Abstract
Background
Antimicrobial resistance poses a huge risk to human health worldwide, while Bangladesh is confronting the most severe challenge between the food supply and the huge consumption of antibiotics annually. More importantly, probiotics containing Bacillus spp. are claimed to be an alternative to antimicrobial stewardship programs. However, their antibiotic resistance remains elusive. Thus, we employed the antimicrobial susceptibility test and PCR to assess the prevalence of resistance, including multidrug resistance (MDR) and resito-genotyping of isolated Bacillus spp.
Results
The phenotypic profile showed that Bacillus spp. were 100% sensitive to gentamicin (2 µg/mL), whereas lowered sensitivity to levofloxacin (67.8%, 0.5–1 µg/mL), ciprofloxacin (62.3%, 0.5–1 µg/mL), clindamycin (52.2%, 0.25–0.5 µg/mL), amoxicillin-clavulanic acid (37.6%, 0.06 µg/mL), azithromycin (33.4%, 1–2 µg/mL), tetracycline (25.6%, 2–4 µg/mL), nitrofurantoin (21.1%, 16–32 µg/mL), co-trimoxazole (19.2%, 2 µg/mL), and erythromycin (18.8%, 0.25–0.5 µg/mL). The strains were completely resistant to penicillin, amoxicillin-clavulanic acid, cefixime, ceftriaxone, vancomycin, and co-trimoxazole, and a species-specific trend was seen in both phenotypic and genotypic resistance patterns. Genotypic resistance indicated prevalence of the bla1 (71.5%), tetA (33%), erm1 (27%), blaTEM (13.1%), blaCTX-M-1/blaCTX-M-2 /sul1 (10.1%), blaSHV (9.6%), and qnrS (4.1%) genes. The β-lactamase resistance gene bla1 was found in all penicillin-resistant (MIC ≥ 32 µg/mL) Bacillus spp. One hundred ninety-one isolates (89.6%) were MDR, with 100% from diarrhea, 90.3% from food, and 88.7% from animal feed.
Conclusion
Based on the MIC value and profile analysis of antibiotic resistance genes, this is the first study that Bacillus spp. antimicrobial susceptibilities have been identified in Bangladesh, and our study will shed light on the adverse effects of feed-borne Bacillus spp. emerging from animal feed to the food chain. A comprehensive investigation is urgently needed by policymakers on tolerance limits and harmful effects in the animal industry.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03199-3.
Keywords: Antimicrobial resistance, Bacillus spp., Food chain, Food security, Multidrug resistant, Resistant gene
Background
Antimicrobial resistance (AMR) is a serious, multifaceted, and complicated healthcare concern worldwide that impacts people, animals, and the environment, resulting in harder-to-treat infections and even death. The “One Health” approach, which incorporates public health and veterinary regulators, the food and agriculture industry, financiers, environmentalists, and customers, is highlighted in the WHO-led Global Action Plan on Antimicrobial Resistance [1, 2]. AMR develops naturally over time, generally through genetic mutations that can transmit from one generation to another or between humans and animals via animal-sourced food. A variety of strategies, including target defence, target replacement, detoxification, and suppression of cellular antibiotic deposition, are used by bacteria to develop antimicrobial resistance (AMR) [2–4]. Although not all resistant bacteria produce diseases, they may initiate the manifestation of a disease or spread the gene encoding AMR to new bacterial pathogens in favorable environments [4]. Consequently, improper and abusive antibiotics might contribute to the development of different drug-resistant bacteria and can disperse antibiotic residues from various settings throughout the food supply chain, acting as a reservoir and propagation matrix for AMR with the potential for antibiotic-resistant gene (ARG) to cross the animal-to-human microbes due to bacterial contamination [5–7]. The transfer of ARGs is a common way that ABR spreads. Once resistant genes are transmitted by plasmids, transposons, or integrons, dispersion is quick, and horizontal gene transfer across bacteria is frequent. Hitherto, it was thought that this type of genetic exchange only occurred among the same bacterial species. Nevertheless, the transmission of ARGs among phylogenetically distinct bacterial clusters, particularly across gram-positive and gram-negative bacteria, has now been proven in natural habitats [8].
Bacillus spp. has long been used as probiotics in human, veterinary, aquaculture, plant, and environmental applications, either directly as microbial food or as food additives heavily contaminated in animal feed and food chains, making a major financial burden for livestock producers and a potential threat to public health [6, 9–11]. B. cereus-caused foodborne diseases are classified into diarrheal (toxico-infections) and emetic (intoxications) syndromes resulting from the formation of several toxins (enterotoxins such as nhe, hbl, cytK, entFM, BceT, HlyII; emetic toxins ces), which occur globally and are becoming a serious challenge [7, 12]. B. cereus exacerbates severe diarrhea and malnutrition in chickens and ducks by causing gizzard erosion and ulceration (GEU) and facilitating recurrent bacterial infections in the lungs by disrupting the gastrointestinal tract and following lung hemorrhagic lesions [13–16]. More interestingly, B. cereus was reported to induce non-gastrointestinal diseases, including bacteremia, septicemia, endophthalmitis, meningitis, endocarditis, urinary tract infections, and lung infections. Furthermore, B. cereus may lead to serious health effects, especially in newborn infants and immunosuppressed individuals [7, 12].
Nonetheless, some B. cereus and other bacteria possess ARG, which may spread among bacteria and ultimately impact humans through the food supply chain or the surroundings [5, 6]. Probiotic strains of B. cereus, B. clausii, B. subtilis, and B. licheniformis have shown resistance markers for β-lactams (blaBCL-1), chloramphenicol (catBcl), aminoglycosides (aadD2), macrolides (erm34), tetracycline (tetM and tetK), and erythromycin (ermD and ermK) [6]. The existence of mobile genetic components in B. cereus enables the uptake and transmission of drug-resistance genes from the environment [12].
Bangladesh, with a significant level of AMR and multidrug-resistant (MDR) bacteria against drugs indicated for use in both animals and people, confronts a local and worldwide hazard [3, 17–19]. However, B. cereus is resistant to numerous antibiotics, posing a global issue [20]. To inhibit the spread of AMR, it is essential to evaluate Bacillus spp. and their AMR profile. Some strains of Bacillus spp. are becoming increasingly resistant to antibiotics, allowing for the acquisition and emergence of new AMR strains. In our prior report, 39% of Bacillus spp. from animal feed and animal-based foods at a contamination level > 105 CFU/g carried 80%, 71%, 55%, and 33% of the entFM, cytK, nheABC, and hblACD enterotoxin genes, respectively, and food-borne Bacillus spp. caused 4.5% of human diarrhea cases in south-eastern Bangladesh [15]. There is a lack of scientific data on the AMR in the livestock sector in Bangladesh. According to available report, resistant bacteria such as E. coli, Salmonella spp., Klebsiella spp., Pseudomonas spp., Staphylococcus spp., and Vibrio spp., were commonly detected in poultry, dairy cattle, raw milk, farm surroundings, and fish items [21, 22]. Nevertheless, there is a dearth of comprehensive data regarding the antibiotic resistance patterns of Bacillus spp. in the human, animal, and environmental sectors in Bangladesh. Thus, to fillin this knowledge gap, this work aimed to determine the prevalence of resistance, particularly MDR, and the correlated genetic factors in isolated Bacillus spp. In the present study, we focused on whether animal feed, food, and human stool could harbor MDR strains and disseminate them through the food supply chain.
Results
Bacterial isolates
The isolates tested in this investigation were chosen from our prior work [15] and identified to be B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium, and B. coagulans, with 152, 56, and 10 strains from animal feed, food, and human diarrheal cases, respectively (Table S1, Figure S1-S4, Table S2-S3). In all analyzed samples, B. cereus was the predominant isolate (49.3–70%), followed by B. subtilis (14.2–30%), B. amyloliquefaciens (5.2–21.4%), B. thuringiensis (3.9–8.9%), B. licheniformis (8.5%), B. megaterium (5.2%), and B. coagulans (1.7–2.6%). Particularly, 7 B. cereus and 3 B. subtilis were isolated and identified from human stool with diarrhea cases.
Phenotypic profile of antimicrobial resistance
MICs and MBCs of isolated bacteria were displayed in the Table 1. As for B. cereus, the MIC and MBC values were determined in our study: PG-NIT-CFM-CTR/VAN-TET/GEN/CMX-AZM-EM/CIP/LEV-CM-AMC and PG-NIT-CFM/CTR/VAN/CMX-TET/GEN-EM/CIP/LEV/CM-AZM-AMC respectively. While the MIC/MBC ratio for B. subtilis was as follows: NIT-CFM-VAN-CTR/TET/GEN/CMX-AZM-PG/CIP/LEV-EM/CM-AMC, B. amyloliquefaciens recorded a similar trend. In contrast, B. licheniformis and B. thuringiensis showed similar patterns: PG-NIT-CFM-VAN- CTR/TET/GEN/CMX-AZM/CIP-LEV/CM-EM-AMC. Interestingly, B. megaterium and B. coagulans followed the same pattern as NIT-TET/VAN-CTR/GEN/CMX-CFM/AZM- EM/CIP/LEV-PG/CM-AMC (Table 1).
Table 1.
