Abstract
Here, we describe a three-step nested-PCR-denaturing gradient gel electrophoresis (DGGE) strategy to detect sulfate-reducing bacteria (SRB) in complex microbial communities from industrial bioreactors. In the first step, the nearly complete 16S rRNA gene was amplified using bacterial primers. Subsequently, this product was used as a template in a second PCR with group-specific SRB primers. A third round of amplification was conducted to obtain fragments suitable for DGGE. The largest number of bands was observed in DGGE patterns of products obtained with primers specific for the Desulfovibrio-Desulfomicrobium group, indicating a large diversity of these SRBs. In addition, members of other phylogenetic SRB groups, i.e., Desulfotomaculum, Desulfobulbus, and Desulfococcus-Desulfonema-Desulfosarcina, were detected. Bands corresponding to Desulfobacterium and Desulfobacter were not detected in the bioreactor samples. Comparative sequence analysis of excised DGGE bands revealed the identity of the community members. The developed three-step PCR-DGGE strategy is a welcome tool for studying the diversity of sulfate-reducing bacteria.
Sulfate-reducing bacteria (SRB) form a phylogenetically diverse and heterogeneous group of anaerobic bacteria using sulfate as a terminal electron acceptor in the degradation of organic matter, resulting in the production of H2S. They are ubiquitous and play an important role in the biogeochemical sulfur cycle. Sulfate reduction dominates the organic matter degradation in environments with high concentrations of sulfate. It has been estimated, for instance, that sulfate reduction accounts for up to 50% of the total organic matter degradation in marine sediments (8). Apart from marine sediments, their presence has also been demonstrated in other environments, such as freshwater lake sediments (30), anaerobic biofilms (3, 26), oil production facilities (24), and wastewater treatment plants (20). Although SRB are generally considered to be obligatory anaerobic bacteria, they have also been detected in aerobic environments, such as the oxic zones of cyanobacterial mats (9) and wastewater biofilms grown under oxic conditions (25).
Because of the importance of SRB to critical processes in ecosystem functioning and environmental remediation, increasing interest in SRB has been shown over the last decade. Different culture-independent methods have been used to study SRB populations in various ecosystems, resulting in an increased knowledge of their diversity. 16S rRNA-targeted oligonucleotide probes specific for SRB have been used in fluorescence in situ hybridization for the detection of these microorganisms in a variety of environments (27). Genes encoding important enzymes in the sulfur cycle have also been used to detect sulfate-reducing bacteria in different environments (14, 33). 16S rRNA-targeted PCR primer sequences specific for SRB subgroups have been designed and used to detect phylogenetic subgroups of SRB (2). Recently, a DNA microarray suitable for SRB diversity analysis has been developed and applied to detect SRB in complex environmental samples (11).
Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified DNA fragments is another molecular tool that has been used to determine the presence and distribution of SRB in natural and engineered environments (29). However, although successful, the banding pattern represents mainly the major constituents of the analyzed community (7). Species that contribute less than 1% of the total population would not be readily detected by this molecular approach (15).
Here, we present a novel strategy to overcome the difficulty in detecting low numbers of sulfate-reducing bacteria in complex microbial communities from natural environments. The strategy consists of a three-step nested-PCR-DGGE approach, i.e., a first amplification step with bacterial primers to amplify the nearly complete 16S rRNA encoding gene, subsequently, a second amplification step using SRB group-specific primers, and a last amplification step to create a DNA fragment suitable for DGGE analysis. Simultaneous DGGE analysis of PCR products obtained by the direct and the indirect approach made it possible to infer the relative abundance of SRB in the samples. The nested-PCR-DGGE approach described herein is a welcome tool in the diversity analysis of SRB in natural samples.
MATERIALS AND METHODS
Reference strains.
