprotectin), a heterodimeric activator of toll-like receptor-4 found at a higher level in the blood of SLE patients that correlates strongly with cardiovascular complications[50]. Formation of platelet-derived microvesicles and platelet disintegration like those we observed upon treatment with anti-dsDNA Ab/dsDNA complexes (Figs. 3,D; 2S,B) provide a huge source of autoantigens. Also, the correlation between a disease activity score (SLEDAI-2k) and platelet count in SLE patients may partially be explained by platelet disintegration caused by anti-dsDNA Ab/dsDNA complexes[51]. In addition, activated platelets sustain complement activation, induce and promote activation and damage of the endothelium, leukocyte recruitment and vasoconstriction[52].
The results of this study that highlight the role of anti-dsDNA Abs in the pathogenesis of SLE may have practical importance. First, a high titer of anti-dsDNA Abs in the blood of SLE patients should be considered as a risk factor for thrombosis because hyper-production of these Abs and formation of dsDNA-containing immune complexes can lead to systemic platelet activation, making platelets highly adhesive to the vessel wall and to each other, which provides a strong predisposition to thrombosis[30,53]. Second, the pathogenic importance of the dsDNA and anti-dsDNA Abs suggests searching for new treatment modalities in autoimmune disorders that would reduce release of dsDNA, immune response to dsDNA, and dsDNA-related platelet activation. An example of such an approach is metformin, a drug used in type 2 diabetes, which turned out to be potentially promising in SLE by decreasing NETosis, release of mitochondrial DNA and production of interferon[54,55].
In summary, this study presents evidence that platelets can be strongly perturbed by anti-dsDNA Abs alone and in complex with dsDNA via the FcγRIIA receptor. The effects of anti-dsDNA Abs on platelets manifest as biochemical and structural alterations that comprise activation followed by energetic exhaustion, dysfunction, and disintegration. The activation of platelets induced by the Ab/DNA complexes is an important part of a more general immune platelet stimulation associated with autoimmune disorders. The activated platelets promote a pro-thrombotic state and underlie a high incidence of arterial and venous thrombosis in SLE patients. Platelet dysfunction resulting from continuous platelet activation can also have a pathogenic significance in SLE, leading to impaired contraction of intravital clots and thrombi, thus affecting their obstructiveness, mechanical and enzymatic stability[30]. Both the immune platelet activation and dysfunction have pathogenic implications beyond thrombosis and contribute to a profound dysregulation of the immune system in SLE and other autoimmune diseases.
Supplementary Material
Highlights:
Anti-dsDNA Abs from the blood of SLE patients activate platelets trough FcγRIIA.
Anti-dsDNA-Abs induce platelet degranulation and P-selectin expression.
Immune platelet activation includes rearrangement of cytoskeletal proteins.
Anti-dsDNA-Abs cause platelet microvesiculation and cytoplasmic disintegration.
Immune platelet activation is a prothrombotic mechanism in SLE.
Acknowledgments and finding sources
The authors thank Drs. Timur Sibgatullin and Adelya Maksudova for providing clinical data. Scanning electron microscopy was performed at the Interdisciplinary Center for Analytical Microscopy of Kazan Federal University. The work was supported by NIH grants UO1HL116330, HL116916, HL139448, HL142122, NSF grant DMR1505662, grant 18-415-160004 of the Russian Foundation for Basic Research and Republic of Tatarstan, and the Program for Competitive Growth at Kazan Federal University.
Abbreviations
- Ab(s)
antibody (antibodies)
- ATP
adenosine triphosphate
- dsDNA
double-stranded DNA
- EDTA
ethylenediaminetetraacetic acid
- ELISA
enzyme-linked immunosorbent assay
- FLS
Forward Light Scatter
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IgG
immunoglobulin G
- mAb
monoclonal antibody
- NETs
neutrophil extracellular traps
- NMWL
Nominal Molecular Weight Limit
- PBS
phosphate-buffered saline
- PBST
phosphate-buffered saline with Twin-20
- PRP
platelet-rich plasma
- SELENA
Safety of Estrogens in Lupus National Assessment study
- SLE
systemic lupus erythematosus
- SLEDAI
Systemic Lupus Erythematosus Disease Activity Index
- SLS
Side Light Scatter
- TMB
3,3′,5,5′-tetramethylbenzidine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
There is no conflict of interest.