Antibiotic MICs and MBC of Bacillus species strains isolated from animal feed, food and diarrhea
| Antibiotics | Bacterial strains → | B. cereus | B. subtilis | B. amyloliquefaciens | B. licheniformis | B. thuringiensis | B. megaterium | B. coagulans | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range tested (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | |
| PG | 0.25–32 | > 32 | > 32 | 0.5 | 1 | 0.5 | 1 | > 32 | > 32 | > 32 | > 32 | 0.25 | 0.5 | 0.25 | 0.5 |
| AMC | 0.01–0.5 | 0.06 | 0.12 | 0.06 | 0.12 | 0.06 | 0.12 | 0.06 | 0.12 | 0.06 | 0.12 | 0.06 | 0.12 | 0.06 | 0.12 |
| CFM | 0.5–8 | > 4 | > 4 | > 4 | > 4 | 1 | 2 | > 4 | > 4 | > 4 | > 4 | 1 | 2 | 1 | 2 |
| CTR | 0.5–8 | 4 | 8 | 2 | 4 | 1 | 2 | 2 | 4 | 2 | 4 | 2 | 4 | 1 | 2 |
| VAN | 0.5–64 | 4 | 8 | 4 | 8 | 4 | 8 | 4 | 8 | 4 | 8 | 4 | 8 | 4 | 8 |
| AZM | 0.5–8 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 4 | 1 | 2 | 1 | 2 |
| EM | 0.25–32 | 0.5 | 1 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 1 | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 |
| TET | 0.25–32 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 4 | 8 | 4 | 8 | 2 | 4 |
| GEN | 0.5–32 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 |
| CM | 0.25–8 | 0.25 | 1 | 0.25 | 1 | 0.25 | 0.5 | 0.5 | 1 | 0.5 | 1 | 0.25 | 0.5 | 0.5 | 1 |
| NIT | 1–128 | 16 | 32 | 16 | 32 | 16 | 32 | 16 | 32 | 32 | 64 | 16 | 32 | 16 | 32 |
| CIP | 0.12–16 | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 | 1 | 2 | 1 | 2 | 0.5 | 1 | 0.5 | 1 |
| LEV | 0.12–16 | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 | 1 | 2 | 0.5 | 1 | 0.5 | 1 |
| CMX | 1–128 | 2 | 8 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 |
MIC Minimum inhibitory concentration, MBC Minimum bactericidal concentration, PG Penicillin G, AMC Amoxicillin-Clavulanic acid, CFM Cefixime, CTR Ceftriaxone, VAN Vancomycin, AZM Azithromycin, EM Erythromycin, TET Tetracycline, GEN Gentamicin, CM Clindamycin, NIT Nitrofurantoin, CIP Ciprofloxacin, LEV Levofloxacin, CMX Co-Trimoxazole
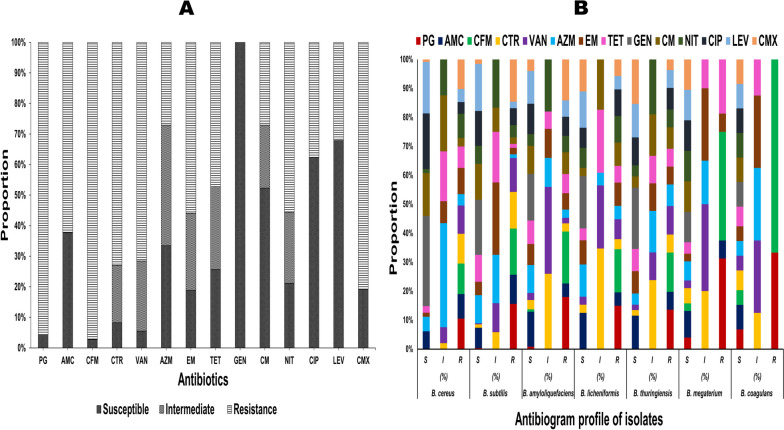
The antibiogram profiles of 14 antibiotics revealed that all isolated Bacillus spp. were generally sensitive to GEN (100%), LEV (67.8%), and CIP (62.3%), 52.2% to CM, 37.6% to AMC, and 33.4% to AZM (Table S4, Supplementary file 1). Further, the Bacillus spp. were generally resistant to β-lactam, glycopeptide, and sulfonamide antibiotics, including CFM (97.2%), PG (95.8%), CMX (81.1%), CTR (72.9%), VAN (71.5%), AMC (62.3%), while 55.9% were resistant to E, 55.5% to NIT, and 47.2% to TET (Fig. 1A, Table S4). The antibiotic-resistant pattern of AMC and AZM in diarrheal cases was significantly higher (p < 0.01) compared to animal feed and food samples. In contrast, the antibiotic-resistant pattern of CIP in animal feed was substantially greater (p < 0.05) than in food and diarrheal cases (Table 2).
Fig. 1.
A Overall antibiogram profile of the isolated Bacillus spp. The bar diagram displayed the proportions of susceptible, intermediate and resistant strains among 218 isolated Bacillus strains to 14 antibiotics. PG: Penicillin G, AMC: Amoxicillin-Clavulanic acid, CFM: Cefixime, CTR: Ceftriaxone, VAN: Vancomycin, AZM: Azithromycin, EM: Erythromycin, TET: Tetracycline, GEN: Gentamicin, CM: Clindamycin, NIT: Nitrofurantoin, CIP: Ciprofloxacin, LEV: Levofloxacin, CMX: Co-Trimoxazole. B Overall antibiogram profile of 7 Bacillus strains
Table 2.
Antibiotic resistance pattern of Bacillus spp. in animal feed, food and diarrhea
| Antibiotic | Antibiotic resistance pattern (%) | |||
|---|---|---|---|---|
| Animal feed | Food | Diarrhea | Level of significance | |
| % (n/N) | % (n/N) | % (n/N) | ||
| PG | 94.1 (143/152) | 98.2 (55/56) | 100 (10/10) | NS |
| AMC | 55.2 (84/152) | 87.5 (49/56) | 100 (10/10) | ** |
| CFM | 95.3 (145/152) | 100 (56/56) | 100 (10/10) | NS |
| CTR | 73.6 (112/152) | 67.8 (38/56) | 100 (10/10) | NS |
| VAN | 69 (105/152) | 73.2 (41/56) | 100 (10/10) | NS |
| AZM | 23 (35/152) | 30.3 (17/56) | 70 (7/10) | ** |
| EM | 53.9 (82/152) | 60.7 (34/56) | 70 (7/10) | NS |
| TET | 48.6 (74/152) | 39.2 (22/56) | 70 (7/10) | NS |
| GEN | 0 | 0 | 0 | - |
| CM | 23.6 (36/152) | 37.5 (21/56) | 20 (2/10) | NS |
| NIT | 57.2 (87/152) | 46.4 (26/56) | 70 (7/10) | NS |
| CIP | 44 (67/152) | 33.9 (19/56) | 0 | * |
| LEV | 33.5 (51/152) | 23.2 (13/56) | 50 (5/10) | NS |
| CMX | 80.2 (122/152) | 85.7 (48/56) | 100 (10/10) | NS |
PG Penicillin G, AMC Amoxicillin-Clavulanic acid, CFM Cefixime, CTR Ceftriaxone, VAN Vancomycin, AZM Azithromycin, EM Erythromycin, TET Tetracycline, GEN Gentamicin, CM Clindamycin, NIT Nitrofurantoin, CIP Ciprofloxacin, LEV Levofloxacin, CMX Co-Trimoxazole, n Number of resistant isolates, N Number of Bacillus isolates, * = significant (p < 0.05), ** = significant (p < 0.01), ns = non-significant
Regarding B. cereus, the isolates were generally sensitive to GEN (100%), CIP (61.6%), and LEV (57.1%) and generally resistant to PG/CFM (100%), CTR (97.4%), CMX (97.3%), VAN (98.2%), EM (85.7%), AMC (80.4%), NIT (79.4%), and TET (70.5%). In the instance of B. subtilis, the isolates were generally sensitive to GEN (100%), LEV (85.7%), CM (65.3%), CIP (63.3%), and AZM (51%). Furthermore, the B. subtilis isolates showed a somewhat similar resistant pattern to that of B. cereus. Interestingly, all B. megaterium and B. coagulans were 100% sensitive to GEN, CM, NIT, CIP, LEV, and CMX (Fig. 1B, Table S5). The ABR profiles of PG, AMC, CFM, CTR, VAN, AZM, EM, TET, CM, NIT, CIP, LEV, and CMX differed significantly (p < 0.01), and species-specific resistance was observed among the seven isolated Bacillus spp. (Table 3).
Table 3.
Antibiotic resistant pattern of seven Bacillus species
| Antibiotics | Fractions of resistant isolates | Level of significance | ||||||
|---|---|---|---|---|---|---|---|---|
| B. cereus | B. subtilis | B. amyloliquefaciens | B. licheniformis | B. thuringiensis | B. megaterium | B. coagulans | ||
| % (n/N) | % (n/N) | % (n/N) | % (n/N) | % (n/N) | % (n/N) | % (n/N) | ||
| PG | 100 (112/112) | 98 (48/49) | 95 (19/20) | 100 (13/13) | 100 (11/11) | 62.5 (5/8) | 20 (1/5) | ** |
| AMC | 80.3 (90/112) | 63.3 (31/49) | 25 (5/20) | 30.7 (4/13) | 45.5 (5/11) | 12.5 (1/8) | 0 | ** |
| CFM | 100 (112/112) | 100 (49/49) | 95 (19/20) | 100 (13/13) | 100 (11/11) | 75.0 (6/8) | 40 (2/5) | ** |
| CTR | 97.4 (109/112) | 79.6 (39/49) | 15 (3/20) | 23.1 (3/13) | 45.5 (5/11) | 0 | 0 | ** |
| VAN | 92.8 (104/112) | 73.5 (36/49) | 10 (2/20) | 46.1 (6/13) | 8/11 (72.7) | 0 | 0 | ** |
| AZM | 37.5 (42/112) | 8.2 (4/49) | 15 (3/20) | 30.8 (4/13) | 54.5 (6/11) | 0 | 0 | ** |
| EM | 85.7 (96/112) | 14.3 (7/49) | 30 (6/20) | 53.8 (7/13) | 45.5 (5/11) | 12.5 (1/8) | 0 | ** |
| TET | 70.5 (79/112) | 8.2 (4/49) | 35 (7/20) | 38.5 (5/13) | 45.5 (5/11) | 37.5 (3/8) | 0 | ** |
| GEN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - |
| CM | 27.7 (31/112) | 14.3 (7/49) | 40 (8/20) | 53.8 (7/13) | 54.5 (6/11) | 0 | 0 | ** |
| NIT | 79.4 (89/112) | 26.5 (13/49) | 30 (6/20) | 61.5 (8/13) | 45.5 (5/11) | 0 | 0 | ** |
| CIP | 38.4 (43/112) | 36.7 (18/49) | 35 (7/20) | 61.5 (8/13) | 54.5 (6/11) | 0 | 0 | * |
| LEV | 42.8 (48/112) | 14.3 (7/49) | 35 (7/20) | 30.8 (4/13) | 54.5 (6/11) | 0 | 0 | ** |
| CMX | 97.3 (109/112) | 91.8 (45/49) | 75 (15/20) | 61.5 (8/13) | 27.3 (3/11) | 0 | 0 | ** |
PG Penicillin G, AMC Amoxicillin-Clavulanic acid, CFM Cefixime, CTR Ceftriaxone, VAN Vancomycin, AZM Azithromycin, EM Erythromycin, TET Tetracycline, GEN Gentamicin, CM Clindamycin, NIT Nitrofurantoin, CIP Ciprofloxacin, LEV Levofloxacin, CMX Co-Trimoxazole. n Number of resistant isolates, N Number of Bacillus isolates, *significant (p < 0.05), **significant (p < 0.01)
Pearson correlation coefficients (ρ) for pairs of antibiotics to assess ABR Bacillus isolates
Bivariate analysis showed a highly significant association (p = < 0.001–0.000) between the resistance patterns of TET and PG, EM/NIT and CFM, AZM/CM and AMC, NIT/CFM/CTR/VAN/EM/CMX and PG, CTR/VAN/EM/TET/NIT/CIP/LEV/CMX and AMC, CTR/VAN/CMX and CFM, VAN/AZM/EM/TET/CM/NIT/CIP/LEV/CMX and CTR, AZM/EM/TET/CM/NIT/CIP/LEV/CMX and VAN, EM/TET/NIT/CIP/CMX and AZM, TET/CM/CIP/LEV/CMX and EM, CM/NIT/CIP/LEV/CMX and TET, NIT/CIP/LEV/CMX/ and CM, CIP/LEV/CMX and NIT, LEV/CMX and CIP, CMX and LEV, and NIT and EM, CM and AZM. A moderate association (p = 0.035–0.017) was found between AMC/CIP/LEV and PG, and TET and CFM. There was a weaker correlation (p > 0.05) between AZM/CM/AMC/CIP/LEV and CFM, and AMC/AZM and PG (Table 4).
Table 4.
