Abstract
Brown rot basidiomycetes have long been thought to lack the processive cellulases that release soluble sugars from crystalline cellulose. On the other hand, these fungi remove all of the cellulose, both crystalline and amorphous, from wood when they degrade it. To resolve this discrepancy, we grew Gloeophyllum trabeum on microcrystalline cellulose (Avicel) and purified the major glycosylhydrolases it produced. The most abundant extracellular enzymes in these cultures were a 42-kDa endoglucanase (Cel5A), a 39-kDa xylanase (Xyn10A), and a 28-kDa endoglucanase (Cel12A). Cel5A had significant Avicelase activity—4.5 nmol glucose equivalents released/min/mg protein. It is a processive endoglucanase, because it hydrolyzed Avicel to cellobiose as the major product while introducing only a small proportion of reducing sugars into the remaining, insoluble substrate. Therefore, since G. trabeum is already known to produce a β-glucosidase, it is now clear that this brown rot fungus produces enzymes capable of yielding assimilable glucose from crystalline cellulose.
Brown rot basidiomycetes are the principal recyclers of lignocellulose in coniferous forest ecosystems, where they make essential contributions to humus formation and soil fertility (10). They also cause extensive and costly decay in wooden structures (37). During brown rot, little lignin is removed from the wood, probably because these fungi do not secrete the ligninolytic peroxidases or laccases that typify white rot fungi. However, brown rot fungi rapidly cleave the cellulose in wood and subsequently remove it so completely that extensively brown-rotted wood consists almost entirely of modified lignin (6). Since most of the cellulose chains in wood are interconnected in a hydrogen-bonded, crystalline form that resists hydrolysis (1, 26), it would appear that brown rot fungi employ an efficient cellulolytic system.
The biodegradation of crystalline cellulose generally involves the action of both endo- and exo-acting cellulases. Classical endoglucanases nick the cellulose internally, thus disrupting its crystallinity and generating new free ends in the polymer. Cellobiohydrolases (exoglucanases) act processively from these free ends, remaining attached to the cellulose and releasing soluble cellobiose molecules, which are subsequently hydrolyzed to assimilable glucose by β-glucosidases (2). In addition, some organisms produce processive endoglucanases, which cleave cellulose internally but also release soluble oligosaccharides before detaching from the polysaccharide (9, 30, 34).
In this connection, it is puzzling that brown rot basidiomycetes are generally thought to lack processive cellulases (6). They are known to produce extracellular reactive oxygen species (ROS) (4) and classical endoglucanases (6), which doubtless can disrupt crystalline cellulose extensively when they act together (29). However, neither of these biodegradative agents can release soluble sugars processively from cellulose, because they both attack the polymer via random, single scissions. It is difficult to see how so inefficient a system could account for the characteristically thorough degradation of wood cellulose by brown rot fungi.
The likely explanation is that processive cellulases occur in brown rot fungi but have been overlooked because they are not produced under most laboratory conditions. There is one report of cellobiohydrolase production by Coniophora puteana (33), but this atypical fungus produces laccase activity on wood, and its decay pattern exhibits features of both brown rot and white rot (23). More recent work has shown that crude extracts from wood colonized by two other brown rot fungi, Wolfiporia cocos and Laetiporius sulfureus, were able to release soluble reducing sugars from crystalline cellulose (24). However, the enzymes responsible for processivity were not characterized.
Gloeophyllum trabeum causes a typical brown rot and is the best-understood fungus in this group. It has been shown to produce a hydroquinone-driven system for the production of extracellular ROS (20, 27), a β-glucosidase (14), a xylanase (31), and two endoglucanases that were reported not to degrade crystalline cellulose (15, 25). If G. trabeum were also to produce a processive cellulase, it would have all of the components currently thought necessary for wood decay. To investigate this possibility, we have grown G. trabeum on microcrystalline cellulose (Avicel) and characterized the principal glycosylhydrolases secreted by the cultures. We report that the major extracellular enzyme produced by G. trabeum on this substrate is a processive endoglucanase with Avicelase activity.
