Abstract
Rhamnolipids, naturally occurring biosurfactants constructed of rhamnose sugar molecules and β-hydroxyalkanoic acids, have a wide range of potential commercial applications. In the course of a survey of 33 different bacterial isolates, we have identified, using a phenotypic assay for rhamnolipid production, a strain of the nonpathogenic bacterial species Pseudomonas chlororaphis that is capable of producing rhamnolipids. Rhamnolipid production by P. chlororaphis was achieved by growth at room temperature in static cultures of a mineral salts medium containing 2% glucose. We obtained yields of roughly 1 g/liter of rhamnolipids, an amount comparable to the production levels reported in Pseudomonas aeruginosa grown with glucose as the carbon source. The rhamnolipids produced by P. chlororaphis appear to be exclusively the mono-rhamnolipid form. The most prevalent molecular species had one monounsaturated hydroxy fatty acid of 12 carbons and one saturated hydroxy fatty acid of 10 carbons. P. chlororaphis, a nonpathogenic saprophyte of the soil, is currently employed as a biocontrol agent against certain types of plant fungal diseases. The pathogenic nature of all bacteria previously known to produce rhamnolipids has been a major obstacle to commercial production of rhamnolipids. The use of P. chlororaphis therefore greatly simplifies this matter by removing the need for containment systems and stringent separation processes in the production of rhamnolipids.
Rhamnolipids were first isolated from Pseudomonas aeruginosa and described by Jarvis and Johnson in 1949 (12). These compounds are predominantly constructed from the union of one or two rhamnose sugar molecules and one or two β-hydroxy (3-hydroxy) fatty acids (13). Rhamnolipids with one sugar molecule are referred to as mono-rhamnolipids, while those with two sugar molecules are di-rhamnolipids. The length of the carbon chains found on the β-hydroxyacyl portion of the rhamnolipid can vary significantly. However, in the case of P. aeruginosa 10-carbon molecule chains are the predominant form (6). P. aeruginosa is an opportunistic human pathogen capable of producing several destructive toxins and causing a range of human diseases primarily in immunocompromised individuals, including cystic fibrosis patients, burn victims, and those suffering from leukemia (18, 29). Primary rhamnolipid production by P. aeruginosa occurs during stationary growth phase in rapidly agitated liquid medium with limiting concentrations of nitrogen or iron (8). P. aeruginosa is capable of growth and rhamnolipid production using a range of different carbon sources; however, the highest levels of rhamnolipid production result from using vegetable-based oils as carbon sources, including soybean oil (13), corn oil (14), canola oil (25), and olive oil (23).
Rhamnolipids exhibit several promising industrial applications (2, 3, 16, 17). They are powerful natural emulsifiers capable of reducing the surface tension of water from roughly 76 mN/m to 25 to 30 mN/m (7). This biosurfactant activity of rhamnolipids makes them excellent candidates for assisting in the breakdown and removal of oil spills. Rhamnolipids also demonstrate antibacterial and antifungal activities, suggesting possible roles in the medical and agricultural fields (3). Since this biosurfactant is derived from a natural source and in a pure form has low toxicity levels, rhamnolipids are an attractive alternative to synthetic compounds. However, since rhamnolipids are produced by P. aeruginosa, a pathogen of humans, animals, and plants, there are safety issues that would have to be addressed before rhamnolipids produced in this manner would be considered safe. Since addressing these safety concerns could prove to be cost prohibitive, the effort to commercialize rhamnolipids would be helped considerably if the rhamnolipids could be produced by a nonpathogenic host. Attempts have been made to clone the necessary genes for rhamnolipid production into a nonpathogenic host with some limited success (20). Also, recently a strain of Pseudomonas putida was found to be naturally capable of rhamnolipid production (28). Although this bacterium belonging to the Pseudomonas species is generally considered harmless, there exists some documentation suggesting a limited pathogenic potential (1, 15). In this work we describe the discovery of a bacterial strain belonging to the nonpathogenic bacterial species P. chlororaphis that is capable of naturally producing rhamnolipids under conditions different from any previously described bacterial production method.
MATERIALS AND METHODS
Microorganism.