Six strains, namely, Desulfotomaculum nigrificans DSM 574, Desulfobulbus propionicus DSM 2032, Desulfobacterium autotrophicum DSM 3382, Desulfobacter postgatei DSM 2034, Desulfosarcina variabilis DSM 2060, and Desulfovibrio vulgaris DSM 644, representing main phylogenetic groups of SRB, were used as reference strains in this study.
Sludge samples.
Samples from two different anaerobic UASB (upflow anaerobic sludge bed) wastewater treatment reactors, i.e., BSD1 and BSD2, were obtained from Biothane Systems Delft, and used to demonstrate the proof-of-principle of this nested-PCR-DGGE strategy. The two reactors treating phthalate- and lactate-containing wastewater were maintained under mesophilic conditions at 37°C and at pH 6.8 to 7.2. The chemical oxygen demand and sulfate in the pthalate-containing wastewater were 9,000 and 90 ppm, respectively, while the same in lactate-containing wastewater were 12,000 and 120 ppm, respectively.
DNA extraction.
Genomic DNA was isolated from the reference strains and the reactor samples using the Ultra Clean Soil DNA isolation kit (MOBIO Laboratories) according to the manufacturer's protocol. The yield and quality of DNA were analyzed electrophoretically on 1% (wt/vol) agarose gel.
Oligonucleotides used in this study.
Details of the different oligonucleotides used in this study are shown in Table 1. Oligonucleotides DFM140 to DSV838 are specific for the 16S rRNA of different phylogenetic groups of SRB designed by Daly et al. (2). The primer pair GM3F/GM4R amplifies the nearly complete sequence of 16S rRNA of members of the bacteria domain (16). The primer 341F(GC) can be used in combination with 518R for the amplification of bacterial 16S rRNA suitable for DGGE analysis (15).
TABLE 1.
Primers used in this study
| Primer set | Target group | Target organism(s) | Positions | Annealing temp | Reference |
|---|---|---|---|---|---|
| DFM140 | 1 | Desulfotomaculum | 140-158 | 58°C | 2 |
| DFM842 | 823-842 | ||||
| DBB121 | 2 | Desulfobulbus | 121-142 | 66°C | 2 |
| DBB1237 | 1215-1237 | ||||
| DBM169 | 3 | Desulfobacterium | 169-183 | 64°C | 2 |
| DBM1006 | 986-1006 | ||||
| DSB127 | 4 | Desulfobacter | 127-148 | 60°C | 2 |
| DSB1273 | 1252-1273 | ||||
| DCC305 | 5 | Desulfonema-Desulfo- sarcina-Desulfococcus | 305-327 | 65°C | 2 |
| DCC1165 | 1144-1165 | ||||
| DSV230 | 6 | Desulfovibrio-Desulfo- microbium | 230-248 | 61°C | 2 |
| DSV838 | 818-838 | ||||
| GM3F | Bacteria | 8-24 | 45°C | 16 | |
| GM4R | 1492-1507 | ||||
| 341F(GC) | Bacteria | 341-357 | 65-55°Ca | 15 | |
| 518R | 518-534 |
These primers were used in a so-called “touchdown” protocol from 65°C to 55°C.
Amplification of the 16S rRNA using group-specific SRB primers.
Amplification of 16S rRNA was attempted on the DNA extracted from the two reactor samples using SRB group-specific primers (Table 1). The reactions were carried out as follows: 5-min initial denaturation of DNA at 95°C, followed by 30 cycles of 1-min denaturation at 94°C, 1-min primer annealing at the temperature specific for each primer pair (Table 1), and 1-min extension at 72°C. Amplification was completed by a final extension step at 72°C for 7 min. A nested amplification of 16S rRNA was also attempted using the universal bacterial primers GM3F and GM4R (annealing temperature 45°C). The product obtained was used as template in the second PCR with group-specific primers.
Three-step nested-PCR-DGGE.