MATERIALS AND METHODS
Reagents.
Chemicals were obtained from Sigma-Aldrich/Fluka (St. Louis, MO) except for cellotriose (V-Labs, Covington, LA), cellotetraose (Seikagaku, Tokyo, Japan), and cellopentaose (Seikagaku). Phosphoric acid swollen cellulose (PASC) was prepared from Avicel as described previously (36).
Cultures.
G. trabeum (ATCC 11539) was maintained on agar plates as described previously (17). For each culture, 7.0 g of Avicel was placed in a 250-ml Erlenmeyer flask and moistened with 12 ml of distilled, deionized water (ddH2O). The flasks were then autoclaved, and 7.5 ml of inoculum was added. To prepare the inoculum, G. trabeum was grown for 4 days in 2.8-liter Fernbach flasks containing 50 ml liquid medium with glucose as previously described (17). Mycelial mats from eight flasks were pooled, washed three times with sterile ddH2O, homogenized in a Waring blender, and added to 800 ml of fresh, sterile medium without glucose. The cultures were capped with aluminum foil and grown statically at 31°C for 10 days. Biomass in the cultures was determined as total ergosterol (5). Extracellular ROS production by the cultures was monitored by assaying the production of 14CO2 from [14C]polyethylene glycol ([14C]PEG) (19).
Extraction of extracellular enzymes.
All steps were done at 0 to 4°C. On day 10, 20 ml of extraction buffer (20 mM sodium acetate and 1.0 mM phenylmethylsulfonyl fluoride at pH 4.0) was added to each of 100 cultures. The contents of all the flasks were pooled and rotary shaken (150 rpm) for 45 min. The slurry was then centrifuged (10 min, 23,000 × g), the supernatant was filtered through a glass fiber filter, and the resulting filtrate was refiltered through a 0.45-μm nylon filter. The pellet was resuspended in fresh extraction buffer and extracted again by the same procedure, after which the pooled filtrates (approximately 3 liters) were concentrated to a volume of about 150 ml with a 10-kDa-cutoff polysulfone hollow fiber ultrafiltration apparatus (Amersham, Piscataway, NJ) that had been washed beforehand for 45 min at ambient temperature with 0.5% (wt/vol) PEG (18.5 kDa) to reduce nonspecific protein binding. The concentrated sample was transferred to an ultrafiltration cell (Millipore, Billerica, MA) fitted with a PEG-washed 10-kDa-cutoff polyethersulfone membrane, dialyzed by repeated buffer exchanges against 20 mM sodium acetate (pH 4.0), and concentrated to a final volume of 15 ml. A protease inhibitor cocktail (Roche, Indianapolis, IN) was added according to the manufacturer's instructions, and the sample was stored for 1 to 3 days at 4°C.
Enzyme purification.
The crude extract was concentrated and dialyzed against 20 mM methylpiperazine buffer (pH 4.7) by repeated centrifugation through a 5-kDa-cutoff polyethersulfone membrane (Centricon Plus-20; Millipore). The membrane was rinsed beforehand with ddH2O according to the manufacturer's instructions and then incubated with PEG solution as described above. The PEG solution was poured off, the crude extract was added, and the Centricon tubes were centrifuged at 4,000 × g and 4°C with three buffer exchanges, followed by concentration to a volume less than 5 ml. All dialysis and sample concentration steps described below were also done by this procedure.
The retentate was loaded onto an ice-jacketed, 5-ml HiTrapQ anion-exchange column (Amersham) that had been equilibrated beforehand with 20 mM methylpiperazine buffer (pH 4.7). The column, which was connected to a fast protein liquid chromatography (FPLC) system (Amersham), was operated at 3 ml/min. Proteins were eluted first with a 60-ml wash of 20 mM methylpiperazine (pH 4.7) and then with a 120-ml linear gradient of NaCl (0 to 0.25 M) in the same buffer. Fractions (3.0 ml) were collected, immediately put on ice, and assayed for activity as described below. Xyn10A was eluted in the wash, whereas Cel5A, Cel12A, and the β-glucosidase activity were eluted together at an NaCl concentration of about 0.15 M.