Thirty-three different bacterial strains were assayed for rhamnolipid production in this study: P. aeruginosa strains PG201, PG201-RhlA::Tn5, and NRRL B-800, Pseudomonas fluorescens strains ATCC 17397, ATCC 17824, ATCC 17577, and ATCC 17816, Pseudomonas chlororaphis strains ATCC 17812, NRRL B-14869, NRRL B-30761, and ATCC 9446, P. putida strains ATCC 17527, W4P396, ATCC 17391, and ATCC 12633, Pseudomonas marginalis strains ATCC 10844 and HT041B, Pseudomonas tolaasii strains ATCC 33618, ATCC 14340, C8, and C9; Pseudomonas syringae strain Cit7; Pseudomonas flavescens strain B62; Pseudomonas oleovorans strains NRRL B-778, NRRL B-14682, and NRRL B-14683; Pseudomonas resinovorans strain NRRL B-2649; Pseudomonas stutzeri strain ATCC 17588; Pseudomonas corrugata strains 388, ATCC 29756, and VC1-VC7; a Ralstonia eutropha strain; and an R. solanacearum strain. P. aeruginosa strains PG201 and its isogenic mutant PG201-RhlA::Tn5 (kindly supplied by George A. O'Toole, Dartmouth Medical School) were used as control strains, serving as a strain capable of rhamnolipid production and one incapable of rhamnolipid production, respectively (19). Strain NRRL B-30761, identified by Stanier et al. (26) as P. chlororaphis, was originally obtained from the Agricultural Research Service culture collection (Northern Regional Research Laboratory [NRRL]). The other bacteria belonging to the genus Pseudomonas, as well as bacteria outside of the genus utilized in this study were from the laboratory collections of D. K. Y. Solaiman and W. Fett.
Media.
Both Kay's minimal medium (0.3% NH4H2PO4, 0.2% K2HPO4, 0.2% glucose, 0.5 mg/liter FeSO4, 0.1% MgSO4) and a mineral salts medium {per liter, 0.7 g KH2PO4, 0.9 g Na2HPO4, 2 g NaNO3, 0.4 g MgSO4 · 7H2O, 0.1 g CaCl2 · 2H2O, 2 ml of trace elements [per liter, 2 g FeSO4 · 7H2O, 1.5 g MnSO4 · H2O, 0.6 g (NH4)6Mo7O24 · 4H2O]} were used in growing P. chlororaphis strain NRRL B-30761 for rhamnolipid production (24, 30). PPGAS medium (0.02 M NH4Cl, 0.02 M KCl, 0.12 M Tris-HCl, 0.0016 M MgSO4, 1% Proteose Peptone [Difco], 0.5% glucose) was used in growing P. aeruginosa strains for rhamnolipid production (30). All Pseudomonas strains were maintained on Pseudomonas isolation agar at 4°C (Difco). The plates used for screening for rhamnolipid production were composed of the mineral salts medium described previously with the addition of 200 μg/ml cetyltrimethylammonium bromide (CTAB; Sigma), 5 μg/ml methylene blue, and 1.5% agar (24).
Cultivation conditions.
P. aeruginosa strains were first grown in Kay's minimal medium for 24 h and then diluted 1:100 into PPGAS medium and incubated for 24 to 72 h. In all cases, incubations were at 37°C with orbital shaking at 250 rpm. P. chlororaphis strain NRRL B-30761, however, produced rhamnolipids when first grown in Kay's minimal medium for 24 to 48 h at 30°C with orbital shaking at 250 rpm, followed by 1:100 dilution in static mineral salts plus glucose (2% final volume) medium in various-size Erlenmeyer flasks at room temperature (20 to 23°C), and incubated between 72 and 120 h.
Analytical methods.
Bacterial strains were initially assayed for rhamnolipid production using the mineral salt-CTAB-methylene blue agar plate method originally developed by Siegmund and Wagner (24). Bacteria were grown for 24 h in Kay's minimal medium under appropriate growth conditions. Shallow wells were cut into the surface of the indicator plates with the heated point of a 10-ml glass pipette. Ten microliters of the appropriate culture was placed into each well. The plates were then incubated at the proper temperature and checked periodically over a 24- to 48-h time period. A positive reaction for rhamnolipids is the formation of a purple-blue haze with a sharply defined edge around the culture well. After incubation, plates are placed at 4°C for a few days. This causes positive reactions to darken significantly and to make visible weak positive reactions that were not apparent upon initial inspection. The filtered supernatants (0.45-μm filter) of bacterial cultures believed to be producing rhamnolipids were measured for changes in surface tension using a DCAT 11 tensiometer (Future Digital Scientific Corp.).