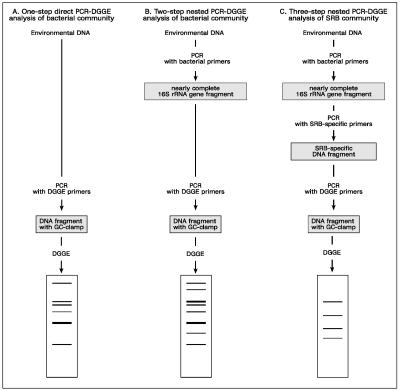
Two strategies were used to analyze the bacterial community in the two reactors (Fig. 1). First, the 16S rRNA fragment was amplified using the primer pair 341F(GC)/518R (Fig. 1A). The PCR was performed using a touchdown annealing protocol (annealing temperature decreased from 65°C to 55°C in 20 cycles). Second, a three-step nested amplification was performed to obtain different SRB group-specific 16S rRNA fragments suitable for DGGE (Fig. 1C). In the first step, a nearly complete 16S rRNA gene fragment was amplified using the universal primer pair GM3F/GM4R. The product obtained was used as a template for a second amplification with SRB group-specific primers. Finally, to generate products suitable for DGGE, a third round of amplification was performed with DGGE primers 341F(GC) and 518R using the product of the second round as template.
FIG. 1.
Schematic overview of the different PCR-DGGE strategies used to study the diversity of sulfate-reducing bacteria in complex microbial communities of anaerobic bioreactors. (A) One-step direct PCR-DGGE strategy; (B) two-step nested-PCR-DGGE strategy; (C) three-step nested-PCR-DGGE strategy. Comparative DGGE analysis of PCR products obtained by strategies A/B and C made it possible to infer the relative abundance of SRB in the samples analyzed.
The PCR products obtained were subjected to DGGE analysis. DGGE was performed as described by Muyzer et al. (17). Briefly, a denaturing gradient of 35 to 75% denaturants (100% denaturants is a mixture of 7 M urea and 40% [vol/vol] formamide) was used in 6% (wt/vol) polyacrylamide gel. DGGE was performed in TAE (Tris-acetate-EDTA) buffer (1×) at 60°C and at a constant voltage of 70 V for 16 h. Following electrophoresis, the gels were incubated in an ethidium bromide solution for 30 min and then rinsed in Milli-Q water before being photographed using a Bio-Rad GelDoc station. Individual bands were excised, reamplified, and run again on a denaturing gradient gel to check their purity. PCR products for sequencing were purified using the Qiaquick PCR purification kit (QIAGEN).
Comparative sequence analysis.
The sequences were first compared to sequences stored in GenBank using the BLAST algorithm. Subsequently, the sequences were imported into the ARB software program, aligned, and added to a phylogenetic tree using the Quick add tool (12). The alignment was further corrected by eye, and a definitive tree was calculated using the neighbor-joining algorithm with Felsenstein correction.
Nucleotide sequence accession numbers.
The sequences were deposited in GenBank under the accession numbers AY817418 to AY817454.
RESULTS AND DISCUSSION
Direct and nested amplification using SRB group-specific primers.
First, a direct amplification of the 16S rRNA encoding gene with genomic DNA extracted from the reactor samples and primers specific for different groups of SRB was attempted. A PCR product was obtained only with genomic DNA from a sample of reactor BSD2 and primers specific for members of the genera Desulfovibrio-Desulfomicrobium (i.e., target group 6). No products were obtained with other group-specific primers and DNA extracted from either of the reactor samples. Nested amplification, however, yielded additional products corresponding to Desulfotomaculum (target group 1), Desulfobulbus (target group 2), and Desulfococcus-Desulfonema-Desulfosarcina (target group 5). A Desulfobulbus group-like product was obtained from the BSD1 sample only, while the other groups were obtained from both the reactor samples. Daly et al. made similar observations while studying the SRB populations in landfill leachate using group-specific SRB primers. They could detect three of the six SRB groups using the direct PCR approach, while the additional two groups were detected only when a nested-PCR approach was applied (2). Nested- or double-PCR protocols have been used before to improve the sensitivity of detection (4, 5, 7, 10). More recently, the nested-PCR approach, when used for the detection of chloroethane reductive dehalogenase genes, was found to increase the sensitivity by reducing the limit of detection to 102 copies/reaction (28).