The pooled ion-exchange fractions containing either (i) Xyn10A or (ii) Cel5A plus Cel12A were concentrated to volumes of 0.5 ml and subjected to gel filtration FPLC on a Superdex 200 column and a Superdex 75 column (each 30 cm in length by 1 cm in diameter; Amersham) that were connected in series and packed in ice. The columns were eluted at 0.3 ml/min with 10 mM sodium acetate buffer (pH 5.0; hereafter called buffer) that contained 0.15 M NaCl. Fractions (0.5 ml) were taken, put on ice, and assayed as described below. Xyn10 was resolved from several unidentified proteins during this step. Cel5A and Cel12A were resolved from the much larger β-glucosidase and some other proteins by gel filtration, but the two cellulases were eluted together in a broad peak followed by a shoulder. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of individual fractions showed that the early-eluting side of the peak consisted mostly of Cel5A, whereas the shoulder consisted mostly of Cel12A.
Gel filtration fractions highly enriched in Cel5A or Xyn10A were pooled, concentrated to volumes of 2.5 ml, made 1.0 M in (NH4)2SO4, and subjected to hydrophobic-interaction FPLC on a 1.0-ml Phenyl Superose column (Amersham). The column, equilibrated beforehand with buffer containing 1.0 M (NH4)2SO4, was operated at 0.5 ml/min and ambient temperature. Proteins were eluted first with a 15-ml wash of buffer plus 1.0 M (NH4)2SO4 followed by a 20-ml decreasing linear gradient of (NH4)2SO4 (1.0 to 0 M) in buffer. Fractions (1.0 ml) were collected, put on ice, and assayed for activity. Xyn10A was eluted at an (NH4)2SO4 concentration of about 0.35 M, whereas Cel5A was eluted at a concentration of about 0.5 M.
Gel filtration fractions enriched in Cel12A were also pooled and subjected to hydrophobic-interaction FPLC, but in this case the sample and column were equilibrated and run in buffer containing 0.75 M (NH4)2SO4. Cel12A was eluted in the wash with several minor contaminants and was immediately dialyzed against buffer to remove the (NH4)2SO4. To resolve Cel12A from the remaining contaminants, an additional gel filtration step was performed in buffer as described above.
All of the purified glycosylhydrolases were concentrated and dialyzed against buffer. They were stored for up to a week at 4°C. Freezing resulted in significant losses of activity.
Assays.
Carboxymethyl cellulase (CMCase) activity was assayed with 250-μl mixtures in 10- to 30-min reactions, using a 1% (wt/vol) solution of carboxymethyl cellulose (CMC) in 50 mM sodium citrate buffer (pH 4.8) at 50°C. Xylanase was assayed likewise with 1% (wt/vol) birch xylan as the substrate. Reducing sugars were determined as glucose equivalents by the dinitrosalicylic acid method (8).
Hydrolytic activity against 4-nitrophenylcellobioside (4NPC) was determined with 50-μl mixtures that contained 50 mM sodium acetate (pH 5.5) in 20- to 30-min reactions at 50°C. Activity against 4-nitrophenylglucoside (β-glucosidase activity) was determined likewise in 50 mM potassium phosphate (pH 6.5). The release of 4-nitrophenol from these substrates was determined by the increase in absorbance at 400 nm (ɛ = 18.3 mM−1 cm−1) after addition of Na2CO3 to the reaction mixtures (32).
Cellulase activity against Avicel was determined in 100-μl reactions that contained 10 to 20 μg Cel5A or Xyn10A, 1% (wt/vol) substrate, 100 mM sodium citrate (pH 4.8), 1 mM CaCl2, 0.02% (wt/vol) serum albumin, and 0.02% (wt/vol) NaN3. The capped mixtures were rotary shaken (200 rpm) at 37°C for 18 h. Reactions with PASC as the substrate were conducted likewise, except that the experiment was also done with Cel12A and the reaction time was 2 h. Soluble reducing sugars were determined as glucose equivalents by the 4-hydroxybenzoic acid hydrazide method (28).