References
- [1].Bertsias G, Cervera R, Boumpas DT, Systemic Lupus Erythematosus: Pathogenesis and Clinical Features, in: EULAR Textb. Rheum. Dis, BMJ / EULAR, London, 2012: pp. 476–505. https://www.eular.ors/myuploaddata/files/samplechapter20_mod17.pdf. [Google Scholar]
- [2].Mok CC, Lau CS, Pathogenesis of systemic lupus erythematosus., J. Clin. Pathol 56 (2003) 481–90. https://jcp.bmj.com/content/56/7/481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Riboldi P, Gerosa M, Moroni G, Radice A, Allegri F, Sinico A, Tincani A, Meroni PL, Anti-DNA antibodies: a diagnostic and prognostic tool for systemic lupus erythematosus?, Autoimmunity. 38 (2005) 39–45. 10.1080/08916930400022616. [DOI] [PubMed] [Google Scholar]
- [4].Pascual V, Farkas L, Banchereau J, Systemic lupus erythematosus: all roads lead to type I interferons, Curr. Opin. Immunol 18 (2006) 676–682. 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- [5].Vallin H, Perers A, V Alm G, Rönnblom L, Anti-double-stranded DNA antibodies and immunostimulatory plasmid DNA in combination mimic the endogenous IFN-alpha inducer in systemic lupus erythematosus, J. Immunol 163 (1999) 6306–6313. https://www.jimmunol.org/content/163/11/6306.long. [PubMed] [Google Scholar]
- [6].Kaplan MJ, Premature vascular damage in systemic lupus erythematosus: an imbalance of damage and repair?, Transl. Res 154 (2009) 61–69. 10.1016/j.trsl.2009.05.005. [DOI] [PubMed] [Google Scholar]
- [7].Boilard E, Blanco P, Nigrovic P. a., Platelets: active players in the pathogenesis of arthritis and SLE, Nat. Rev. Rheumatol 8 (2012) 534–542. https://www.nature.com/articles/nrrheum.2012.118. [DOI] [PubMed] [Google Scholar]
- [8].Zhang W, Dang S, Wang J, Nardi MA, Zan H, Casali P, Li Z, Specific crossreaction of anti-dsDNA antibody with platelet integrin GPIIIa49–66, Autoimmunity. 43 (2010) 682–9. doi:10.3109/08916934.2010.506207. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4702250&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Afeltra A, Vadacca M, Conti L, Galluzzo S, Mitterhofer AP, Ferri GM, Del Porto F, Caccavo D, Gandolfo GM, Amoroso A, Thrombosis in systemic lupus erythematosus: congenital and acquired risk factors, Arthritis Rheum. 53 (2005) 452–9. http://doi.wiley.com/10.1002/art.21172. [DOI] [PubMed] [Google Scholar]
- [10].Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, Mejía JC, Aydintug AO, Chwalinska-Sadowska H, de Ramon E, Fernández-Nebro A, Galeazzi M, Valen M, Mathieu A, Houssiau F, Caro N, Alba P, Ramos-Casals M, Ingelmo M, Hughes GRV, Morbidity and mortality in systemic lupus erythematosus during a 10-year period, Medicine (Baltimore). 82 (2003) 299–308. https://insights.ovid.com/crossref?an=00005792-200309000-00002. [DOI] [PubMed] [Google Scholar]
- [11].Palatinus A, Adams M, Thrombosis in systemic lupus erythematosus, Semin. Thromb. Hemost 35 (2009) 621–629. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=20013529&retmode=ref&cmd=prlinks%5Cnpapers3://publication/doi/10.1055/s-0029-1242716. [DOI] [PubMed] [Google Scholar]
- [12].Driest KD, Sturm MS, O’Brien SH, Spencer CH, Stanek JR, Ardoin SP, Factors associated with thrombosis in pediatric patients with systemic lupus erythematosus, Lupus. 25 (2016) 749–753. http://journals.sagepub.com/doi/10.1177/0961203316638164. [DOI] [PubMed] [Google Scholar]
- [13].Duffau P, Seneschal J, Nicco C, Richez C, Lazaro E, Douchet I, Bordes C, Viallard J-F, Goulvestre C, Pellegrin J-L, Weil B, Moreau J-F, Batteux F, Blanco P, Platelet CD154 potentiates interferon-α secretion by plasmacytoid dendritic cells in systemic lupus erythematosus, Sci. Transl. Med 2 (2010) 47ra63. http://stm.sciencemag.org/cgi/doi/10.1126/scitranslmed.3001001. [DOI] [PubMed] [Google Scholar]
- [14].Lood C, Amisten S, Gullstrand B, Jonsen A, Allhorn M, Truedsson L, Sturfelt G, Erlinge D, Bengtsson AA, Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease, Blood. 116 (2010) 1951–1957. https://www.ncbi.nlm.nih.gov/pubmed/20538795. [DOI] [PubMed] [Google Scholar]