The PCR products obtained by the direct and nested amplification indicated that only members of the Desulfovibrio-Desulfomicrobium group were dominant in the sample of the BSD2 reactor. Members belonging to the Desulfotomaculum, Desulfobulbus, and Desulfococcus-Desulfonema-Desulfosarcina group could be present in low numbers, as these were detected only after using the nested PCR. The enhanced detection signal in the nested PCR may be due to the first-round PCR resulting in the amplification of sufficient amounts of DNA even from the groups present in low numbers and also due to the dilution of inhibitory substances such as humic acids present in the sample. SRB belonging to the Desulfobacter or Desulfobacterium group were not detected in these reactor samples with either of the approaches. This corresponds to some of the previous studies that have found Desulfobacter (21, 34) and Desulfobacterium (6, 23) associated with marine environments.
SRB diversity analysis using a nested-PCR-DGGE strategy.
The molecular detection of microorganisms by DGGE may become difficult if they are present in low numbers, even more so in case of sludge samples of wastewater treatment plants, which consist of a complex mixtures of microorganisms. The DGGE with eubacterial primers mainly detects the major constituents of the analyzed community overlooking the less abundant but potentially very important species (15). The same problem was envisaged while analyzing the SRB populations in the sludge samples of BSD1 and BSD2 reactors, thus necessitating the use of nested-PCR-DGGE with SRB group-specific primers. A similar strategy has been successfully used in the detection of ammonia oxidizers, when the abundance of these microorganisms was 0.01% of the total bacterial population (22). Boon et al. (1) analyzed the diversity of bacterial groups from the wastewater treatment plants using the nested-PCR-DGGE approach, while the same approach was used for the species-specific analysis of bifidobacterial communities by Temmerman et al. (32).
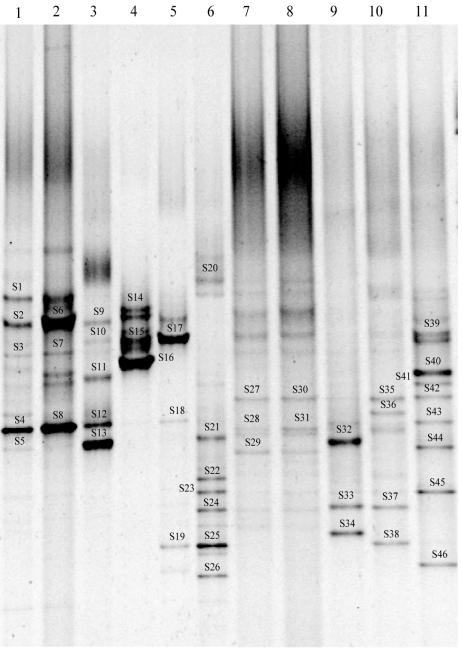
Two different strategies were applied to analyze the SRB diversity in the bioreactors. In the first approach (Fig. 1A), the 16S rRNA products generated directly with the DGGE primers 341F(GC) and 518R from the DNA extract of the samples BSD1 and BSD2 were used for analysis in DGGE. The DGGE pattern obtained gave an overall bacterial diversity within the reactors. However, the pattern resulted in only a few bands that may be representing the dominant bacterial groups in the ecosystem. Only one weak band could be identified as that belonging to SRB (Fig. 2, band 5), indicating that the SRB are not among the dominant community members in the reactors. This seems consistent with the results of direct PCR amplification performed to generate the SRB-specific 16S rRNA fragments. In the second approach (Fig. 1C), the pattern derived from the products of three-step PCR consisted only of the SRB bands, since by using SRB-specific primers in the second step, this approach excludes the amplification of non-SRB bacteria (Fig. 2, lanes 3 through 6 and 9 through 11; Table 2). This approach also enabled the comparison of the group-specific pattern of SRB to the bacterial community pattern of the same sample.