For all glycosylhydrolase assays, 1 U of activity was defined as the amount of enzyme that converts 1 nmol of substrate per min. Protein concentrations were determined by the method of Bradford with serum albumin as the standard (3).
Oligoglucoside analysis.
Thin-layer chromatography of the oligoglucosides released from Avicel was done on silica gel (LK5D 150A plates; Whatman, Clifton, NJ). Samples from Avicelase assays or G. trabeum cultures were concentrated under a vacuum, and aliquots containing 5 μg total sugars were applied. The plate was developed with ethyl acetate/water/methanol (40:15:20 vol/vol), and the spots were visualized with anisaldehyde reagent as described previously (18).
Electrophoresis.
SDS-PAGE of crude and purified glycosylhydrolases was done on 4 to 15% gradient gels (Ready Gel precast gels; Bio-Rad). The gels were calibrated with the following molecular mass standards: phosphorylase b (97 kDa), serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), trypsin inhibitor (20 kDa), and α-lactalbumin (14 kDa). The gels were stained with Coomassie blue R-250.
Two-dimensional isoelectric focusing (IEF)/SDS-PAGE was performed by Kendrick Laboratories (Madison, WI). Sample preparation and electrophoresis were done as outlined on the Kendrick website (http://www.kendricklabs.com). One μg of an internal standard, tropomyosin, was added to each sample. After the first dimension was run, the pH gradient in the gel was recorded with a surface electrode. The following molecular mass standards were run down one side of the gel in the second dimension: myosin (220 kDa), phosphorylase a (94 kDa), catalase (60 kDa), actin (43 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa). The gels were stained with Coomassie blue R-250.
Protein sequencing.
Purified proteins were subjected to SDS-PAGE, stained with Coomassie blue, and cut from the gel. The bands were submitted for tryptic digestion and internal Edman sequencing of selected peptides at the University of Texas Protein Chemistry Laboratory (Galveston, TX) as outlined on their website (http://www2.utmb.edu/proch/Default.htm). For N-terminal sequencing at the same facility, the protein on the gel was transferred electrophoretically to a polyvinylidene difluoride membrane (Bio-Rad) at 100 V for 1 h with 25 mM Tris buffer (pH 8.3) that contained 192 mM glycine. The protein was then stained with Coomassie blue, excised, and submitted.
Protein sequence accession numbers.
The partial amino acid sequences of the G. trabeum glycosylhydrolases Cel5A, Xyn10A, and Cel12A have been deposited in the Swiss-Prot database and assigned the accession numbers P84194, P84195, and P84196, respectively.
RESULTS
Fungal growth and production of extracellular proteins.
G. trabeum grew rapidly on Avicel, producing a thick mycelial mat on the surface of the substrate after 10 days. The biomass increased 23-fold during this time, as shown by the difference in ergosterol content between the inoculum (4.3 ± 2.4 μg/culture; n = 5) and day 10 cultures (98.2 ± 14.4 μg/culture; n = 7). The cultures also produced extracellular ROS, as shown by their ability to mineralize [14C]PEG (4.4 ± 1.5% in 9 days, 6.2 ± 1.9% in 12 days; n = 6).
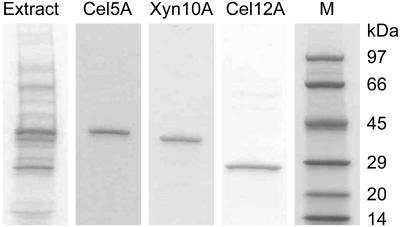
SDS-PAGE analyses of crude extracellular extracts from 10-day cultures showed the presence of about a dozen protein bands. The three most prominent of these proteins had apparent molecular masses of 42 kDa, 39 kDa, and 28 kDa (Fig. 1). Densitometric analyses of the gels indicated that the 42-kDa band accounted for about 20% of the total protein, the 39-kDa band accounted for about 10%, and the 28-kDa band accounted for about 15%. To investigate the possibility that our extraction procedure failed to release some proteins efficiently, we added ethylene glycol (75%, vol/vol) to some of the extractions to disrupt the binding of glycosylhydrolase cellulose-binding modules to the Avicel. However, SDS-PAGE of these extracts showed no significant change in the relative intensities of the protein bands (data not shown).