- [15].Lood C, Eriksson S, Gullstrand B, Jönsen A, Sturfelt G, Truedsson L, Bengtsson A, Increased C1q, C4 and C3 deposition on platelets in patients with systemic lupus erythematosus – a possible link to venous thrombosis?, Lupus. 21 (2012) 1423–1432. http://journals.sagepub.com/doi/10.1177/0961203312457210. [DOI] [PubMed] [Google Scholar]
- [16].Nhek S, Clancy R, Lee KA, Allen NM, Barrett TJ, Marcantoni E, Nwaukoni J, Rasmussen S, Rubin M, Newman JD, Buyon JP, Berger JS, Activated platelets induce endothelial cell activation via an interleukin-1β pathway in systemic lupus erythematosus, Arterioscler. Thromb. Vasc. Biol 37 (2017) 707–716. https://www.ahajournals.org/doi/10.1161/ATVBAHA.116.308126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pfeiler S, Stark K, Massberg S, Engelmann B, Propagation of thrombosis by neutrophils and extracellular nucleosome networks, Haematologica. 102 (2017) 206–213. http://www.haematologica.org/cgi/doi/10.3324/haematol.2016.142471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hoppenbrouwers T, Autar ASA, Sultan AR, Abraham TE, van Cappellen WA, Houtsmuller AB, van Wamel WJB, van Beusekom HMM, van Neck JW, de Maat MPM, In vitro induction of NETosis: Comprehensive live imaging comparison and systematic review, PLoS One. 12 (2017) e0176472. http://dx.plos.org/10.1371/journal.pone.0176472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rauova L, Hirsch JD, Greene TK, Zhai L, Hayes VM, Kowalska MA, Cines DB, Poncz M, Monocyte-bound PF4 in the pathogenesis of heparin-induced thrombocytopenia, Blood. 116 (2010) 5021–5031. 10.1182/blood-2010-03-276964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tokhtaeva E, Sachs G, Vagin O, Diverse Pathways for Maturation of the Na,K-ATPase β 1 and β 2 Subunits in the Endoplasmic Reticulum of Madin-Darby Canine Kidney Cells, J. Biol. Chem 285 (2010) 39289–39302. http://www.jbc.org/lookup/doi/10.1074/jbc.M110.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Litman RM, A deoxyribonucleic acid polymerase from Micrococcus luteus (Micrococcus lysodeikticus) isolated on deoxyribonucleic acid-cellulose, J. Biol. Chem 243 (1968) 6222–6233. http://www.jbc.org/cgi/content/short/243/23/6222. [PubMed] [Google Scholar]
- [22].Nevzorova TA, Temnikov DA, Vinter VG, Special features of the DNA-hydrolyzing activity of the antibodies in systemic lupus erythematosus, Biochem. 68 (2003) 1300–1306. https://link.springer.com/article/10.1023/B:BIRY.0000011650.48894.7e. [DOI] [PubMed] [Google Scholar]
- [23].Arnout J, Vermylen J, Current status and implications of autoimmune antiphospholipid antibodies in relation to thrombotic disease, J. Thromb. Haemost 1 (2003) 931–942. http://doi.wiley.com/10.1046/j.1538-7836.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- [24].Forastiero RR, Martinuzzo ME, Broze GJ, High titers of autoantibodies to tissue factor pathway inhibitor are associated with the antiphospholipid syndrome, J. Thromb. Haemost 1 (2003) 718–724. http://doi.wiley.com/10.1046/j.1538-7836.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- [25].Esmon CT, Esmon NL, The link between vascular features and thrombosis, Annu. Rev. Physiol 73 (2011) 503–514. http://www.annualreviews.org/doi/10.1146/annurev-physiol-012110-142300. [DOI] [PubMed] [Google Scholar]
- [26].Tamaki H, Khasnis A, Venous thromboembolism in systemic autoimmune diseases: A narrative review with emphasis on primary systemic vasculitides, Vasc. Med 20 (2015) 369–376. http://journals.sagepub.com/doi/10.1177/1358863X15573838. [DOI] [PubMed] [Google Scholar]
- [27].de Moerloose P, Boehlen F, Neerman-Arbez M, Fibrinogen and the risk of thrombosis, Semin. Thromb. Hemost 36 (2010) 007–017. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0030-1248720. [DOI] [PubMed] [Google Scholar]