Pearson correlation coefficients for pairs of antibiotics to assess antibiotic-resistant Bacillus isolates from animal feed, food and diarrhea
| Statistical analysis | PG | AMC | CFM | CTR | VAN | AZM | EM | TET | GEN | CM | NIT | CIP | LEV | CMX | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PG | Pearson Correlation Coefficient | 1 | |||||||||||||
| p-value (two tailed) | - | ||||||||||||||
| AMC | Pearson Correlation Coefficient | 0.148* | 1 | ||||||||||||
| p-value (two tailed) | 0.029 | ||||||||||||||
| CFM | Pearson Correlation Coefficient | 0.811** | 0.120 | 1 | |||||||||||
| p-value (two tailed) | < 0.001 | 0.077 | |||||||||||||
| CTR | Pearson Correlation Coefficient | 0.341** | 0.434** | 0.276** | 1 | ||||||||||
| p-value (two tailed) | < 0.001 | < 0.001 | < 0.001 | ||||||||||||
| VAN | Pearson Correlation Coefficient | 0.329** | 0.450** | 0.267** | 0.966** | 1 | |||||||||
| p-value (two tailed) | < 0.001 | < 0.001 | < 0.001 | < 0.001 | |||||||||||
| AZM | Pearson Correlation Coefficient | 0.126 | 0.190** | 0.102 | 0.371** | 0.384** | 1 | ||||||||
| p-value (two tailed | 0.062 | 0.005 | 0.131 | < 0.001 | < 0.001 | ||||||||||
| EM | Pearson Correlation Coefficient | 0.234** | 0.473** | 0.190** | 0.687** | 0.711** | 0.540** | 1 | |||||||
| p-value (two tailed | < 0.001 | < 0.001 | 0.005 | < 0.001 | < 0.001 | < 0.001 | |||||||||
| TET | Pearson Correlation Coefficient | 0.196** | 0.379** | 0.159* | 0.576** | 0.597** | 0.644** | 0.840** | 1 | ||||||
| p-value (two tailed | 0.004 | < 0.001 | 0.019 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | ||||||||
| GEN | Pearson Correlation Coefficient | -c | -c | -c | -c | -c | -c | -c | -c | -c | |||||
| p-value (two tailed | - | - | - | - | - | - | - | - | - | ||||||
| CM | Pearson Correlation Coefficient | 0.126 | 0.190** | 0.102 | 0.371** | 0.384** | 1.000** | 0.540** | 0.644** | -c | 1 | ||||
| p-value (two tailed | 0.062 | 0.005 | 0.131 | < .001 | < .001 | 0.000 | < .001 | < .001 | -c | ||||||
| NIT | Pearson Correlation Coefficient | 0.232** | 0.468** | 0.188** | 0.680** | 0.704** | 0.545** | 0.991** | 0.847** | -c | 0.545** | 1 | |||
| p-value (two tailed | < .001 | < .001 | 0.005 | < .001 | < .001 | < .001 | < .001 | < .001 | -c | < .001 | |||||
| CIP | Pearson Correlation Coefficient | 0.161* | 0.288** | 0.131 | 0.473** | 0.490** | 0.784** | 0.689** | 0.820** | -c | 0.784** | 0.695** | 1 | ||
| p-value (two tailed | 0.017 | < .001 | 0.054 | < .001 | < .001 | < .001 | < .001 | < .001 | -c | < .001 | < .001 | ||||
| LEV | Pearson Correlation Coefficient | 0.143* | 0.237** | 0.166 | 0.419** | 0.434** | 0.886** | 0.610** | 0.727** | -c | 0.886** | 0.616** | 0.886** | 1 | |
| p-value (two tailed | 0.035 | < .001 | 0.088 | < .001 | < .001 | < .001 | < .001 | < .001 | -c | < .001 | < .001 | < .001 | |||
| CMX | Pearson Correlation Coefficient | 0.431** | 0.343** | 0.350** | 0.790** | 0.763** | 0.293** | 0.543** | 0.455** | -c | 0.293** | 0.538** | 0.374** | 0.331** | 1 |
| p-value (two tailed | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 | < .001 | -c | < .001 | < .001 | < .001 | < .001 | - | |
*Correlation is significant at the 0.05 level (2-tailed), ** Correlation is significant at the 0.01 level (2 tailed), CCannot be computed because at least one of the variables is constant, PG Penicillin G, AMC Amoxicillin-Clavulanic acid, CFM Cefixime, CTR Ceftriaxone, VAN Vancomycin, AZM Azithromycin, EM Erythromycin, TET Tetracycline, GEN Gentamicin, CM Clindamycin, NIT Nitrofurantoin, CIP Ciprofloxacin, LEV Levofloxacin, CMX Co-Trimoxazole
Genotyping profile of antibiotic resistance
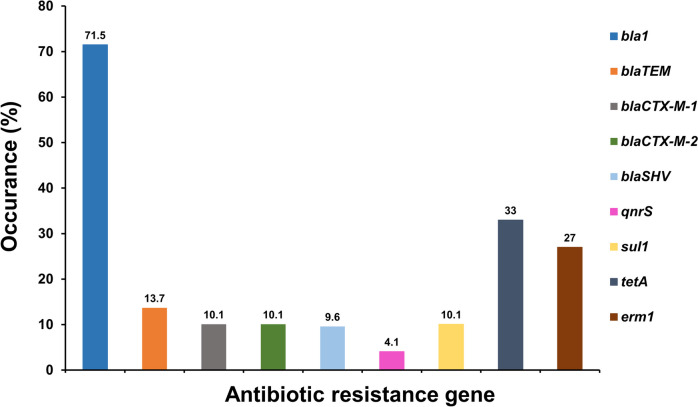
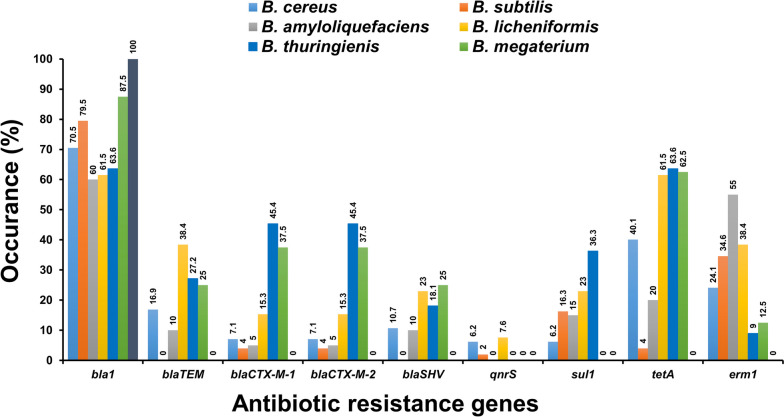
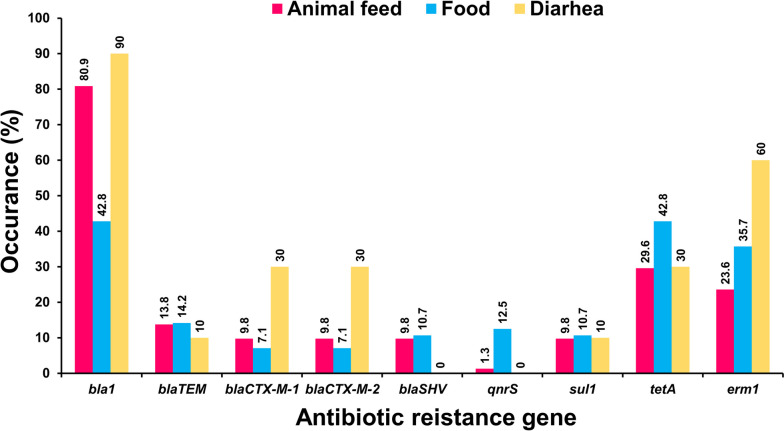
Antibiotic resistance genes (ARGs) of isolated Bacillus spp. showed unique amplified target genes (Fig. S5A-S5I). The ARG profiles exhibited by Bacillus spp. were categorized into ten discrete types of isolates, indicating a considerable degree of genetic heterogeneity (Table 5). Of the 9 ARGs, the bla1 gene was the most frequent (71.5%), followed by tetA (33%), erm1 (27%), blaTEM (13.7%), blaSHV (9.6%), and qnrS (4.1%). The blaCTX-M-1, blaCTX-M-2, and sul1 genes were observed with an equivalent frequency of 10.1% (Fig. 2, Table S6). According to sample-wise distribution, the prevalence of bla1 was highest (80.9%) and qnrS was lowest (1.3%) in animal feed, whereas bla1 and tetA were highest (42.8%) and blaCTX-M-1 and blaCTX-M-2 were lowest (7.1%) in food, and bla1 was highest (90.0%) and blaTEM and sul1 were lowest (10.0%) in diarrheal cases. The bla1 and erm1 genes were significantly higher in the diarrheal case (p < 0.01 and p < 0.05, respectively) compared to the animal feed and food samples. In contrast, the qnrS gene was significantly greater (p < 0.01) in animal feed compared to both food and diarrheal cases (Table S7). The distribution of ARGs among 7 Bacillus strains is shown in Fig. 3 and Table S8. Obviously, bla1 and tetA were predominantly distributed among Bacillus isolates, accounting for 40.1–100%. Furthermore, B. cereus and B. licheniformis harbored the remaining ARGs (blaTEM, blaCTX-M-1, blaCTX-M-2, blaSHV, qnrS, sul1, and erm1) at a rate of 6.2–38.4%. Interestingly, B. coagulans lacked almost all of the ARGs except for the bla1 gene. The distribution of ARGs among animal feed, food and diarrheal cases is depicted in Fig. 4 and Table S9. Importantly, there were substantial variations in the prevalence of 6 ARGs among 7 isolated Bacillus spp., including blaTEM/blaCTX-M-1/blaCTX-M-2/tetA (p < 0.01), sul1/erm1 (p < 0.05), and species-specific occurrence was observed (Table S8).
Table 5.
Antibiotic resistance gene profiling by PCR
| Profile | Antibiotic resistance gene | Origin | Total (%) (n = 218) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bla1 | blaTEM | blaCTX-M-1 | blaCTX-M-2 | blaSHV | qnrS | sul1 | tetA | erm1 | Animal feed | Food | Diarrhea | |||||||
| LF (n = 42) | BF (n = 37) | DF (n = 26) | CF (n = 28) | FF (n = 19) | E (n = 25) | M (n = 31) | HS (n = 10) | |||||||||||
| I | + | + | + | + | - | - | + | + | + | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 10 (4.6) |
| II | + | + | - | - | + | - | - | + | - | 6 | 2 | 4 | 1 | 2 | 3 | 3 | 21 (9.6) | |
| III | + | - | + | + | - | - | - | + | - | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 13 (5.9) |
| IV | + | - | - | - | - | - | - | - | + | 10 | 4 | 9 | 3 | 5 | 3 | 4 | 38 (17.4) | |
| V | + | - | - | - | - | - | - | - | - | 21 | 19 | 10 | 8 | 5 | 2 | 9 | 2 | 76 (34.8) |
| VI | - | - | - | - | - | - | - | + | + | 5 | 5 | 10 (4.5) | ||||||
| VII | - | - | - | - | - | - | - | + | - | 1 | 1 | 10 | 3 | 4 | 19 (8.7) | |||
| VIII | - | - | - | - | - | + | - | - | - | 1 | 1 | 5 | 2 | 9 (4.1) | ||||
| IX | - | - | - | - | - | - | - | - | + | 1 | 3 | 1 | 1 | 6 (2.7) | ||||
| X | - | - | - | - | - | - | + | - | - | 5 | 3 | 1 | 5 | 2 | 16 (7.3) | |||
LF Layer feed, BF Broiler feed, DF Duck feed, CF Cattle feed, FF Fish feed, E Egg, M Milk, HS Human stool, + positive, -negative
Fig. 2.