FIG. 1.
SDS-PAGE analysis of extracellular proteins in the G. trabeum crude extract (4 μg) and of the purified Cel5A, Xyn10A, and Cel12A samples (0.5 μg each) that were submitted for partial amino acid sequencing. Lane M contained molecular mass standards.
Purification of the major glycosylhydrolases.
The crude extracellular extracts contained hydrolase activity towards Avicel as well as high activities towards a variety of standard glycosylhydrolase substrates: CMC, 4NPC, xylan, and PASC (Table 1). We fractionated the extract proteins by ion-exchange, gel-filtration, and hydrophobic-interaction chromatography. To track the purification of potentially interesting cellulases, we assayed the column fractions for CMCase and 4-nitrophenylcellobiosidase (4NPCase) activity and analyzed fractions from each protein peak by SDS-PAGE. Almost all of the CMCase activity was contained in the 42-kDa and 28-kDa proteins. By contrast, almost all of the 4NPCase activity was contained in the 39-kDa protein and in a β-glucosidase (data not shown). The β-glucosidase activity did not survive attempts to purify it, but the purified 42-kDa CMCase, 39-kDa 4NPCase, and 28-kDa CMCase were stable for at least a week at 4°C.
TABLE 1.
Purification and substrate specificities of G. trabeum glycosylhydrolases
| Enzyme (mass in kDa) | Amt of protein (μg) | Yield of activity in extract (%) | Sp act (U/mg) on substrate:
|
||||
|---|---|---|---|---|---|---|---|
| CMC | 4NPC | Xylan | PASC | Avicel | |||
| Extract | 11,600 | 100 | 27,000 | 4,900 | 26,000 | 670 | 7.3c |
| Cel5A (42) | 82 | 3.1a | 120,000 | 65 | 770 | 54 | 4.5d |
| Xyn10A (39) | 123 | 2.6b | 950 | 12,000 | 390,000 | 9.2 | 1.8 |
| Cel12A (28) | 4 | 0.1a | 42,000 | 69 | 880 | 49 | -e |
Percent of CMCase activity in the crude extract.
Percent of 4NPCase activity in the crude extract.
Mean specific activity of five separate preparations (standard deviation = 0.8 U/mg).
Mean specific activity of three separate preparations (standard deviation = 0.4 U/mg).
-, not determined.
A variable mixture of minor proteins, some having low levels of CMCase or 4NPCase activity, was present in all of the crude extracts. We surmise that some of these were not native G. trabeum proteins but rather products of random ROS attack on the major extracellular enzymes. It would be surprising if such modifications did not occur, because G. trabeum produces high levels of ROS on cellulose (4), and oxidative damage to proteins by ROS is a well-known phenomenon (12). The presence of so many minor proteins complicated the purification protocols, and to obtain highly purified glycosylhydrolases we had to pool the chromatographic fractions very selectively, with the result that final yields of the 42-kDa and 39-kDa enzymes were only several percent of the activities present in the crude extract (Table 1). Yields of the 28-kDa enzyme were even lower, because it was unstable in the concentrated (NH4)2SO4 needed to make proteins adsorb to the hydrophobic-interaction column.
Identification of the glycosylhydrolases.
The resulting preparations of the three enzymes were apparently homogeneous by SDS-PAGE (Fig. 1). Assays for glycosylhydrolase activity showed that the 42-kDa and 28-kDa enzymes exhibited relatively high specific activities on CMC and PASC (noncrystalline cellulose), and therefore can be classified as endoglucanases (Table 1). The 39-kDa enzyme exhibited a relatively high specific activity on 4NPC while lacking activity on 4-nitrophenylglucoside and therefore appeared at first to be a cellobiohydrolase. However, its higher activity on xylan showed that it is better classified as a xylanase.