Rhamnolipids were purified by first separating the cells from supernatant by centrifugation (6,800 × g). The supernatant was then acidified using 12 M hydrochloric acid to pH 2.0, and the precipitated rhamnolipids were collected by centrifugation (12,100 × g). Rhamnolipids were extracted three times with a chloroform-ethanol (2:1) mixture, which was then evaporated away leaving behind relatively pure rhamnolipids having an oil-like appearance (30). The oily residues were dissolved in an appropriate volume of methanol and transferred to a previously weighed container. The methanol was evaporated under a stream of nitrogen, and the weight of the recovered rhamnolipids was determined to calculate the total rhamnolipid yield. Rhamnolipid preparations were separated, visualized, and compared to known rhamnolipid samples (JBR599; Jeneil Biosurfactant Co., LCC) using thin-layer chromatography (TLC; silica gel 60 plates, with a carrier solution of chloroform-methanol-water, 65:15:2 by volume), and developed using a 50:1:0.05 mixture of the solution glacial acetic acid-sulfuric acid-anisaldehyde. Finally, rhamnolipid preparations were analyzed using high-performance liquid chromatography-mass spectrometry (HPLC/MS). A Waters 2690 separation module (Waters Co., Milford, MA) fitted with 5 cm by 2.1 mm and 15 cm by 2.1 mm Symmetry C18 3.5-μm columns linked in series was used for the HPLC separation portion. A Micromass ZMD mass spectrometer containing atmospheric pressure chemical ionization (APCI) and electrospray ionization (ESI) probes (Waters Co.) was next used for structure elucidation of the chromatograph-separated products.
RESULTS AND DISCUSSION
A wide range of different Pseudomonas species, as well as other types of bacteria, were screened for rhamnolipid production using CTAB-methylene blue indicator plates. Of the bacterial strains assayed by this method, one of the strains demonstrated a weak positive reaction for rhamnolipid production. The strain with the positive reaction was identified as P. chlororaphis strain NRRL B-30761. In the case of NRRL B-30761, the indicator plates gave an initially weak positive reaction after 48 h of incubation at 30°C. However, after the plates were placed at 4°C for 48 h a still weak but decidedly positive reaction for rhamnolipids was clearly visible. No other strains assayed demonstrated a positive reaction after placing the plates at 4°C for 48 h.
Attempts to isolate rhamnolipids directly from the liquid cultures of NRRL B-30761 giving positive reactions on the indicator plates failed. In an attempt to increase the level of production by strain B-30761 of the suspected rhamnolipids, experiments using different growth conditions were performed. The surface tensions of the cultures of strain B-30761 grown under the different conditions were measured using the DCAT 11 tensiometer. Since rhamnolipids are surfactants, a culture with a large decrease in surface tension should be making a reasonable quantity of rhamnolipids. The growth conditions for strain B-30761 that resulted in the greatest decrease in surface tension were initial growth for 24 to 48 h at 30°C and 250 rpm in Kay's minimal medium, followed by 1:100 dilution in mineral salts medium and incubation for 48 h statically at room temperature. The resulting spent culture lowered the surface tension of the mineral salts medium from 65 mN/m to 25 to 30 mN/m, consistent with the effects of rhamnolipids upon surface tension.
TLC results suggested that the isolated surface-active product from P. chlororaphis strain NRRL B-30761 was composed of rhamnolipids. The product was separated on TLC plates alongside a sample of a commercially available purified rhamnolipid from P. aeruginosa. When the two samples were visualized, similarities in the separation profiles were observed. The commercial rhamnolipid sample had two predominant characteristic spots. The lower spot consisted of di-rhamnolipids (Rf = 0.16), while the higher spot consisted of mono-rhamnolipids (Rf = 0.37). The product from the P. chlororaphis strain was observed to have three predominant spots. Spot 1 (Rf = 0.39) migrated at a similar mobility to the mono-rhamnolipids from the commercial rhamnolipid sample. The remaining two unknown spots, 2 (Rf = 0.62) and 3 (Rf = 0.84), did not migrate in a manner similar to the known sample (it was later determined that spots 1 and 2 consisted of various mono-rhamnolipid forms, while spot 3 consisted of the methyl ester forms of the rhamnolipids).