Distribution of antibiotic resistance genes of Bacillus spp. isolated from animal feed, food and diarrheal cases in Bangladesh. The numerical value displayed above each bar shows the positive rate associated with the respective antibiotic resistance genes
Fig. 3.
Species wise distribution of antibiotic resistance gene of Bacillus spp. isolated from animal feed, food and diarrheal cases in Bangladesh. The numerical value displayed above each specific color bar shows the positive rate associated with antibiotic resistance genes of the respective Bacillus strains
Fig. 4.
Sample wise distribution of antibiotic resistance gene of Bacillus spp. isolated from animal feed, food and diarrheal cases in Bangladesh. The numerical value displayed above each specific color bar shows the positive rate associated with respective antibiotic resistance genes in animal feed, food and diarrhea
Pearson correlation coefficients (ρ) for pairs of ARGs of Bacillus isolates
A bivariate analysis conducted on ARGs of Bacillus isolates showed a highly significant correlation (p = < 0.001–0.000) between bla1 and blaSHV/blaCTX-M-1/blaCTX-M-2, sul1 and blaTEM, sul1 and blaSHV, bla1 and blaTEM/tetA/erm1, blaTEM and blaCTX-M-1/blaCTX-M-2/blaSHV/tetA/erm1, blaCTX-M-1 and sul1/tetA/erm1, blaCTX-M-2 and blaSHV/sul1/tetA/erm1, blaSHV and tetA/erm1, and erm1 and tetA, blaCTX-M-1 and blaCTX-M-2. A moderate association (p = 0.027–0.015) was observed between qnrS/sul1 and bla1, tetA/erm1, and sul1. There were weaker correlations (p > 0.05) between qnrS and sul1/blaTEM/erm1/blaCTX-M-1/blaCTX-M-2/blaSHV/ tetA (Table 6).
Table 6.
Pearson correlation coefficients for pairs of ARGs of Bacillus isolates from animal feed, food and diarrhea
| Statistical analysis | bla1 | blaTEM | blaCTX-M-1 | blaCTX-M-2 | blaSHV | qnrS | sul1 | tetA | erm1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| bla1 | Pearson Correlation Coefficient | 1 | ||||||||
| p-value (two tailed) | - | |||||||||
| blaTEM | Pearson Correlation Coefficient | 0.252** | 1 | |||||||
| p-value (two tailed) | < 0.001 | - | ||||||||
| blaCTX-M-1 | Pearson Correlation Coefficient | 0.211** | 0.839** | 1 | ||||||
| p-value (two tailed) | 0.002 | < 0.001 | - | |||||||
| blaCTX-M-2 | Pearson Correlation Coefficient | 0.211** | 0.839** | 1.000** | 1 | |||||
| p-value (two tailed) | 0.002 | < 0.001 | 0.000 | - | ||||||
| blaSHV | Pearson Correlation Coefficient | 0.206** | 0.817** | 0.975** | 0.975** | 1 | ||||
| p-value (two tailed) | 0.002 | < 0.001 | < 0.001 | < 0.001 | - | |||||
| qnrS | Pearson Correlation Coefficient | 0.165* | 0.092 | 0.096 | 0.096 | 0.097 | 1 | |||
| p-value (two tailed | 0.015 | 0.176 | 0.157 | 0.157 | 0.153 | - | ||||
| sul1 | Pearson Correlation Coefficient | 0.150* | 0.208** | 0.239** | 0.239** | 0.233** | 0.016 | 1 | ||
| p-value (two tailed | 0.026 | 0.002 | < 0.001 | < 0.001 | < 0.001 | 0.815 | - | |||
| tetA | Pearson Correlation Coefficient | 0.443** | 0.569** | 0.477** | 0.477** | 0.465** | 0.098 | 0.150* | 1 | |
| p-value (two tailed | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.151 | 0.027 | - | ||
| erm1 | Pearson Correlation Coefficient | 0.384** | 0.656** | 0.550** | 0.550** | 0.536** | 0.093 | 0.159* | 0.867** | 1 |
| p-value (two tailed | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.169 | 0.019 | < 0.001 | - | |
ARGs Antibiotic resistance genes, *Correlation is significant at the 0.05 level (2-tailed), **Correlation is significant at the 0.01 level (2 tailed)
MDR and MAR resistance profiles of Bacillus spp.
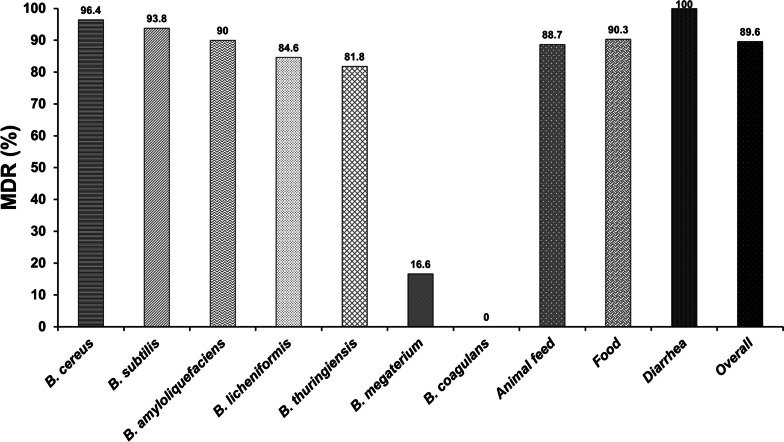
Antibiogram typing revealed that 89.6% of isolated Bacillus strains were MDR (Fig. 5). B. cereus exhibited a higher MDR (96.4%), followed by B. subtilis (93.8%), B. amyloliquefaciens (90%), B. licheniformis (84.6%), B. thuringiensis (81.8%), and B. megaterium (16.6%), while B. coagulans had no MDR. Moreover, MDR was found in 100% of diarrheal isolates, followed by food (90.3%) and animal feed (88.7%) isolates. Interestingly, the species-wise MDR patterns differed significantly (p < 0.01); in contrast, the sample-wise distribution was not statistically different (p > 0.05) (Table 7, Table S10). Among the antibiogram types, pattern PG-AMC-CFM-CTR-VAN-CMX showed the highest prevalence (12 isolates) in animal feed. On the contrary, the PG-AMC-CFM-CTR-EM-VAN-NIT-CMX pattern revealed the highest prevalence (9 isolates) in food, whereas the PG-AMC-CFM-CTR-VAN-CMX and PG-AMC-CFM-CTR-AZM-EM-TET-LEV-VAN-NIT-CMX patterns revealed the highest prevalence (3 isolates) in diarrhea (Table 6).
Fig. 5.
Distributions of multidrug resistant pattern in isolated Bacillus spp., animal feed, food, diarrhea and overall. The numerical value displayed above each specific bar shows the positive rate of the respective multidrug resistance pattern
Table 7.
MDR profile of the isolated Bacillus spp
| Source | Pattern No | Antibiotic Resistance Pattern | No. of antibiotics (classes) | No of resistance isolates | MDR isolates (%) | MAR index | Level of significance |
|---|---|---|---|---|---|---|---|
| Animal feed | 1 | CFM | 1(1) | 1 | 0 (0) | 0.071 | NS (p = 0.517) |
| 2 | PG-CFM | 2 (1) | 9 | 0 (0) | 0.142 | ||
| 3 | PG-AMC-CFM | 3(1) | 1 | 0 (0) | 0.214 | ||
| 4 | PG-CFM-TET | 3(2) | 2 | 0 (0) | 0.214 | ||
| 5 | PG-CFM-CTR-CMX | 4 (1) | 2 | 0 (0) | 0.285 | ||
| 6 | PG-CFM-CTR-NIT | 4 (2) | 2 | 0 (0) | 0.285 | ||
| 7 | PG-CFM-TET-EM | 4(3) | 1 | 1 (0.6) | 0.285 | ||
| 8 | PG-CFM-CTR-TET-NIT | 5 (3) | 1 | 1 (0.6) | 0.357 | ||
| 9 | PG-CFM-CTR-TET-CMX | 5 (3) | 1 | 1 (0.6) | 0.357 | ||
| 10 | PG-CFM-CTR-NIT-CMX | 5 (3) | 3 | 3 (1.9) | 0.357 | ||
| 11 | PG-AMC-CFM-CTR-VAN-CMX | 6 (3) | 12 | 12 (7.7) | 0.428 | ||
| 12 | PG-CTR-AZM-NIT-CMX | 5 (4) | 1 | 1 (0.6) | 0.357 | ||
| 13 | PG-CFM-TET-CIP-LEV-VAN | 6 (4) | 3 | 3 (1.9) | 0.428 | ||
| 14 | PG-CFM-CTR-EM-VAN-CMX | 6 (4) | 2 | 2 (1.2) | 0.428 | ||
| 15 | PG-CFM-CTR-CIP-NIT-CMX | 6 (4) | 4 | 4 (2.5) | 0.428 | ||
| 16 | PG-CFM-EM-CIP-LFV-VAN | 6 (4) | 1 | 1 (0.6) | 0.357 | ||
| 17 | PG-CFM-LEV-VAN-NIT-CMX | 6 (5) | 1 | 1 (0.6) | 0.428 | ||
| 18 | PG-CFM-CIP-VAN-NIT-CMX | 6 (5) | 2 | 2 (1.2) | 0.428 | ||
| 19 | CFM-EM-CIP-CL-NIT-CMX | 6 (6) | 1 | 1 (0.6) | 0.428 | ||
| 20 | PG-AMC-CFM-CTR-EM-TET-CMX | 7 (4) | 1 | 1 (0.6) | 0.5 | ||
| 21 | PG-AMC-CFM-CTR-EM-VAN-CMX | 7 (4) | 2 | 2 (1.2) | 0.5 | ||
| 22 | PG-AMC-CFM-CTR-CIP-VAN-CMX | 7 (4) | 1 | 1 (0.6) | 0.5 | ||
| 23 | PG-CFM-CIP-LEV-VAN-NIT-CMX | 7 (5) | 1 | 1 (0.6) | 0.5 | ||
| 24 | PG-CFM-CTR-EM-VAN-NIT-CMX | 7 (5) | 1 | 1 (0.6) | 0.5 | ||
| 25 | PG-CFM-AZM-CIP-LEV-CM-VAN | 7 (5) | 2 | 2 (1.2) | 0.5 | ||
| 26 | PG-CFM-CTR-EM-TET-VAN-CMX | 7 (5) | 2 | 2 (1.2) | 0.5 | ||
| 27 | PG-CFM-TET-CIP-LEV-VAN-CMX | 7 (5) | 1 | 1 (0.6) | 0.5 | ||
| 28 | PG-CFM-CTR-EM-CIP-VAN-CMX | 7 (5) | 2 | 2 (1.2) | 0.5 | ||