- [28].Petri MA, Conklin J, O’Malley T, Dervieux T, Platelet-bound C4d, low C3 and lupus anticoagulant associate with thrombosis in SLE, Lupus Sci. Med 6 (2019) e000318. http://lupus.bmj.com/lookup/doi/10.1136/lupus-2019-000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Natorska J, Celińska-Lowenhoff M, Undas A, High prevalence of antinuclear antibodies in patients following venous thromboembolism, Adv. Clin. Exp. Med 27 (2018) 827–832. http://www.advances.umed.wroc.pl/en/article/2018/27/6/827/. [DOI] [PubMed] [Google Scholar]
- [30].Le Minh G, Peshkova AD, Andrianova IA, Sibgatullin TB, Maksudova AN, Weisel JW, Litvinov RI, Impaired contraction of blood clots as a novel prothrombotic mechanism in systemic lupus erythematosus, Clin. Sci 132 (2018) 243–254. https://www.ncbi.nlm.nih.gov/pubmed/29295895. [DOI] [PubMed] [Google Scholar]
- [31].Pokrovskaya ID, Aronova MA, Kamykowski JA, Prince AA, Hoyne JD, Calco GN, Kuo BC, He Q, Leapman RD, Storrie B, STEM tomography reveals that the canalicular system and α-granules remain separate compartments during early secretion stages in blood platelets, J. Thromb. Haemost 14 (2016) 572–584. http://doi.wiley.com/10.1111/jth.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gibbins JM, Mahaut-Smith MP, Platelets and megakaryocytes, Humana Press Inc., Totowa, 2004. [Google Scholar]
- [33].Neubauer K, Zieger B, The mammalian septin interactome, Front. Cell Dev. Biol 5 (2017) 1–9. http://journal.frontiersin.org/article/10.3389/fcell.2017.00003/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schmidt K, Nichols BJ, Functional interdependence between septin and actin cytoskeleton., BMC Cell Biol. 5 (2004) 43. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=535351&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chattopadhyay M, Dahiya N, Atreya C, MicroRNA-223 regulates septin-2 and septin-6 in stored platelets, MicroRNA. 7 (2018) 223–228. http://www.eurekaselect.com/163220/article. [DOI] [PubMed] [Google Scholar]
- [36].Kaplan C, Steinmann M, Zapiorkowska NA, Ewers H, Functional redundancy of septin homologs in dendritic branching, Front. Cell Dev. Biol 5 (2017) 1–11. http://journal.frontiersin.org/article/10.3389/fcell.2017.00011/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thiele T, Steil L, Gebhard S, Scharf C, Hammer E, Brigulla M, Lubenow N, Clemetson KJ, Völker U, Greinacher A, Profiling of alterations in platelet proteins during storage of platelet concentrates, Transfusion. 47 (2007) 1221–1233. http://doi.wiley.com/10.1111/j.1537-2995.2007.01255.x. [DOI] [PubMed] [Google Scholar]
- [38].Bartsch I, Bläser S, Röseler S, Sandrock K, Busse A, Huber M, Rempp H, Lieber M, Horn J, Brendle C, Zieger B, Human endothelial and platelet septin SEPT11: Cloning of novel variants and characterisation of interaction partners, Thromb. Haemost 104 (2010) 1201–1210. http://www.thieme-connect.de/DOI/DOI?10.1160/TH10-07-0472. [DOI] [PubMed] [Google Scholar]
- [39].Holmsen H, Robkin L, Hydrogen peroxide lowers ATP levels in platelets without altering adenylate energy charge and platelet function, J. Biol. Chem 252 (1977) 1752–1757. https://www.ncbi.nlm.nih.gov/pubmed/838739. [PubMed] [Google Scholar]
- [40].Arman M, Krauel K, Human platelet IgG Fc receptor FcγRIIA in immunity and thrombosis, J. Thromb. Haemost 13 (2015) 893–908. http://doi.wiley.com/10.1111/jth.12905. [DOI] [PubMed] [Google Scholar]
- [41].Ponomareva AA, Nevzorova TA, Mordakhanova ER, Andrianova IA, Rauova L, Litvinov RI, Weisel JW, Intracellular origin and ultrastructure of platelet-derived microparticles, J. Thromb. Haemost 15 (2017) 1655–1667. http://doi.wiley.com/10.1111/jth.13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Qiao J, Al-Tamimi M, Baker RI, Andrews RK, Gardiner EE, The platelet Fc receptor, FcγRIIa, Immunol. Rev 268 (2015) 241–252. http://doi.wiley.com/10.1111/imr.12370. [DOI] [PubMed] [Google Scholar]