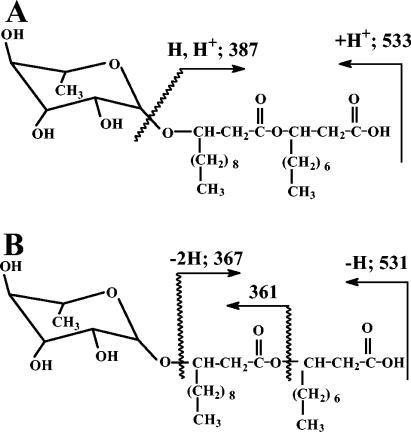
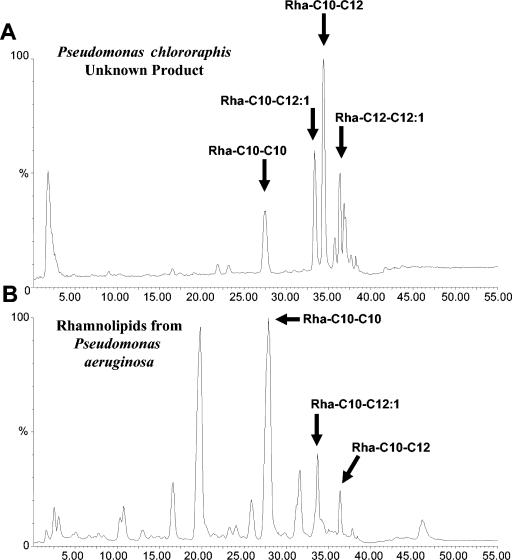
The product from P. chlororaphis was next submitted to HPLC/MS analysis to confirm the presence of rhamnolipids. Rhamnolipid structural information was obtained through the use of mass detector equipment with an APCI and/or ESI probe. Figure 1A presents the expected positive APCI fragmentation sites of a C12-C10 rhamnolipid molecule and the sizes of the corresponding product. An ion was observed at m/z 387 in the APCI spectrum consistent with the acyl moiety fragment of the molecule, but no direct evidence for the individual hydroxy fatty acid could be obtained from that ion fragment; however, positive APCI provided a clean chromatogram with molecular ions resulting from proton addition. Positive ESI showed the addition of sodium to the molecular ion and a noisier chromatogram. Alternatively, negative ESI produced a spectrum documenting ions of m/z 367 and m/z 361 that contained specific information on the fatty acid and the linkage order to the rhamnose ring and corresponded nicely to the products expected from negative ESI fragmentation of a C12-C10 rhamnolipid molecule, shown in Fig. 1B. Negative ESIs have been previously reported by Déziel et al. (5) for the analysis and structural elucidation of rhamnolipids. Accordingly, the rhamnolipid structures under the eluting peaks in the chromatogram in Fig. 2A were determined using negative ESI. Commercial samples with known compositions of rhamnolipids produced by P. aeruginosa were also analyzed by the HPLC/MS method used for the products secreted by P. chlororaphis (Fig. 2B). The comparison of the products from these two strains indicates that P. chlororaphis did not secreted detectable amount of di-rhamnolipids as P. aeruginosa, eluting between 10 and 25 min in the chromatogram in Fig. 2B. The fatty acid moiety for the mono-rhamnolipid consisted of two saturated hydroxy fatty acids of 10- and 12-carbon lengths with a minor composition of product carrying a double bond on the C12 and C14 fatty acids (Fig. 2A). Also, the rhamnolipids did not show the reverse order of linkage observed with P. aeruginosa (5) and most of the rhamnolipids have the long-chain fatty acid glycosylated to the sugar ring and esterified to the second short fatty acid. No evidence for a single fatty acid moiety was found for the rhamnolipids secreted by P. chlororaphis.
FIG. 1.
The structure of a C12-C10 rhamnolipid showing the molecule fragmentation sites and fragment sizes expected to result from the application of a positive APCI probe (A) or a negative ESI probe (B).
FIG. 2.
Chromatographs resulting from HPLC/MS analysis of the P. chlororaphis surface-active product (A) and commercially produced rhamnolipids from P. aeruginosa (B). Arrows indicate peaks common to both samples.