| 29 | PG-CFM-AZM-EM-CIP-NIT-CMX | 7 (5) | 1 | 1 (0.6) | 0.5 | ||
| 30 | PG-CFM-AZM-EM-TET-CM-VAN | 7 (5) | 1 | 1 (0.6) | 0.5 | ||
| 31 | PG-CFM-EM-CIP-CM-NIT-CMX | 7 (6) | 2 | 2 (1.2) | 0.5 | ||
| 32 | PG-CFM-CTR-CIP-LEV-VAN-NIT-CMX | 8 (5) | 1 | 1 (0.6) | 0.571 | ||
| 33 | PG-CFM-CTR-TET-CIP-LEV-VAN-NIT | 8 (5) | 2 | 2 (1.2) | 0.571 | ||
| 34 | PG-AMC-CFM-CIP-LEV-CM-NIT-CMX | 8 (5) | 1 | 1 (0.6) | 0.571 | ||
| 35 | PG-AMC-CFM-CTR-EM-TET-VAN-CMX | 8 (5) | 2 | 2 (1.2) | 0.571 | ||
| 36 | PG-AMC-CFM-CTR-LEV-VAN-NIT-CMX | 8 (5) | 3 | 3 (1.9) | 0.571 | ||
| 37 | PG-AMC-CFM-CTR-TET-VAN-NIT-CMX | 8 (5) | 2 | 2 (1.2) | 0.571 | ||
| 38 | PG-AMC-CFM-CTR-TET-LEV-VAN-CMX | 8 (5) | 1 | 1 (0.6) | 0.571 | ||
| 39 | PG-AMC-CFM-CTR-EM-VAN-NIT-CMX | 8 (5) | 3 | 3 (1.9) | 0.571 | ||
| 40 | PG-CFM-AZM-EM-TET-CM-VAN-CMX | 8 (6) | 2 | 2 (1.2) | 0.571 | ||
| 41 | PG-AMC-CFM-EM-CIP-CM-NIT-CMX | 8 (6) | 3 | 3 (1.9) | 0.571 | ||
| 42 | PG-CFM-TET-CIP-LEV-VAN-NIT-CMX | 8 (6) | 2 | 2 (1.2) | 0.571 | ||
| 43 | PG-CFM-CTR-AZM-TET-CIP-NIT-CMX | 8 (6) | 2 | 2 (1.2) | 0.571 | ||
| 44 | PG-CFM-CTR-EM-TET-VAN-NIT-CMX | 8 (6) | 2 | 2 (1.2) | 0.571 | ||
| 45 | PG-CFM-EM-TET-CIP-VAN-NIT-CMX | 8 (7) | 1 | 1 (0.6) | 0.571 | ||
| 46 | PG-AMC-CFM-CTR-AZM-EM-CM-NIT-CMX | 9 (5) | 2 | 2 (1.2) | 0.642 | ||
| 47 | PG-AMC-CFM-CTX-AZM-EM-TET-NIT-CMX | 9 (5) | 1 | 1 (0.6) | 0.642 | ||
| 48 | PG-AMC-CFM-CTR-TET-CIP-LEV-VAN-CMX | 9 (5) | 1 | 1 (0.6) | 0.642 | ||
| 49 | PG-AMC-CFM-CTX-EM-CM-VAN-NIT-CMX | 9 (6) | 2 | 2 (1.2) | 0.642 | ||
| 50 | PG-AMC-CFM-CTR-EM-TET-VAN-NIT-CMX | 9 (6) | 4 | 4 (2.5) | 0.642 | ||
| 51 | PG-AMC-CFM-CTR-EM-TET-CIP-LEV-VAN-CMX | 9 (6) | 1 | 1 (0.6) | 0.642 | ||
| 52 | PG-AMC-CFM-CTR-TET-CIP-VAN-NIT-CMX | 9 (6) | 1 | 1 (0.6) | 0.642 | ||
| 53 | PG-AMC-CFM-TET-CIP-LEV-VAN-NIT-CMX | 9 (6) | 1 | 1 (0.6) | 0.642 | ||
| 54 | PG-CFM-CTR-EM-TET-LEV-VAN-NIT-CMX | 9 (7) | 1 | 1 (0.6) | 0.642 | ||
| 55 | PG-CFM-CTX-EM-TET-CIP-VAN-NIT-CMX | 9 (7) | 1 | 1 (0.6) | 0.642 | ||
| 56 | PG-AMC-CFM-CTR-AZM-EM-CM-VAN-NIT-CMX | 10 (6) | 4 | 4 (2.5) | 0.714 | ||
| 57 | PG-AMC-CFM-CTR-AZM-ER-TET-CIP-LEV-VAN-CMX | 11 (6) | 4 | 4 (2.5) | 0.785 | ||
| 58 | PG-AMC-CFM-CTR-EM-TET-CIP-LEV-VAN-NIT-CMX | 11 (7) | 5 | 5 (3.2) | 0.785 | ||
| 59 | PG-AMC-CFM-CTR-AZM-EM-TET-CIP-LEV-CM-VAN-NIT | 12 (7) | 4 | 4 (2.5) | 0.857 | ||
| 60 | PG-AMC-CFM-CTR-AZM-EM-TET-CIP-LEV-VAN-NIT-CMX | 12 (7) | 7 | 7 (4.5) | 0.857 | ||
| 61 | PG-CFM-CTR-EM-TET-LEV-CM-VAN-NIT-CMX | 10 (8) | 2 | 2 (1.2) | 0.714 | ||
| 62 | PG-CFM-CTR-EM-TET-CIP-LEV-CM-VAN-NIT-CMX | 11 (8) | 1 | 1 (0.6) | 0.785 | ||
| 63 | PG-AMC-CFM-CTR-EM-TET-CIP-LEV-CM-VAN-NIT-CMX | 12 (8) | 5 | 5 (3.2) | 0.857 | ||
| 64 | PG-AMC-CFM-CTR-AZM-EM-TET-CIP-CM-VAN-NIT-CMX | 12 (8) | 3 | 3 (1.9) | 0.857 | ||
| 65 | PG-AMC-CFM-CTR-AZM-EM-TET-CIP-LEV-CM-VAN-NIT-CMX | 13 (8) | 7 | 7 (4.5) | 0.928 | ||
| Total | 151 | 134 (88.7) | |||||
| Food | 1 | PG | 1 (1) | 1 | 0 (0) | 0.071 | |
| 2 | PG-CFM | 2 (1) | 3 | 0 (0) | 0.142 | ||
| 3 | PG-AMC-CFM-LEV | 4 (2) | 1 | 0 (0) | 0.285 | ||
| 4 | PG-AMC-CFM-LEV-NIT | 5 (3) | 1 | 1 (0.6) | 0.357 | ||
| 5 | PG-AMC-CFM-LEV-CMX | 5 (3) | 2 | 2 (1.2) | 0.357 | ||
| 6 | PG-CFM-CIP-CM-CMX | 5 (4) | 1 | 1 (2.1) | 0.357 | ||
| 7 | PG-CFM-TET-CIP-LEV-VAN | 6 (4) | 3 | 3 (6.2) | 0.428 | ||
| 8 | PG-AMC-CFM-CIP-LEV-CM-CMX | 8 (4) | 1 | 1 (2.1) | 0.571 | ||
| 9 | CFM-EM-TET-CM-CMX | 5 (5) | 1 | 1 (2.1) | 0.357 | ||
| 10 | PG-CFM-EM-TET-CM-CMX | 6 (5) | 3 | 3 (6.2) | 0.428 | ||
| 11 | PG-CFM-EM-CIP-CM-CMX | 6 (5) | 2 | 2 (4.1) | 0.428 | ||
| 12 | PG-AMC-CFM-CTR-VAN-CMX | 6 (3) | 1 | 1 (2.1) | 0.428 | ||
| 13 | PG-AMC-CFM-CTR-VAN-NIT-CMX | 7 (4) | 2 | 2 (4.1) | 0.5 | ||
| 14 | PG-AMC-CFM-CTR-CIP-LEV-VAN-CMX | 8 (4) | 3 | 3 (6.2) | 0.571 | ||
| 15 | PG-AMC-CFM-CTR-AZM-CM-VAN-CMX | 8 (5) | 4 | 4 (8.3) | 0.571 | ||
| 16 | PG-AMC-CFM-CTR-EM-CM-VAN-CMX | 8 (5) | 1 | 1 (2.1) | 0.571 | ||
| 17 | PG-AMC-CFM-CTR-EM-VAN-NIT-CMX | 8 (5) | 9 | 9 (18.7) | 0.571 | ||
| 18 | PG-AMC-CFM-CTR-AZM-EM-CM-VAN-CMX | 9 (5) | 3 | 3 (6.2) | 0.642 | ||
| 19 | PG-AMC-CFM-CTR-EM-TET-VAN-NIT-CMX | 9 (6) | 5 | 5 (10.4) | 0.642 | ||
| 20 | PG-AMC-CFM-CTR-AZM-EM-TET-VAN-NIT-CMX | 10 (6) | 5 | 5 (10.4) | 0.714 | ||
| Total | 52 | 47 (90.3) | |||||
| Diarrhea | 1 | PG-AMC-CFM-CTR-VAN-CMX | 6 (3) | 3 | 3 (30.0) | 0.428 | |
| 2 | PG-AMC-CFM-CTR-AZM-EM-TET-VAN-NIT-CMX | 10 (6) | 2 | 2 (20.0) | 0.714 | ||
| 3 | PG-AMC-CFM-CTR-AZM-EM-TET-LEV-VAN-NIT-CMX | 11 (7) | 3 | 3 (30.0) | 0.785 | ||
| 4 | PG-AMC-CFM-CTR-AZM-EM-TET-LEV-CM-VAN-NIT-CMX | 12 (8) | 2 | 2 (20.0) | 0.857 | ||
| Total | 10 | 10 (100) | |||||
| Grand Total (overall MDR isolates) | 213 | 191 (89.6) | |||||
MDR Multidrug resistance, PG Penicillin G, AMC Amoxicillin-Clavulanic acid, CFM Cefixime, CTR Ceftriaxone, VAN Vancomycin, AZM Azithromycin, EM Erythromycin, TET Tetracycline, CM Clindamycin, NIT Nitrofurantoin, CIP Ciprofloxacin, LEV Levofloxacin, CMX Co-Trimoxazole, NS Not significant
Furthermore, the MAR index of Bacillus spp. was arranged from 0.071–0.928, while B. cereus yielded the highest MAR index, ranging from 0.285–0.928, followed by B. thuringiensis (0.142–0.857), B. subtilis (0.142–0.642), B. amyloliquefaciens/B. licheniformis (0.142–0.571), B. megaterium (0.142–0.285), and B. coagulans (0.071–0.142). Regarding sample type, animal feed isolates had the highest MAR index (0.428–0.928), followed by diarrheal isolates (0.428–0.857) and food isolates (0.071–0.714) (Table S10, Table 6). In our study, 90.3% of isolates showed a MAR index > 0.2, with 100, 94.6, and 88.1% from diarrhea, food, and animal feed, respectively. Furthermore, there was a substantial variation (p < 0.05) among the MAR index having > 0.2 of seven Bacillus strains, while no significant difference (p > 0.05) was observed among different sample types (Tables S11 and S12).
Discussion
In the current study, the isolated Bacillus strains were sensitive to GEN, CIP, LEV, CM, AMC, TET, EM, AZM, NIT, and CMX, which was consistent with the previous reports [4, 5, 7, 9, 12, 20, 23, 24]. However, there were species-specific sensitivities of the Bacillus strains to CIP, LEV, CM, TET, EM, AZM, and NIT at various doses. Regarding the resistant pattern, diarrheal strains were completely resistant to PG, AMC, CFM, CTR, VAN, and CMX, whereas animal feed-borne strains were generally resistant to PG, AMC, CFM, VAN, and CMX, and food-originated strains were generally resistant to PG, CFM, CTR, and CMX. As for different species, B. cereus, B. thuringiensis, and B. licheniformis strains were all completely resistant to the beta-lactam antibiotics of PG and CFM, while B. subtilis strains were completely resistant to CFM. Moreover, they were generally resistant to PG, AML, CFM, CTR, VAN, EM, TET, and NIT compared to AZM, CM, CIP, and LEV, which was compatible with the other studies [1, 3, 5, 7, 8, 20, 23–25].