- [43].Rauova L, Poncz M, McKenzie SE, Reilly MP, Arepally G, Weisel JW, Nagaswami C, Cines DB, Sachais BS, Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia, Blood. 105 (2005) 131–138. https://www.ncbi.nlm.nih.gov/pubmed/15304392. [DOI] [PubMed] [Google Scholar]
- [44].Reilly MP, Heparin-induced thrombocytopenia/thrombosis in a transgenic mouse model requires human platelet factor 4 and platelet activation through FcgammaRIIA, Blood. 98 (2001) 2442–2447. https://www.ncbi.nlm.nih.gov/pubmed/11588041. [DOI] [PubMed] [Google Scholar]
- [45].Frampton G, Perl S, Bennett A, Cameron JS, Platelet-associated DNA and anti-DNA antibody in systemic lupus erythematosus with nephritis., Clin. Exp. Immunol 63 (1986) 621–8. http://www.ncbi.nlm.nih.gov/pubmed/3486735. [PMC free article] [PubMed] [Google Scholar]
- [46].Gao C, Boylan B, Bougie D, Gill JC, Birenbaum J, Newman DK, Aster RH, Newman PJ, Eptifibatide-induced thrombocytopenia and thrombosis in humans require FcγRIIa and the integrin β3 cytoplasmic domain, J. Clin. Invest 119 (2009) 504–511. http://www.jci.org/articles/view/36745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Linge P, Fortin PR, Lood C, Bengtsson AA, Boilard E, The non-haemostatic role of platelets in systemic lupus erythematosus, Nat. Rev. Rheumatol 14 (2018) 195–213. http://www.nature.com/articles/nrrheum.2018.38. [DOI] [PubMed] [Google Scholar]
- [48].Etulain J, Martinod K, Wong SL, Cifuni SM, Schattner M, Wagner DD, P-selectin promotes neutrophil extracellular trap formation in mice, Blood. 126 (2015) 242–246. https://www.ncbi.nlm.nih.gov/pubmed/25979951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Viallard J-F, Increased soluble and platelet-associated CD40 ligand in essential thrombocythemia and reactive thrombocytosis, Blood. 99 (2002) 2612–2614. https://www.ncbi.nlm.nih.gov/pubmed/11895803. [DOI] [PubMed] [Google Scholar]
- [50].Lood C, Tydén H, Gullstrand B, Jönsen A, Källberg E, Mörgelin M, Kahn R, Gunnarsson I, Leanderson T, Ivars F, Svenungsson E, Bengtsson AA, Platelet-derived S100A8/A9 and cardiovascular disease in systemic lupus erythematosus, Arthritis Rheumatol. 68 (2016) 1970–1980. http://doi.wiley.com/10.1002/art.39656. [DOI] [PubMed] [Google Scholar]
- [51].Abdel Galil SM, Edrees AM, Ajeeb AK, Aldoobi GS, El-Boshy M, Hussain W, Prognostic significance of platelet count in SLE patients, Platelets. 28 (2017) 203–207. https://www.tandfonline.com/doi/full/10.1080/09537104.2016.1214253. [DOI] [PubMed] [Google Scholar]
- [52].Scherlinger M, Guillotin V, Truchetet M-E, Contin-Bordes C, Sisirak V, Duffau P, Lazaro E, Richez C, Blanco P, Systemic lupus erythematosus and systemic sclerosis: All roads lead to platelets, Autoimmun. Rev 17 (2018) 625–635. http://linkinghub.elsevier.com/retrieve/pii/S1568997218300910. [DOI] [PubMed] [Google Scholar]
- [53].Zhi H, Dai J, Liu J, Zhu J, Newman DK, Gao C, Newman PJ, Platelet activation and thrombus formation over IgG immune complexes requires integrin αIIbβ3 and lyn kinase, PLoS One. 10 (2015) e0135738. https://dx.plos.org/10.1371/journal.pone.0135738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Xin G, Wei Z, Ji C, Zheng H, Gu J, Ma L, Huang W, Morris-Natschke SL, Yeh J-L, Zhang R, Qin C, Wen L, Xing Z, Cao Y, Xia Q, Lu Y, Li K, Niu H, Lee K-H, Huang W, Metformin uniquely prevents thrombosis by inhibiting platelet activation and mtDNA release, Sci. Rep 6 (2016) 36222. http://www.nature.com/articles/srep36222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang H, Li T, Chen S, Gu Y, Ye S, Neutrophil extracellular trap mitochondrial DNA and its autoantibody in systemic lupus erythematosus and a proof-of-concept trial of metformin, Arthritis Rheumatol. 67 (2015) 3190–3200. http://doi.wiley.com/10.1002/art.39296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.