FIG. 2.
DGGE patterns of 16S rRNA fragments obtained after enzymatic amplification using different primer pairs and DNA from two anaerobic bioreactors. Lanes: 1 to 6, DGGE pattern of samples from BSD1; 7 to 11, DGGE pattern of samples from BSD2; 1 and 7, DGGE pattern obtained with PCR products amplified using strategy A; 2 and 8, DGGE pattern obtained from the product amplified using the two-step nested approach (strategy B); 3 and 9, pattern obtained when primers specific to the Desulfotomaculum group were used in the three-step nested approach; 4, pattern obtained when primers specific to the Desulfobulbus group were used in the three-step nested approach (strategy C); 5 and 10, pattern obtained when primers specific to the Desulfococcus-Desulfonema-Desulfosarcina group were used in the three-step nested approach; 6 and 11, pattern obtained when primers specific to the Desulfovibrio-Desulfomicrobium group were used in the three-step nested approach. Bands that were excised for sequence analysis are numbered.
TABLE 2.
Sequence similarity of excised DNA fragments
| Band no.a | Database match with accession no. in parentheses | % Similarity | Phylogenetic group |
|---|---|---|---|
| BSD-S1 | Chryseobacterium sp. (AY244770) | 96 | Flavobacteria |
| BSD-S2 | Pseudomonas aeruginosa (X06684) | 99 | γ-Proteobacteria |
| BSD-S4 | Syntrophus buswellii (X85131) | 99 | δ-Proteobacteria |
| BSD-S5 | Desulfotomaculum sp. 175 (AF295656) | 95 | Firmicutes |
| BSD-S6 | Pseudomonas aeruginosa (X06684) | 99 | γ-Proteobacteria |
| BSD-S8 | Syntrophus buswellii (X85131) | 99 | δ-Proteobacteria |
| BSD-S12 | Syntrophus buswellii (X85131) | 99 | δ-Proteobacteria |
| BSD-S13 | Desulfotomaculum sp. 175 (AF295656) | 100 | Firmicutes |
| BSD-S14 | Desulfobulbus elongatus (X95180) | 98 | δ-Proteobacteria |
| BSD-S15 | Desulfobulbus rhabdoformis (U12253) | 98 | δ-Proteobacteria |
| BSD-S16 | Desulfobulbus propionicus (AY548789) | 95 | δ-Proteobacteria |
| BSD-S17 | Sulfate-reducing bacterium R-ButA1 (AJ012596) | 100 | δ-Proteobacteria |
| BSD-S19 | Uncultured Desulfosarcina sp. (AY177791) | 98 | δ-Proteobacteria |
| BSD-S20 | Desulfovibrio aminophilus (AF067964) | 96 | δ-Proteobacteria |
| BSD-S23 | Desulfovibrio longus (AY359867) | 98 | δ-Proteobacteria |
| BSD-S24 | Desulfovibrio indonesiensis (Y09504) | 100 | δ-Proteobacteria |
| BSD-S25 | Desulfovibrio sp. Met 82 (AY062930) | 100 | δ-Proteobacteria |
| BSD-S26 | Desulfovibrio aminophilus (AF067964) | 100 | δ-Proteobacteria |
| BSD-S27 | Flavobacterium-like sp. (AY353822) | 97 | Flavobacteria |
| BSD-S29 | Uncultured bacterial clone TDC-S1 (AF447150) | 99 | Unknown |
| BSD-S30 | Flavobacterium-like sp. (AY353822) | 95 | Flavobacteria |
| BSD-S31 | Uncultured bacterium SJA-152 (AJ009496) | 99 | unknown |
| BSD-S32 | Desulfotomaculum sp. 175 (AF295656) | 96 | Firmicutes |
| BSD-S33 | Uncultured Desulfotomaculum sp. (AY261820) | 97 | Firmicutes |
| BSD-S34 | Desulfotomaculum sp. OX39 (AJ577273) | 95 | Firmicutes |