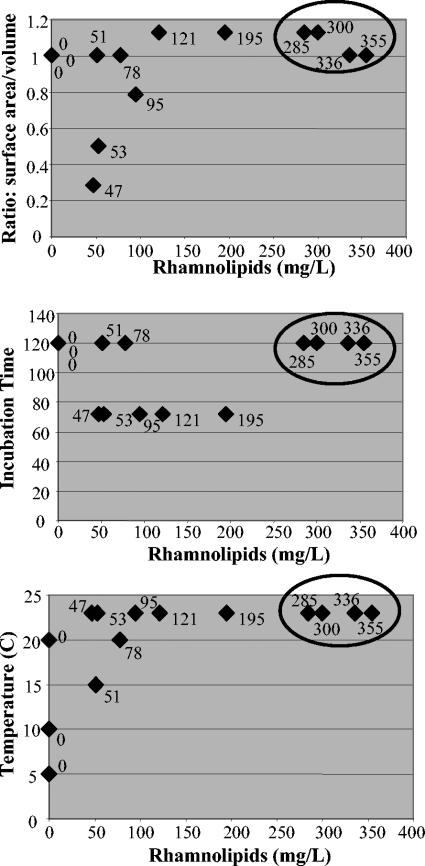
In an attempt to increase rhamnolipid production by NRRL B-30761, various incubation times, temperatures, and surface areas of the growth containers were investigated as to their effects on production. The results indicated that incubations in Kay's minimal medium could be reduced to 24 h without affecting production levels. Incubations of 120 h in mineral salts medium compared to 72 h resulted in significant increases in rhamnolipid production. Room temperature (20 to 23°C) incubation appeared to result in the highest level of rhamnolipid production. Finally, some argument can be made that the surface area per medium volume correlates to the rhamnolipid production levels. A large surface area of the growth container per volume of growth medium often resulted in an increase in rhamnolipid production. This effect is most clearly visible when comparisons are only made between growth containers of similar types, for example, when only data from culture flasks are compared. The three parameters of temperature, incubation time, and the ratio of surface area to medium volume were each compared against the resulting amounts of rhamnolipids produced from samples grown in culture flasks (Fig. 3). The top four rhamnolipid-producing samples in each comparison were grown at temperatures above 20°C, with incubation times of at least 120 h, and had the largest surface area to medium volume ratios.
FIG. 3.
A comparison of the three growth conditions (temperature, incubation time [hours], and surface area to medium volume ratio in flask) and their effects on rhamnolipid production. The positions of the top four rhamnolipid-producing cultures are circled for easy comparison in each of the three graphs. The amount of rhamnolipid produced (milligrams/liter) for each sample is noted beside the respective datum point.
The lengths of the fatty acid chains of rhamnolipids can vary significantly, resulting in a multitude of different rhamnolipid compositions. Through HPLC/MS, we have observed a range of rhamnolipid variants made by P. chlororaphis strain NRRL B-30761 and contrasted them with the published lists of forms produced by P. aeruginosa and Burkholderia pseudomallei (Table 1). Fatty acyl chains composed of 8, 10, 12, and 14 carbons in length, as well as 12- or 14-carbon chains with double bonds (12:1, 14:1), have been observed in rhamnolipids produced by P. aeruginosa or B. pseudomallei. The current body of research has shown that the pathogen P. aeruginosa is capable of making both mono-rhamnolipids and di-rhamnolipids, while B. pseudomallei, also a pathogen, makes only di-rhamnolipids (9, 11). Our research indicates that P. chlororaphis strain NRRL B-30761, in contrast, makes only mono-rhamnolipids, suggesting that it lacks a homologue of the rhlC gene, which in P. aeruginosa is responsible for the biosynthesis of di-rhamnolipids (21). B. pseudomallei makes rhamnolipids with fatty acyl chains only 14 carbons in length, whereas P. aeruginosa is capable of making rhamnolipids containing fatty acyl chains with carbon lengths of 8, 10, 12, 14, 12:1, and 14:1 (5, 9). Of the observed fatty acid moieties, the combinations that P. aeruginosa has not been observed to make are: C10-C14 or C14-C10, and C14-C14. Our research has shown that P. chlororaphis strain NRRL B-30761 is also capable of making rhamnolipids with fatty acids containing carbon lengths of 8, 10, 12, 14, 12:1, and 14:1. Likewise, strain B-30761 has not been observed to make the C14-C14 combination. However, in contrast to P. aeruginosa, we observed the production of the C14-C10 form in strain B-30761, but not the production of the C8-C8, C8-C12:1, or C12:1-C8 combination. In the majority of cases for P. aeruginosa, the major form of rhamnolipid purified is the C10-C10 form, with the other forms constituting minor fractions of the product (6). This differs from P. chlororaphis strain NRRL B-30761, where the major forms of rhamnolipids appear to be C12-C10 or C12:1-C10, comprising, on average, 30% and 40% of the total rhamnolipid preparations, respectively (Fig. 2). While the P. chlororaphis minor rhamnolipid forms, C10-C10, C12-C12, and C12:1-C12, on average, constitute slightly less than 10% each of the total rhamnolipid preparations.