The B. cereus strains were generally sensitive to GN, CIP, and LEV, while intermediately sensitive to AZM, CM, and TET. In contrast, they were generally resistant to PG, CFM, CTR, CFM, VAN, EM, AMC, NIT, and TET. This result agreed with earlier findings [4, 7, 20, 23, 24, 26–28], which were isolated from rice, cereals, chicken meat, fresh vegetables, edible fungi, powdered milk, foodstuffs, human stool, and clinical samples. The isolated B. subtilis was generally sensitive to GEN, LEV, CM, and CIP; intermediately sensitive to EM, TET, AZM, and NIT; and generally resistant to CFM, PG, CMX, CTR, VAN, and AMC. However, this bacteria strain detected in bread, powdered milk, soil, and shrimp culture ponds showed sensitivity to GEN, VAN, CM, EM, TET, and CMX while being resistant to PG, ampicillin, cefpodoxime, and cefepime [4, 9, 29, 30]. These two dominant species had species-specific responses to AMC, AZM, EM, TET, NIT, and LEV. The main factor might be large abuse, which prolongs time. The high sensitivity to GEN, CIP, and LEV could be attributed to the limited administration of CIP and LEV, while GEN is not absorbed via oral application. Consequently, CTR, CFM, AMC, CMX, and NIT were largely abused in the animal industry and added frequently against infectious diseases. On the other hand, antibiotic regulation contributes to AMR. In several countries, CIP and LEV are prohibited for use on animals due to human drugs.
It is worth mentioning that 89.6% of Bacillus isolates were MDR, with 100% of isolates from diarrhea, 90.3% from food, and 88.7% from animal feed exhibiting the MDR pattern. These findings were consistent with other reports that found MDR Bacillus spp. in several sources, including food [4, 5, 7, 31, 32]. In our investigation, 100% B. cereus and 91.8% B. subtilis isolates yielded MAR indexes > 0.2, indicating plasmid-mediated resistance and a significant risk of contamination. This implies a high inclination and trend for antibiotic resistance among the MDR bacterial isolates [33]. In Bangladesh, MDRs of E. coli, Salmonella spp., Campylobacter spp., and Enterobacter spp. were detected in livestock populations due to contamination of animal-derived food and food products [3]. However, MDR in Bacillus spp. has not been reported. Our data indicated Bacillus strains might produce extended-spectrum beta-lactamase (ESBL) and resistance to third-generation cephalosporins (CFM and CTR), macrolide (EM), tetracycline, second- and third-generation quinolones (CIP and LEV), and sulfonamides (CMX).
In this study, genes encoding β-lactamase (bla1, blaTEM, blaCTX-M-1, blaCTX-M-2, blaSHV), fluroquinolone (qnrS), sulfonamide (sul1), tetracycline (tetA), and macrolide (erm1) were derived from animal feed, food, and diarrhea. The most commonly observed β-lactamase genes were bla1 than blaTEM, blaCTX-M-1, blaCTX-M-2, and blaSHV among Bacillus strains. Similar detection of the bla1 gene in Bacillus strains was reported by other isolates from chicken meat, meat products, human stool, food, and the environment [27, 34, 35]. However, some of our findings were inconsistent with the previous studies [23, 36, 37]. They observed a higher prevalence of blaTEM, blaCTX-M-1, blaCTX-M-2, and blaSHV in waste water, clinical samples, and food samples. The second dominant gene was the tetA gene in our study, which was consistent with prior studies [8, 18, 25], but other authors reported a higher occurrence [28, 35]. The prevalence of the erythromycin-resistant gene, erm1, was low compared to earlier reports [9, 25, 35, 37]. Furthermore, our study revealed a low prevalence of sulfonamide, sul1 and quinolone, and qnrS resistance genes. The sul1 gene in Bacillus strains isolated from aquaculture ponds was lower than our report [1], whereas the sul1 gene in wastewater was higher than our report [37]. Bacillus spp. are often used as food microbial additives and spread ARGs by horizontal transfer of plasmids, leading to the failure of antibiotic treatment and dramatically altering their phenotypes [4, 5, 38]. The Bacillus anthrax-associated plasmids pXO1 and pXO2 have been detected in certain B. cereus strains with pathogenic potential resembling B. anthracis [39]. For instance, atypical B. cereus strains such as B. cereus G9241, B. cereus biovar anthracis CA, Bcbva, and Bcbva-like strain BC-AK were linked to anthrax-like disease in mammals, livestock, and humans in the United States, China, and West Africa, implying that it may be widespread [40]. The highly efficient mobilization capacities and horizontal gene transfer may pose a serious threat to gene circulation, particularly ARGs. Probiotic Bacillus spp. have already been connected to clinical infections, as well as β-lactams, aminoglycosides, macrolides, chloramphenicol, tetracycline, and erythromycin resistance genes, which may contribute to the spread of ABR in animal microbiota and the possible transmission of ARG to humans [10]. Nevertheless, abusive antibiotic use can transmit antibiotic residues in foods derived from animals, like milk, meat, and eggs, as well as in the environment [6]. As a potential driver of both genes and bacteria resistant to antibiotics, the ARG may be further transmitted to people directly through the food chain [10].
The high resistance of Bacillus strains to CFM, CTR, AMC, and PG might be caused by β-lactamase and the presence of ABC (ATP binding cassette) efflux transporters from B. subtilis that are tolerant to lincosamide [25, 26]. The different antibiotic mechanisms might be associated with inherent resistance, built-up resistance, gene modification, and DNA transfer that aid in bacterial survival by manipulating the penicillin binding protein (PBP), enzymatic blockage, porin mutations, and efflux pumps [6]. The current study revealed that Bacillus isolates primarily carried the β-lactamase resistance genes bla1, blaTEM, blaCTX-M-1, blaCTX-M-2, blaSHV, tetracycline resistance gene tetA, and erythromycin resistance gene erm1, respectively. It was confirmed that certain ARG classes could be acquired by the majority of antibiotics and evaluated in various Bacillus strains. According to resistant gene distribution, 10 distinct ARG patterns were detected in the isolates. The association of the above β-lactamase and other antibiotic genes within the same isolate has been reported [9, 23, 28, 36, 37]. However, the most common associations were bla1 + erm1, bla1 + tetA, bla1 + blaTEM, blaTEM + tetA, and blaCTX-M-1 + tetA (Table S2). This occurrence indicates a greater spread of β-lactamase, tetracycline, and erythromycin genes, most likely owing to a genetic component in their mobilization as well as the horizontal transfer of ABR determinants between Bacillus strains or from other bacteria into Bacillus spp. [7, 8, 36].
In our prior study, we revealed a high Bacillus spp. contamination level with significant toxigenic potential in several resources, including animal feed, animal-derived goods, and regular food items [15], where 90.3% of isolates displayed > 0.2 MAR index, indicating a high-risk source of contamination [41]. The presence of MDR and MAR Bacillus in animal feed, food, and diarrhea indicated that the abuse of antibiotics poses a severe public health hazard by transmitting AMR to people through the food supply chain.
There is a dearth of research data on the transmission of ARGs and the AMR of Bacillus strains in Bangladesh. According to a review report, the emergence of AMR is mainly attributed to antibiotic misuse or overuse by broiler (> 60%) and layer (94.6%) farmers as over-the-counter medication and failure to maintain the drug withdrawal period [3]. Empirical data suggests that antibiotic residues against Bacillus spp. exist in the liver and kidney as well as in commercially available feed in Bangladesh, acting as a subtherapeutic dose that hastens the emergence of AMR [15, 42]. Unhygienic livestock and poultry farming in Bangladesh is a significant risk indicator for spreading zoonotic bacteria and antibiotic resistance to people and the environment [3]. Our data confirmed that AMR- B. cereus strains prevailed in all analyzed samples. The significance of this finding is underlined by earlier research [4, 5, 7, 20, 24, 25] that showed Bacillus spp. can transfer ARGs. Bangladesh urgently requires the development of effective surveillance and control plans for the identification and prevention of ABR bacteria utilizing standard antibiotic susceptibility tests in regular animal and human microbiological laboratory settings.
Conclusion
It is the first investigation of the presence of ARGs of Bacillus spp. with public health significance in animal feed, food, and human stool in Bangladesh. The feed- and food-borne Bacillus spp. exhibited species-specific trends in both phenotypic and genotypic resistance patterns with respect to antibiotic resistance. The associations of various antibiotic-resistant genes indicated a greater spread of β-lactamase, tetracycline, and erythromycin genes across the food chain. Animal feed and animal-derived products might serve as a channel for B. cereus propagation regarding their potential pathogenicity and the development of AMR in humans. This work validates the sources examined as major outlets for the spread of MDR bacteria and ARGs in the food chain of Bangladesh and once again highlights the urgency of a global campaign to combat AMR.
Materials and methods
Sampling, selection, isolation, storage, and molecular characterization of Bacillus spp. isolates
A total of 218 Bacillus spp. isolates were examined and retrieved in our previous study (Table S1), including animal feed (n = 90), food (n = 40), and human stool (n = 50) in southeast Bangladesh [15]. These Bacillus spp. strains were initially detected and isolated through cultivation on MYPA (HiMedia, Mumbai, Maharashtra, India) plates, grams staining, biochemical assays, and PCR targeting 16srDNA, nheABC, hblACD, cytK, and entFM genes [15, 43, 44] The strains were preserved in TSB medium (HiMedia, Mumbai, Maharashtra, India) containing 15% glycerol at -800C. The bacterial strains were cultivated aerobically in TSB at 37 °C with agitation at 225 rpm. The Ethical Reviewing Board on Institutional Animal Care and Use Committee at Noakhali Science and Technology University, Bangladesh, granted approval for the experimental protocols. An Informed Consent Form (ICF) was obtained prior to initiating research activities and collecting human stool samples.
Antibiotic susceptibility test of the Bacillus isolates
Minimum inhibitory concentration (MIC)
To assess any link between antimicrobial resistance patterns and the chosen antibiotic category, the antibiogram profile of isolates was tested by estimating the MIC in appropriate broth using sterile U bottom 96-well plates with lids (SPL Life Science, Pochon, Kyonggi-do, South Korea) employing the microtiter broth dilution method [45]. The MIC represented the lowest level of antimicrobial that completely inhibited the growth of the organism. According to 2020 Clinical and Laboratory Standards Institute (CLSI) criteria, susceptible, intermediate, and resistant MIC (μg/ml) interpretation was done ([45], Table S13). Briefly, 2–3 single pure fresh colonies of Bacillus spp. grown on NA (24 h old) were inoculated into 5 ml of MHB (HiMedia, Mumbai, Maharashtra, India) and kept at 370C for up to 8 h. To standardize the turbidity of bacterial suspension, MHB was used to achieve a turbidity of 0.5 McFarland concentration (1 × 105 CFU/mL) through a two-fold serial dilution of antibiotics through visual evaluation with a card featuring a white backdrop and distinct black lines. The microtiter plates were incubated in a shaking incubator at 370C. The lowest concentration that inhibited indicator strains growth was noted. All MIC tests were done in triplicate. Staphylococcus aureus ATCC 29213 was used as a positive control.