| BSD-S35 | Uncultured Desulfosarcina (AY177788) | 98 | δ-Proteobacteria |
| BSD-S36 | Uncultured δ-proteobacterium (AJ240977) | 99 | δ-Proteobacteria |
| BSD-S37 | Desulfonema ishimotonii (U45991) | 98 | δ-Proteobacteria |
| BSD-S38 | Desulfonema ishimotonii (U45991) | 96 | δ-Proteobacteria |
| BSD-S39 | Desulfovibrio alcoholivorans (AF053751) | 94 | δ-Proteobacteria |
| BSD-S40 | Desulfovibrio fructosivorans (AF050101) | 95 | δ-Proteobacteria |
| BSD-S41 | Desulfovibrio burkinensis (AF053752) | 97 | δ-Proteobacteria |
| BSD-S42 | Desulfovibrio sp. BG6 (U85468) | 98 | δ-Proteobacteria |
| BSD-S43 | Desulfovibrio fructosivorans (AF050101) | 95 | δ-Proteobacteria |
| BSD-S44 | Desulfovibrio sp. BG6 (U85468) | 98 | δ-Proteobacteria |
| BSD-S45 | Desulfovibrio indonesiensis (Y09504) | 95 | δ-Proteobacteria |
| BSD-S46 | Desulfovibrio sp. BG6 (U85468) | 96 | δ-Proteobacteria |
Bands BSD-S1 to BSD-S46 are the same bands as S1 to S46 in the denaturing gradient gel (Fig. 2), BSD-S1 to BSD-S28 were obtained from bioreactor BSD1, and BSD-S29 to BSD-S46 were obtained from bioreactor BSD2.
Initially, the two-step nested amplification products (Fig. 1B) from the samples were compared to the one-step direct amplification products and the DGGE pattern obtained (Fig. 2, lanes 1, 2, 7, and 8) was found to be similar, if not identical. The bands in the DGGE pattern of nested products were found to be of increased intensity. The results of this comparison more or less ruled out the possible bias of preferential amplification from successive PCRs (31). Moreover, according to previous studies (7), this bias of preferential amplification may be overestimated, and the intensity of the bands in DGGE corresponds semiquantitatively with the abundance of species.
The highest number of bands was observed in the DGGE pattern of the Desulfovibrio-Desulfomicrobium group, indicating a very high diversity within this group (Fig. 2, lanes 6 and 11). Although the Desulfovibrio-Desulfomicrobium-like products could be amplified through direct amplification, indicating their abundance in the overall bacterial community, no DGGE band in the overall bacterial community pattern could be identified as belonging to the Desulfovibrio-Desulfomicrobium group. The reason may be the high diversity within this group, resulting in too little PCR product per species to give visible bands.
The choice of the primer pair 341F(GC) and 518R, which generates DNA fragments suitable for DGGE analysis, was made because all DNA fragments obtained with the SRB group-specific primers in the second amplification step included the target sites for these DGGE primers. However, primer pair 341F(GC) and 907R, which amplified a larger gene fragment with more sequence information, gave good results as well, although only for those SRB group-specific primers that included the target sites for these primers, i.e., primers for the Desulfobulbus and Desulfococcus-Desulfonema-Desulfosarcina group (results not shown).
Comparative sequence analysis.