TABLE 1.
Comparison of the compositions of the various fatty acyl chains and sugar moieties of the rhamnolipid molecules produced by the three organisms known to be capable of rhamnolipid production
| Product(s) | Pseudomonas aeruginosaa | Burkholderia pseudomalleib | Pseudomonas chlororaphis NRRL B-30761 |
|---|---|---|---|
| Mono-rhamnolipids | Yes | NDc | Yes |
| Di-rhamnolipids | Yes | Yes | ND |
| C8-C8 | Yes | ND | ND |
| C10-C8 | Yes | ND | Yes |
| C8-C12:1, C12:1-C8 | Yes | ND | ND |
| C10-C10 | Yes | ND | Yes |
| C12-C10 | Yes | ND | Yes |
| C12:1-C10 | Yes | ND | Yes |
| C12-C12 | Yes | ND | Yes |
| C12:1-C12 | Yes | ND | Yes |
| C14-C10 | ND | ND | Yes |
| C14:1-C10 | Yes | ND | Yes |
| C14-C14 | ND | Yes | ND |
Our work describes, for the first time, production of rhamnolipids by a member of the species P. chlororaphis, a nonpathogenic bacterium. In fact, members of the P. chlororaphis species are regularly used as biocontrol strains and sprayed directly onto plant seeds to protect the seeds against fungal pathogens (27). Prior to this publication, only P. aeruginosa and B. pseudomallei, two pathogenic bacterial species, were reported to produce rhamnolipids. A single strain of P. putida was also reported recently to produce rhamnolipids (28). The use of a P. chlororaphis strain as an industrial producer of rhamnolipids would eliminate the problems that a pathogenic producer presents, such as concerns about the toxins so readily produced and released by P. aeruginosa and B. pseudomallei finding their way into any rhamnolipid-based product (4, 18, 22).
The research in this paper focused on rhamnolipid production by P. chlororaphis using glucose as the sole carbon source for growth. In our study the highest level of rhamnolipid production by P. chlororaphis was 0.355 g/liter (Fig. 3), but recent changes in our extraction method have resulted in rhamnolipid yields of roughly 1 g/liter (data not shown). The rhamnolipid production levels achieved by cultures of P. aeruginosa using glucose as the sole carbon source result in yields of 1.0 to 1.6 g/liter (7, 25). While the production levels of P. chlororaphis are less than those of P. aeruginosa, they do not appear to differ significantly.
Rhamnolipid production in P. chlororaphis is best achieved through incubation at room temperature, with static growth in a minimal medium. Such production conditions, as opposed to the optimal conditions for P. aeruginosa, including rapid mechanical agitation at 37°C, should save a producer of rhamnolipids energy costs during the production process. The potential for energy savings in concert with the simplifying nature of a nonpathogenic organism should serve to make P. chlororaphis a more attractive candidate for commercial rhamnolipid production.
Acknowledgments
We thank Marshall Reed, Loida Cruz-Bass, and Bun-Hong Lai for assistance with this project.