To check these assignments, we submitted SDS-PAGE bands of the three proteins for tryptic digestion and partial amino acid sequencing (Fig. 2). The results, when aligned with known glycosylhydrolase sequences available through the CAZY website (http://afmb.cnrs-mrs.fr/CAZY) (13), showed that the 42-kDa enzyme is a family 5 endoglucanase (Cel5A), the 39-kDa enzyme is a family 10 xylanase (Xyn10A), and the 28-kDa enzyme is a family 12 endoglucanase (Cel12A).
FIG. 2.
Peptide sequence comparisons between glycosylhydrolases of G. trabeum and other organisms. A. G. trabeum Cel5A (GtCel5A) compared with Hypocrea jecorina Cel5A (HjCel5A, National Center for Biotechnology Information [http://ncbi.nlm.nih.gov/entrez/query.fcgi?] accession no. P07982) and Macrophomina phaseolina Cel5 (MpCel5, accession no. 1085763). B. G. trabeum Xyn10A (GtXyn10A) compared with Agaricus bisporus Xyn10 (AbXyn10, accession no. CAB05886) and C. fimi Xyn10A (CfXyn10A, accession no. P07986). C. G. trabeum Cel12A (GtCel12A) compared with Aspergillus aculeatus F-50 Cel12 (AaCel12, accession no. P22669) and A. aculeatus KSM 510 Cel12 (AaCel12′, accession no. AAD02275). All sequences are internal except the first Cel5A sequence of G. trabeum, which is N-terminal.
Activity of the enzymes on crystalline cellulose.
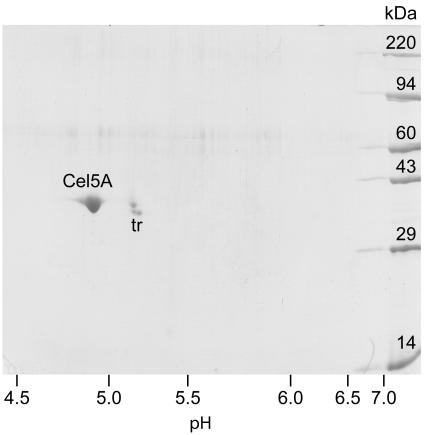
Purified Cel5A and Xyn10A from G. trabeum released soluble reducing sugars from Avicel (Table 1). This result prompted us to recheck the purity of the enzymes, because the processivity of a cellulase might be increased if another cellulase is present as a contaminant and the two enzymes act synergistically. Two-dimensional IEF/SDS-PAGE showed that Cel5A was apparently pure and that it had a pI of 4.9 (Fig. 3). Our Xyn10A preparation was about 95% pure by two-dimensional IEF/SDS-PAGE and had a pI of 4.8 (data not shown). To look for possible synergism between Cel5A and Xyn10A, we assayed Avicel hydrolysis by a combination (1:1, wt/wt) of the enzymes. The results showed that Cel5A and Xyn10A acted additively (data not shown). We did not obtain enough Cel12A to assay it for Avicelase activity or recheck its purity, but in any case it is unlikely to hydrolyze crystalline cellulose because the family 12 endoglucanases of fungi lack cellulose-binding modules (11).
FIG. 3.
Two-dimensional IEF/SDS-PAGE of purified G. trabeum Cel5A. The locations of Cel5A and the tropomyosin internal standard (tr) are indicated.
Mode of Avicel hydrolysis by Cel5A.