Separated DNA fragments were excised and sequenced to substantiate the presence of particular SRB groups in the two reactors. A total of 46 bands (Fig. 2) were excised from the DGGE patterns, out of which 9 yielded ambiguous sequences, which were not further analyzed. By using the BLAST search algorithm, high similarity values were found with sequences of SRB for which the primers were designed (Table 2). Phylogenetic analysis confirmed these results (Fig. 3). The sequences of DGGE bands BSD-S14 to BSD-S16 nicely grouped with the sequence of Desulfobulbus propionicus. The sequences of band BSD-S19 and BSD-S35 to BSD-S38 grouped together and were affiliated with Desulfosarcina variabilis. The sequences of bands BSD-S39 to BSD-S44 formed a coherent group with the closest relative, Desulfovibrio fructosivorans, while the sequences of bands BSD-S20 and BSD-S23 to BSD-S26 were grouping with the sequence of Desulfovibrio aminophilus. The sequences of BSD-S5, BSD-S6, and BSD-S32 to BSD-S34 formed a group distantly related to Desulfotomaculum geothermicum, while the sequence of bands BSD-S45 and BSD-S46 were more related to Desulfotomaculum nigrificans.
FIG. 3.
Phylogenetic tree showing the affiliation of sulfate-reducing bacteria from two different anaerobic bioreactors. The numbers of the sequences in this tree (e.g., BSD-S16) refer to the numbers in the denaturing gradient gel (i.e., S16 [Fig. 2]). The bar indicates 10% sequence variation.
The bacterial pattern obtained from the sludge samples of the two reactors showed the presence of only a few dominant species, none of which belonged to SRB. Detection of SRB-related bands from the same samples when SRB-specific primers were used confirmed that the DGGE pattern does not reflect the actual diversity of, but only numerically dominant species in, the sample.
Sequencing of DGGE bands from the bacterial pattern of the two samples showed that the bacterial populations belonged to γ- and δ-proteobacteria as well as flavobacteria. DGGE analysis of one-step PCR products from the BSD1 sample revealed three dominant bands of which band BSD-S4 (Fig. 2, lane 1) seemed to be the most intense. The sequence of this band was found to be affiliated with the bacterial genus Syntrophus, which forms syntrophic relationships with methanogens. The presence of aromatic compounds like phthalate and benzoate in the wastewater fed to the reactor BSD1 explains the predominance of Syntrophus buswellii in this reactor. These bacteria degrade aromatic compounds such as benzoate to acetate and hydrogen. In the terephthalate-degrading anaerobic sludge system, δ-proteobacteria closely affiliated with the bacterial genera Syntrophus that form syntrophic relationships with methanogens to degrade aromatic compounds have been found (35).
The other two sequences (bands BSD-S1 and BSD-S2) also seemed predominant members of the microbial community. Band BSD-S1 had 96% sequence similarity to Flavobacterium strains, and band BSD-S2 had 99% sequence similarity to γ-proteobacteria. Populations related to Chryseobacterium spp. represented band BSD-S1, and Pseudomonas spp. represented band BSD-S2. Anaerobic metabolism of phthalate and other aromatic compounds by denitrifying bacteria like Pseudomonas has been already established (18, 19).
The bacterial pattern in reactor BSD2 (Fig. 2, lanes 7 and 8) did not show many intense bands. Only a few weak bands were visible, of which two could be sequenced. The sequencing showed that the bands belonged to either uncultured bacteria or Flavobacterium-like species. Members of the Cytophaga-Flavobacterium group are ubiquitous microorganisms and are known to have a diverse physiology. Previous studies (13) have found Cytophaga-Flavobacterium among the dominant members in industrial treatment facilities.
In conclusion, the described three-step nested-PCR-DGGE approach makes it possible to study the diversity of sulfate-reducing bacteria with high resolution in samples from mixed microbial communities containing SRB in low number. However, the specificity of the primers targeting different phylogenetic groups of SRB is of prime importance for the success of this approach. The method is robust, reproducible, and rapid and reveals sequences for phylogenetic analysis and probe design.
Acknowledgments
We are grateful to Marc de Pijper (Biothane Systems Delft) for providing bioreactor samples.
This work was supported financially by the Dutch Science Foundation—Earth and Life Sciences (NWO-ALW).