Minimum Bactericidal Concentration (MBC)
Five microliters of inoculum from the MIC experiment's well that had no bacterial growth after 24 h were spotted on NA (HiMedia, Mumbai, Maharashtra, India). The plates were incubated at 370C for 16 to 24 h in order to evaluate the MBC described earlier [46]. MBC was set as the lowest level on a NA plate where there was no visual growth. Bacillus spp. was detected, and it was determined that the growth of bacteria was bacteriostatic, while the absence of growth indicated bactericidal effects. The NA plate was cultivated with the indicated inoculum dilution to test for contamination and cell viability [45]. All analyses were performed in triplicate.
Determination of multidrug resistant (MDR) and multiple antibiotic resistance (MAR) index
MDR was considered to have at least one agent that was resistant to three or more types of antibiotics [47]. The following 14 antibiotics procured in powdered form from Sisco Research Laboratories Pvt. Ltd. (SRL, E, Mumbai, Maharashtra 400,099, India) were used: Penicillin G, PG (0.25–32 µg/mL); Amoxicillin + Clavulanic acid, AMC (0.01–0.5 µg/mL); Cefixime, CFM (0.5–4 µg/mL); Ceftriaxone, CTR (1–8 µg/mL); Azithromycin, AZM (0.5–8 µg/mL); Erythromycin, EM (0.25–32 µg/mL); Tetracycline, TET (0.25–32 µg/mL); Ciprofloxacin, CIP (0.12–16 µg/mL); Levofloxacin, LEV (0.12–16 µg/mL); Clindamycin, CM (0.25–8 µg/mL); Vancomycin, VAN (0.5–32 µg/mL); Gentamicin, GEN (0.5–32 µg/mL); Nitrofurantoin, NIT (32–128 µg/mL), and Co-Trimoxazole, CMX (1–128 µg/mL). The MAR index of isolated Bacillus spp. was determined as a/b, where “a” is the number of antibiotics to which a strain is resistant and “b” is the total number tested [41].
Resistance genotyping
The genomic DNA of various Bacillus species isolated from animal feed, food, and stool samples was extracted utilizing the TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit Ver.3.0 (GW Vitek, Seoul, Korea). Following the manufacturer's instructions, pure colonies of Bacillus spp. isolates were prepared for DNA extraction from Tryptic Soy Broth (TSB) and Luria Bertani (LB) broth. A NanoDropTM 8000 spectrophotometer (Thermo Scientific, California, USA) was utilized to measure the concentration and purity of the eluted DNA.
Phenotypically resistant Bacillus spp. were screened by PCR for 9 ARGs, including 5 β-lactamase (bla1, blaTEM, blaCTX-M-1, blaCTX-M-2, and blaSHV), single fluroquinolone (qnrS), sulfonamide (sul1), tetracycline (tetA), and macrolide (erm1). PCR protocols were followed exactly as reported earlier [1, 8, 23, 36, 48–50] (Table 8, Table S14). B. cereus ATCC 14579 and E. coli ATCC 25922 served as positive controls, while sterile Milli-Q water (Sigma Aldrich, Bengaluru, Karnataka 560,099, India) served as a negative control. The PCR reaction was performed in a 25 µl volume with OneTaqQuick-Load 2 × Master Mix (New England Biolabs Inc., United States), 0.2 µmol L−1 final concentration of each primer, and 2.5 µl of ready DNA template. PCR was conducted on a T100 Thermal cycler (Bio-Rad, United States). The PCR products were analyzed on 1.5% agarose gel (MP Biomedicals LLC, United States) with a Mini-Sub Cell GT Horizontal Electrophoresis System (Bio-Rad, United States), stained with Ethidium bromide (EtBr), displayed with a UV transilluminator (Gel Doc EZ), and imaged using a gel documentation system.
Table 8.
Primer used in this study
| Primer | Sequence (5´ → 3´) | Annealing temperature (oC) | Product size (bp) | Reference |
|---|---|---|---|---|
| bla1 | F = CATTGCAAGTTGAAGCGAAA | 50 | 680 | [27] |
| R = TGTCCCGTAACTTCCAGCTC | ||||
| blaTEM | F = ATGAGTATTCAACATTTCCG | 55 | 850 | [25] |
| R = CCAATGCTTAATCAGTGAGG | ||||
| blaCTX-M-1 | F = AAAAATCACTGCGCCAGTTC | 52 | 415 | [25] |
| R = AGCTTATTCATCGCCACGTT | ||||
| blaCTX-M-2 | F = CGACGCTACCCCTGCTATT | 52 | 552 | [25] |
| R = CCAGCGTCAGATTTTTCAGG | ||||
| blaSHV | F = GCGAAAGCCAGCTGTCGGGC | 62 | 304 | [26] |
| R = GATTGGCGGCGCTGTTATCGC | ||||
| qnrS | F = GCAAGTTCATTGAACAGGGT | 54 | 428 | [28] |
| R-TCTAAACCGTCGAGTTCGGCG | ||||
| sul1 | F = CGGCGTGGGCTACCTGAACG | 57 | 433 | [1] |
| R = GCCGATCGCGTGAAGTTCCG | ||||
| tetA | F = GGCGGTCTTCTTCATCATGC | 58 | 502 | [8] |
| R = CGGCAGGCAGAGCAAGTAGA | ||||
| ermA | F = TCTAAAAAGCATGTAAAAGAA | 52 | 645 | [50] |
| R = CTTCGATAGTTTATTAATATTAGT |
Statistical analysis
The antibiotic susceptibility results were provided in MS-2016 Excel sheets and analyzed using IBM SPSS Statistics version 24 (SPSS Inc., Chicago, IL, USA). The prevalence was computed by a descriptive study and the Chi-square test, and the degree of significance was established using Pearson correlation coefficients. The statistical significance was calculated as ∗p < 0.05 and ∗∗p < 0.01.
Supplementary Information
Additional file 1: Figure S1. Cultural morphology and Grams staining of isolated Bacillus spp. Figure S2.16srDNA gene of the isolated Bacillus spp. by PCR test. Figure S3. Toxin genes (nheA, nheB, nheC, cytK) of isolated Bacillus spp. by PCR test. Figure S4. Toxin genes (hblA, hblC, hblD, entFM) of isolated Bacillus spp. by PCR test. Figure S5. Antibiotic resistance genes of isolated Bacillus spp. by PCR test. Table S1. Strains and source of selected Bacillus spp. Table S2. Biochemical characteristics of Bacillus spp. Table S3. Primers of toxin gene and 16srRNA gene and PCR protocol used in this study. Table S4. Overall antimicrobial susceptibility profile of Bacillus spp. Table S5. Overall antimicrobial susceptibility profiles of 7 Bacillus species. Table S6. Distributions of antibiotic genes among animal feed, food and diarrhea. Table S7. Prevalence of ARGs of Bacillus spp. in animal feed, food and diarrhea. Table S8. Prevalence of ARGs of 7 Bacillus species. Table S9. Distributions of ARGs among 7 Bacillus species. Table S10. MDR profiles of B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium and B. coagulans. Table S11. Species wise percentage of MAR index >0.2. Table S12. Sample wise percentage of MAR index >0.2. Table S13. MIC break point of Antibiotic used. Table S14. PCR protocol of antibiotic resistant gene primer used in this study.
Acknowledgements
The Microbiology Laboratory, Department of Microbiology, Noakhali Science and Technology University, Noakhali, Bangladesh, supported and carried out the research. The technical assistance of Md Amzad Hossain and Md Ruhul Amin is kindly acknowledged.
Abbreviations
- AMR
Antimicrobial resistance
- ARG
Antibiotic resistant gene
- CDC
Center for Disease Control and Prevention
- CFU
Colony-forming unit
- CLSI
Clinical and Laboratory Standards Institute
- DNA
Deoxyribonucleic acid
- EtBr
Ethidium bromide
- LB
Luria-Bertani
- MAR
Multiple antibiotic resistance
- MBC
Minimum bactericidal concentration
- MDR
Multidrug-resistant
- MHB
Muller Hinton Broth
- MIC
Minimum inhibitory concentration
- MYPA
Mannitol-Egg-Yolk-Polymyxin-Agar
- NA
Nutrient Agar
- PCR
Polymerase chain reaction
- TSB
Tryptic soy broth
- WHO
World Health Organization
Authors’ contributions
M.AH. acquisition of data, formal analysis, wrote the main manuscript text and prepared figure; H.HL. and L.JQ. acquisition of data and writing the original draft. M.AL, F.H, M.AR. interpretation of data, writing–review and editing; F.A. and C.H. concept and design. All authors reviewed the manuscript.
Funding
This study was funded by the Science and Technology Partnership Program, Ministry of Science and Technology of China [grant No. KY202204001]. This study was supported in part by the Ministry of Science and Technology (MoST), China [grant No.2022YFC2304000].
Availability of data and materials
The dataset can be accessed upon a reasonable request made to the Corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Md Atiqul Haque and Huilong Hu contributed equally to this work.
Contributor Information
Firoz Ahmed, Email: firoz@nstu.edu.bd.
Cheng He, Email: hecheng@cau.edu.cn.
References
- 1.Adekanmbi AO, Adejoba AT, Banjo OA, Saki M. Detection of sul1 and sul2 genes in sulfonamide-resistant bacteria (SRB) from sewage, aquaculture sources, animal wastes and hospital wastewater in South-West Nigeria. Gene Rep. 2020;20:100742. doi: 10.1016/j.genrep.2020.100742. [DOI] [Google Scholar]
- 2.World Health Organization (WHO), Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed on 25 May 2022.