REFERENCES
- 1.Anaissie, E., V. Fainstein, P. Miller, H. Kassamali, S. Pitlik, G. P. Bodey, and K. Rolston. 1987. Pseudomonas putida. Newly recognized pathogen in patients with cancer. Am. J. Med. 82:1191-1194. [DOI] [PubMed] [Google Scholar]
- 2.Banat, I. M., R. S. Makkar, and S. S. Cameotra. 2000. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 53:495-508. [DOI] [PubMed] [Google Scholar]
- 3.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081-1100. [DOI] [PubMed] [Google Scholar]
- 5.Déziel, E., F. Lepine, D. Dennie, D. Boismenu, O. A. Mamer, and R. Villemur. 1999. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1440:244-252. [DOI] [PubMed] [Google Scholar]
- 6.Déziel, E., F. Lepine, S. Milot, and R. Villemur. 2000. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim. Biophys. Acta 1485:145-152. [DOI] [PubMed] [Google Scholar]
- 7.Guerra-Santos, L., O. Kappeli, and A. Fiechter. 1984. Pseudomonas aeruginosa biosurfactant production in continuous culture with glucose as carbon source. Appl. Environ. Microbiol. 48:301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerra-Santos, L. H., O. Kappeli, and A. Fiechter. 1986. Dependence of Pseudomonas aeruginosa continuous culture biosurfactant production on nutritional and environmental factors. Appl. Microbiol. Biotechnol. 24:443-448. [Google Scholar]
- 9.Haussler, S., M. Nimtz, T. Domke, V. Wray, and I. Steinmetz. 1998. Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect. Immun. 66:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haussler, S., M. Rohde, N. von Neuhoff, M. Nimtz, and I. Steinmetz. 2003. Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect. Immun. 71:2970-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh, S., H. Honda, F. Tomita, and T. Suzuki. 1971. Rhamnolipids produced by Pseudomonas aeruginosa grown on n-paraffin. J. Antibiot. (Tokyo) 24:855-859. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis, F. G., and M. J. Johnson. 1949. A glyco-lipid produced by Pseudomonas aeruginosa. J. Am. Chem. Soc. 71:4124-4126. [Google Scholar]
- 13.Lang, S., and D. Wullbrandt. 1999. Rhamnose lipids—biosynthesis, microbial production and application potential. Appl. Microbiol. Biotechnol. 51:22-32. [DOI] [PubMed] [Google Scholar]
- 14.Linhardt, R. J., R. Bakhit, L. Daniels, F. Mayerl, and W. Pickenhagen. 1989. Microbially produced rhamnolipid as a source of rhamnose. Biotech. Bioeng. 33:365-368. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlane, L., B. A. Oppenheim, and P. Lorrigan. 1991. Septicaemia and septic arthritis due to Pseudomonas putida in a neutropenic patient. J. Infect. 23:346-347. [DOI] [PubMed] [Google Scholar]
- 16.Maier, R. M., and G. Soberon-Chavez. 2000. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl. Microbiol. Biotechnol. 54:625-633. [DOI] [PubMed] [Google Scholar]
- 17.Mulligan, C. N. 2005. Environmental applications for biosurfactants. Environ. Pollut. 133:183-198. [DOI] [PubMed] [Google Scholar]
- 18.Nicas, T. I., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 19.Ochsner, U. A., A. Fiechter, and J. Reiser. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269:19787-19795. [PubMed] [Google Scholar]
- 20.Ochsner, U. A., J. Reisner, A. Fiechter, and B. Witholt. 1995. Production of Pseudomonas aeruginosa rhamnolipid biosurfactant in heterologous hosts. Appl. Environ. Microbiol. 61:3503-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahim, R., U. A. Ochsner, C. Olvera, M. Graninger, P. Messner, J. S. Lam, and G. Soberon-Chavez. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708-718. [DOI] [PubMed] [Google Scholar]
- 22.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert, M., M. E. Mercade, M. P. Bosch, J. L. Parra, M. J. Espuny, M. A. Manresa, and J. Guinea. 1989. Effect of the carbon source on biosurfactant production by Pseudomonas aeruginosa 44T1. Biotechnol. Lett. 11:871-874. [Google Scholar]
- 24.Siegmund, I., and F. Wagner. 1991. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 5(4):265-268. [Google Scholar]
- 25.Sim, L., O. P. Ward, and Z. Y. Li. 1997. Production and characterisation of a biosurfactant isolated from Pseudomonas aeruginosa UW-1. J. Ind. Microbiol. Biotechnol. 19:232-238. [DOI] [PubMed] [Google Scholar]
- 26.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 27.Tombolini, R., D. J. van der Gaag, B. Gerhardson, and J. K. Jansson. 1999. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA 342 on barley seeds visualized by using green fluorescent protein. Appl. Environ. Microbiol. 65:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuleva, B. K., G. R. Ivanov, and N. E. Christova. 2002. Biosurfactant production by a new Pseudomonas putida strain. Z. Naturforsch. C 57(3-4):356-360. [DOI] [PubMed] [Google Scholar]
- 29.Woods, D. E., M. S. Schaffer, H. R. Rabin, G. D. Campbell, and P. A. Sokol. 1986. Phenotypic comparison of Pseudomonas aeruginosa strains isolated from a variety of clinical sites. J. Clin. Microbiol. 24:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y., and R. M. Miller. 1992. Enhanced octadecane dispersion and biodegradation by a Pseudomonas rhamnolipid surfactant (biosurfactant). Appl. Environ. Microbiol. 58:3276-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]