We selected Cel5A for further investigation because it was the major extracellular enzyme in Avicel-grown G. trabeum cultures and because it had the highest activity on crystalline cellulose among the enzymes we could identify. The specific rate at which Cel5A introduced insoluble reducing sugars into Avicel was about 0.4 U/mg protein, less than 10% of the specific rate at which it generated soluble reducing sugars from this substrate (Table 1). This low ratio of insoluble to soluble hydrolysis products indicates that G. trabeum Cel5A is a processive endoglucanase (9, 30). Nonprocessive endoglucanases yield a higher proportion of insoluble reducing sugars when they cleave cellulose—typically 30 to 50% (16).
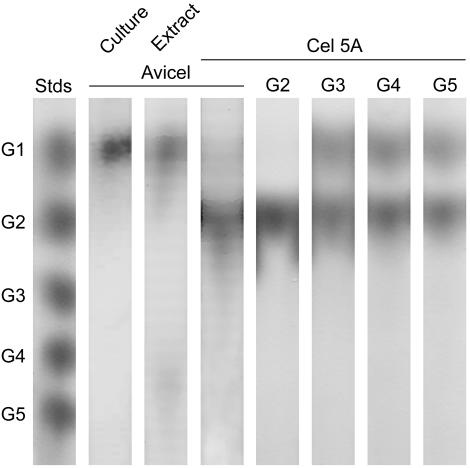
Cellobiose was the only major reducing sugar released from Avicel by Cel5A, although a trace of glucose was apparent (Fig. 4). When recalibrated against a cellobiose standard curve, the average specific Avicelase activity of Cel5A was 4.1 U/mg, slightly lower than the value obtained with a glucose standard curve (Table 1). Cel5A did not cleave cellobiose, but it cleaved larger oligomers quantitatively to a mixture of cellobiose and glucose. Intact G. trabeum cultures and crude protein extracts contained a complete cellulase system, giving glucose as the sole soluble product from Avicel (Fig. 4).
FIG. 4.
Thin layer chromatography showing products released from Avicel or from oligoglucosides. The leftmost lane shows standards of glucose (G1), cellobiose (G2), cellotriose (G3), cellotetraose (G4), and cellopentaose (G5). The other lanes show, from left to right, products released by G. trabeum cultures from Avicel, products released by the crude extracellular extract from Avicel, products released by purified Cel5A from Avicel, and products released by purified Cel5A from the oligoglucosides G2 through G5.
DISCUSSION
A rough estimate of the Cel5A contribution to crystalline cellulose hydrolysis by G. trabeum extracts can be obtained from its mean specific activity on Avicel (4.1 U cellobiose per mg) and abundance relative to all extracellular proteins in the crude extracellular extract (about 20%). This calculation yields a Cel5A contribution of 0.8 U cellobiose released per mg crude protein. Since the crude extract also contained β-glucosidase, the total Cel5A contribution under these conditions was roughly 1.6 U glucose released per mg crude protein. The crude protein fraction had a mean Avicelase specific activity of 7.3 U glucose per mg (Table 1), and therefore Cel5A by itself can account for about a fifth of the total Avicelase activity we found in G. trabeum extracts. This calculation suggests that Cel5A had a significant role in enabling the fungus to grow on Avicel.
The missing activity is presumably attributable to enzymes that did not survive purification or to synergism between enzymes in the crude extract. It seems likely that synergism between some of the G. trabeum glycosylhydrolases does occur, because the crude extract exhibited a much higher specific activity on PASC than did the purified enzymes (Table 1). Since we found no evidence for synergism between Cel5A and Xyn10A, we suspect that interactions involving Cel12A or other enzymes in the extract were responsible. So far, we have not obtained enough of these enzymes to pinpoint the source of synergism.
A protein with the same N-terminal sequence as that of Cel5A was recently detected in wood colonized by G. trabeum (35), but its identity remains uncertain, because another G. trabeum endoglucanase with the same N-terminal sequence has also been reported (25). This 40.5-kDa endoglucanase is significantly different from Cel5A in three respects: (i) it hydrolyzes amorphous cellulose but not crystalline cellulose; (ii) it does not hydrolyze cellotriose, one of the products it yields from amorphous cellulose; and (iii) it has a much lower pI of 3.1. Further work is needed to determine the relationship between Cel5A and this other G. trabeum endoglucanase.