- 3.Al Amin M, Hoque MN, Siddiki AZ, Saha S, Kamal MM. Antimicrobial resistance situation in animal health of Bangladesh. Vet World. 2020;13:2713–2727. doi: 10.14202/vetworld.2020.2713-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alanber MN, Alharbi NS, Khaled JM. Evaluation of multidrug-resistant Bacillus strains causing public health risks in powdered infant milk formulas. J Infect Public Health. 2020;13:1462–1468. doi: 10.1016/j.jiph.2019.11.013. [DOI] [PubMed] [Google Scholar]
- 5.György É, Laslo É, Antal M, András CD. Antibiotic resistance pattern of the allochthonous bacteria isolated from commercially available spices. Food Sci Nutr. 2021;9:4551–4561. doi: 10.1002/fsn3.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haque MA, Khan A, He C. Feed-borne Bacillus cereus: an emerging threat to food chain related hazard, safety and pathogenic potentiality. In: Abbas RZ, Khan A, editors. Veterinary Pathobiology and Public Health. 1. Faisalabad: Unique Scientific Publishers®; 2021. pp. 251–269. [Google Scholar]
- 7.Sornchuer P, Tiengtip R. Prevalence, virulence genes, and antimicrobial resistance of Bacillus cereus isolated from foodstuffs in Pathum Thani Province. Thailand Pharm Sci Asia. 2021;48:194–203. doi: 10.29090/psa.2021.02.19.119. [DOI] [Google Scholar]
- 8.Rather MA, Aulakh RS, Gill JPS, Mir AQ, Hassan MN. Detection and sequencing of plasmid encoded tetracycline resistance determinants (tetA and tetB) from food–borne Bacillus cereus isolates. Asian Pac J Trop Med. 2012;5:709–712. doi: 10.1016/S1995-7645(12)60111-4. [DOI] [PubMed] [Google Scholar]
- 9.Adimpong DB, Sorensen KI, Thorsen L, Stuer-Lauridsen B, Abdelgadir WS, Nielsen DS, et al. Antimicrobial susceptibility of Bacillus strains isolated from primary starters for African traditional bread production and characterization of the bacitracin operon and bacitracin biosynthesis. Appl Environ Microbiol. 2012;78:7903–7914. doi: 10.1128/AEM.00730-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haque MA, Quan H, Zuo Z, Khan A, Siddique N, He C. Pathogenicity of feed-borne Bacillus cereus and its implication on food safety. Agrobiol Records. 2021;3:1–16. doi: 10.47278/journal.abr/2020.015. [DOI] [Google Scholar]
- 11.Sultana N, Haque MA, Rahman MM, Akter MR, Begum MD, Fakhruzzaman M, et al. Microbiological quality of commercially available poultry feeds sold in Bangladesh. Asian J Med Biol Res. 2017;3:52–60. doi: 10.3329/ajmbr.v3i1.32036. [DOI] [Google Scholar]
- 12.Navaneethan Y, Esah EM. Prevalence, toxigenic profiles, multidrug resistance, and biofilm formation of Bacillus cereus isolated from ready-to eat cooked rice in Penang, Malaysia. Food Control. 2020;121:107553. doi: 10.1016/j.foodcont.2020.107553. [DOI] [Google Scholar]
- 13.Zhang Q, Zuo Z, Guo Y, Zhang T, Han Z, Huang S, et al. Contaminated feed-borne Bacillus cereus aggravates respiratory distress post avian influenza virus H9N2 infection by inducing pneumonia. Sci Rep. 2019;9:7231. doi: 10.1038/s41598-019-43660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo Z, Li Q, Guo Y, Li X, Huang S, Hegemann JH, et al. Feed-borne Bacillus cereus exacerbates respiratory distress in birds infected with Chlamydia psittaci by inducing hemorrhagic pneumonia. Avian Pathol. 2020;49:251–260. doi: 10.1080/03079457.2020.1716940. [DOI] [PubMed] [Google Scholar]
- 15.Haque MA, Fei W, Chen Y, Ahmed F, Hossen F, Islam MA, et al. Bacillus spp. contamination: a novel risk originated from animal feed to human food chains in south-eastern Bangladesh. Front Microbiol. 2022;12:783103. doi: 10.3389/fmicb.2021.783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque MA, Zuo Z, He C. Pathogenesis of gizzard erosion and ulcerations and comprehensive control strategy in poultry. Vol. 2. In: Abbas RZ, Saeed NM, Younus M, Aguilar-Marcelino L, Khan A, editors. One Health Triad. Unique Scientific Publishers: Faisalabad; 2023. pp. 129–13. [Google Scholar]
- 17.Akter S, Haque MA, Jahan I. Epidemiological investigation, prevalence & antibiogram study of potential zoonotic bacterial pathogens of household pets at Dinajpur district of Bangladesh. Int J Innov Sci Res Technol. 2020;5:1630–1641. doi: 10.38124/IJISRT20AUG795. [DOI] [Google Scholar]
- 18.Habib A, Afroz F, Rahman MM, Begum MD, Kamruzzaman M, Haque MA. Molecular characterization and antimicrobial resistance of Escherichia coli isolated from retail chicken meat. South Asian J Biol Res. 2021;4:38–47. [Google Scholar]
- 19.Nupur MN, Afroz F, Hossain MK, Harun-ur-Rashid SM, Rahman MG, Kamruzzaman M, et al. Prevalence of potential zoonotic bacterial pathogens isolated from household pet birds and their antimicrobial profile in northern Bangladesh. Agrobiological Records. 2023;11:28–38. doi: 10.47278/journal.abr/2023.005. [DOI] [Google Scholar]
- 20.Liu C, Yu P, Yu S, Wang J, Guo H, Zhang Y, et al. Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiol. 2020;20:310. doi: 10.1186/s12866-020-01996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: a systematic review. Int J Infect Dis. 2019;80:54–61. doi: 10.1016/j.ijid.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Islam MM. Bacterial resistance to antibiotics: access, excess, and awareness in Bangladesh. Expert Rev Anti Infect Ther. 2021;19:973–981. doi: 10.1080/14787210.2021.1865804. [DOI] [PubMed] [Google Scholar]
- 23.Torkar KG, Bedenić B. Antimicrobial susceptibility and characterization of metallo-β-lactamases, extended-spectrum β-lactamases, and carbapenemases of Bacillus cereus isolates. Microb Pathog. 2018;118:140–145. doi: 10.1016/j.micpath.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Fiedler G, Schneider C, Igbinosa EO, Kabisch J, Brinks E, Becker B, et al. Antibiotics resistance and toxin profiles of Bacillus cereus-group isolates from fresh vegetables from German retail markets. BMC Microbiol. 2019;19:250. doi: 10.1186/s12866-019-1632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Q, Fang Y, Zhu J, Xu W, Zhu K. Characterization of Bacillus species from market foods in Beijing. China Processes. 2021;9:866. doi: 10.3390/pr9050866. [DOI] [Google Scholar]
- 26.Park YB, Kim JB, Shin SW, Kim JC, Cho SH, Lee BK, et al. Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J Food Prot. 2009;72:612–617. doi: 10.4315/0362-028X-72.3.612. [DOI] [PubMed] [Google Scholar]
- 27.Tahmasebi H, Talebi RB, Zarif RB. Isolated of Bacillus cereus in chicken meat and investigation β-lactamase antibiotic-resistant in Bacillus cereus from chicken meat. Adv life sci. 2014;4:200–206. [Google Scholar]
- 28.Avşar C, Civek S, Aras ES. Phenotypic and genotypic characterization of foodborne bacteria isolated from Sinop Province, Turkey. Food Biotechnol. 2017;31:141–161. doi: 10.1080/08905436.2017.1331450. [DOI] [Google Scholar]
- 29.Aslim B, Saglam N, Beyatli Y. Determination of some properties of Bacillus isolated from soil. Turk J Biol. 2002;26:41–48. [Google Scholar]
- 30.Balakrishnan S, John KR, George MR. Antibiotic susceptibility of Bacillus spp. isolated from shrimp (Penaeus monodon) culture ponds. Indian J Mar Sci. 2003;32:81–84. [Google Scholar]
- 31.Meena BS, Kapoor KN, Agarwal RK. Occurrence of multi-drug resistant Bacillus cereus in foods. J Food Sci Technol. 2000;37:289–291. [Google Scholar]
- 32.Khasnabis J, Adhikari P, Chowdhury D, Rai C, Roy A. Incidence of multiple drug resistant Bacillus cereus in some popular snacks and sweets sold in Kolkata city India. Indian J Microbiol Res. 2017;4:14–19. [Google Scholar]
- 33.Bayode MT, Olalemi AO, Babayemi Olawale Oladejo BO. Multiple antibiotic resistant index and detection of qnrS and qnrB genes in bacterial consortium of urine samples from clinical settings. Eur J Biol Res. 2021;11:45–56. [Google Scholar]
- 34.Savić D, Miljković-Selimović B, Lepšanović Z, Tambur Z, Konstantinović S, Stanković N, et al. Antimicrobial susceptibility and β-lactamase production in Bacillus cereus isolates from stool of patients, food and environment samples. Vojnosanit Pregl. 2016;73:904–909. doi: 10.2298/VSP150415134S. [DOI] [PubMed] [Google Scholar]
- 35.Mousa D, Abd El Tawab A, El Hofy F, Maarouf A. Molecular studies on antibiotic resistant Bacillus cereus isolated from meat products and human in Kaliobia, Egypt. Benha Vet Med J. 2020;38:125–130. doi: 10.21608/bvmj.2020.25802.1187. [DOI] [Google Scholar]
- 36.Adesoji AT, Ogunjobi AA. Detection of extended spectrum beta-lactamases resistance genes among bacteria isolated from selected drinking water distribution channels in southwestern Nigeria. BioMed Res Int. 2016;2016:7149295. doi: 10.1155/2016/7149295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obayiuwana A, Ibekwe A. Antibiotic resistance genes occurrence in wastewaters from selected pharmaceutical facilities in Nigeria. Water. 2020;12:1897. doi: 10.3390/w12071897. [DOI] [Google Scholar]
- 38.Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, et al. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis–one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/AEM.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Böhm ME, Huptas C, Krey VM, Scherer S. Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol Biol. 2015;15:246. doi: 10.1186/s12862-015-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldwin VM. You can’t B. cereus – a review of Bacillus cereus strains that cause anthrax-like disease. Front Microbiol. 2020;11:1731. doi: 10.3389/fmicb.2020.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molla MAM. 2019. Poultry feed laced with antibiotics. The Daily Star, 13 April 2019. Available online: https://www.thedailystar.net/frontpage/news/poultry-feed-laced-antibiotics-1729231 (Accessed on 27 July 2022).
- 43.Microbiology Laboratory Guidebook [MLG]. United States Department of Agriculture, Food Safety and Inspection Service, MLG 3.02. 2015
- 44.Saeed ZK, Abbas BA, Othman RM. Molecular identification and phylogenetic analysis of lactic acid bacteria isolated from goat raw milk. Iraqi J Vet Sci. 2020;34:259–263. doi: 10.33899/ijvs.2019.125896.1176. [DOI] [Google Scholar]
- 45.CLSI. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute. 2020.
- 46.Poh-Hwa T, Yoke-Kqueen C, Indu Bala J, Son R. Bioprotective properties of three Malaysia Phyllanthus species: an investigation of the antioxidant and antimicrobial activities. Int Food Res J. 2011;18:887–893. [Google Scholar]
- 47.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Tenover FC, Koehler TM. Beta-lactamase gene expression in a penicillin-resistant Bacillus anthracis strain. Antimicrob Agents Chemother. 2004;48:4873–4877. doi: 10.1128/AAC.48.12.4873-4877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 50.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/AAC.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Cultural morphology and Grams staining of isolated Bacillus spp. Figure S2.16srDNA gene of the isolated Bacillus spp. by PCR test. Figure S3. Toxin genes (nheA, nheB, nheC, cytK) of isolated Bacillus spp. by PCR test. Figure S4. Toxin genes (hblA, hblC, hblD, entFM) of isolated Bacillus spp. by PCR test. Figure S5. Antibiotic resistance genes of isolated Bacillus spp. by PCR test. Table S1. Strains and source of selected Bacillus spp. Table S2. Biochemical characteristics of Bacillus spp. Table S3. Primers of toxin gene and 16srRNA gene and PCR protocol used in this study. Table S4. Overall antimicrobial susceptibility profile of Bacillus spp. Table S5. Overall antimicrobial susceptibility profiles of 7 Bacillus species. Table S6. Distributions of antibiotic genes among animal feed, food and diarrhea. Table S7. Prevalence of ARGs of Bacillus spp. in animal feed, food and diarrhea. Table S8. Prevalence of ARGs of 7 Bacillus species. Table S9. Distributions of ARGs among 7 Bacillus species. Table S10. MDR profiles of B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium and B. coagulans. Table S11. Species wise percentage of MAR index >0.2. Table S12. Sample wise percentage of MAR index >0.2. Table S13. MIC break point of Antibiotic used. Table S14. PCR protocol of antibiotic resistant gene primer used in this study.
Data Availability Statement
The dataset can be accessed upon a reasonable request made to the Corresponding author.