REFERENCES
- 1.Boon, N., W. De Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group- specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 2.Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693-1705. [DOI] [PubMed] [Google Scholar]
- 3.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.el Fantroussi, S., J. Mahillon, H. Naveau, and S. N. Agathos. 1997. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested-PCR monitoring. Appl. Environ. Microbiol. 63:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erb, R. W., and I. Wagner-Döbler. 1993. Detection of polychlorinated biphenyl degradation genes in polluted sediments by direct DNA extraction and polymerase chain reaction. Appl. Environ. Microbiol. 59:4065-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauque, G. D. 1995. Ecology of sulfate-reducing bacteria, p. 217-241. In L. L. Barton (ed.), Sulfate-reducing bacteria. Plenum, New York, N.Y.
- 7.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities, p. 353-373. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 8.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea bed—the role of sulfate reduction. Nature (London) 296:643-645. [Google Scholar]
- 9.Krekeler, D., P. Sigalevich, A. Teske, H. Cypionka, and Y. Cohen. 1997. A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai), Desulfovibrio oxyclinae sp. nov. Arch. Microbiol. 167:369-375. [Google Scholar]
- 10.Levesque, M. J., S. La Boissière, J. C. Thomas, R. Beaudet, and R. Villemur. 1997. Rapid method for detecting Desulfitobacterium frappieri strain PCP-1 in soil by the polymerase chain reaction. Appl. Microbiol. Biotechnol. 47:719-725. [DOI] [PubMed] [Google Scholar]
- 11.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manz, W., M. Wagner, R. Amann, and K. -H. Schleifer. 1994. In situ characterization of the microbial consortia active in two wastewater treatment plants. Water Res. 28:1715-1723. [Google Scholar]
- 14.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 17.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Nozawa, T., and Y. Maruyama. 1988. Denitrification by a soil bacterium with phthalate and other aromatic compounds as substrates. J. Bacteriol. 170:2501-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nozawa, T., and Y. Maruyama. 1988. Anaerobic metabolism of phthalate and other aromatic compounds by a denitrifying bacterium. J. Bacteriol. 170:5778-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analysis of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oude Elferink, S. J. W. H., H. T. S. Boschker, and A. J. M. Stams. 1998. Identification of sulfate reducers and Syntrophobacter sp. in anaerobic granular sludge by fatty-acid biomarkers and 16S rRNA probing. Geomicrobiol. J. 15:3-17. [Google Scholar]
- 22.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postgate, J. R. 1984. The sulfate reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, England.
- 24.Rabus, R., M. Fukui, H. Wilkis, and F. Widdel. 1996. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic environment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsing, N., M. Kühl, and B. B. Jorgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probe and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raskin, L., R. I. Amann, L. K. Poulsen, B. E. Rittmann, and D. A. Stahl. 1995. Use of ribosomal RNA-based molecular probes for characterization of complex microbial communities in anaerobic biofilms. Water Sci. Technol. 31:261-272. [Google Scholar]
- 27.Ravenschlag, K., K. Sahm, C. Knoblauch, B. Jørgensen, and R. I. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regeard, C., J. Maillard, and C. Holliger. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107-118. [DOI] [PubMed] [Google Scholar]
- 29.Santegoeds, C. M., L. R. Damgaard, G. Hesselink, J. Zopfi, P. Lens, G. Muyzer, and D. de Beer. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analysis. Appl. Environ. Microbiol. 65:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sass, H., H. Cypionka, and H.-D. Babenzien. 1997. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol. Ecol. 22:245-255. [Google Scholar]
- 31.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Temmerman, R., L. Masco, T. Vanhoutte, G. Huys, and J. Swings. 2003. Development and validation of a nested-PCR-denaturing gradient gel electrophoresis method for taxonomic characterization of bifidobacterial communities. Appl. Environ. Microbiol. 69:6380-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469-585. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, Inc., New York, N.Y.
- 35.Wu, J. H., W. T. Liu, I. C. Tseng, and S. S. Cheng. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373-382. [DOI] [PubMed] [Google Scholar]