Our data indicate that the contribution Xyn10A made to crystalline cellulose hydrolysis was relatively small, because its specific activity on Avicel and relative abundance in cultures were considerably lower than what we found for Cel5A. The presence of minor impurities in the Xyn10A preparation also leaves open the possibility that some of its Avicelase activity was attributable to another enzyme. Nevertheless, it appears likely that G. trabeum Xyn10A can cleave crystalline cellulose, because it resembles Xyn10A (Cex) from Cellulomonas fimi (Fig. 2), which has already been shown to hydrolyze Avicel slowly (34).
Cel12A, which may be the same as a G. trabeum endoglucanase already reported (15), probably participates in the degradation of noncrystalline cellulose. It has long been known that brown rot fungi degrade the amorphous regions of wood cellulose rapidly, leaving behind crystalline cellulose that is degraded slowly (6, 21). ROS are probably responsible for the initial attack on these amorphous regions, because the porosity of wood is too low during early brown rot to admit enzymes (7). However, the relatively small size of Cel12A and some other brown rot endoglucanases (22) may allow them to penetrate and contribute to cellulose depolymerization before decay is extensive.
Larger processive cellulases, such as Cel5A, probably act later on the more recalcitrant crystalline cellulose, after the pore size of the wood has increased. It is likely that they act in concert with ROS, since these nonspecific oxidants are expected to reduce the crystallinity of the remaining cellulose (29). Synergism with ROS or other cellulases would be beneficial, because G. trabeum Cel5A is not particularly efficient at degrading crystalline cellulose by itself—its specific activity on Avicel is 20% of the activity reported for the processive endoglucanase CenC of C. fimi (34). Nevertheless, Cel5A is a complete cellulase, capable of degrading crystalline cellulose to glucose when β-glucosidase is also present. The old view that brown rot fungi lack cellulases with activity on crystalline cellulose must now be considered incorrect.
Acknowledgments
We are grateful to Diane Dietrich for the ergosterol analyses and to Shawn Mansfield for valuable advice on methods for cellulase purification.
This work was supported by grant DE-FG02-94ER20140 to K.E.H. from the U.S. Department of Energy.
REFERENCES
- 1.Atalla, R. H., and D. L. Vanderhart. 1984. Native cellulose: a composite of two distinct crystalline forms. Science 223:283-285. [DOI] [PubMed] [Google Scholar]
- 2.Beguin, P., and J. P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, R., K. A. Jensen, Jr., C. J. Houtman, and K. E. Hammel. 2002. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett. 531:483-488. [DOI] [PubMed] [Google Scholar]
- 5.Davis, M. W., and R. T. Lamar. 1992. Evaluation of methods to extract ergosterol for quantitation of soil fungal biomass. Soil Biol. Biochem. 24:189-198. [Google Scholar]
- 6.Eriksson, K.-E. L., R. A. Blanchette, and P. Ander. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany.
- 7.Flournoy, D. S., T. K. Kirk, and T. L. Highley. 1991. Wood decay by brown rot fungi: changes in pore structure and cell wall volume. Holzforschung 45:383-388. [Google Scholar]
- 8.Ghose, T. K. 1987. Measurement of cellulase activities. Pure Appl. Chem. 59:257-268. [Google Scholar]
- 9.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. Ce1I, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbertson, R. L., and L. Ryvarden. 1986. North American polypores. Fungiflora, Oslo, Norway.
- 11.Goedegebuur, F., T. Fowler, J. Phillips, P. van der Kley, P. van Solingen, L. Dankmeyer, and S. D. Power. 2002. Cloning and relational analysis of 15 novel fungal endoglucanases from family 12 glycosyl hydrolase. Curr. Genet. 41:89-98. [DOI] [PubMed] [Google Scholar]
- 12.Halliwell, B., and J. M. C. Gutteridge. 1999. Free radicals in biology and medicine, 3rd ed. Oxford University Press, Oxford, England.
- 13.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herr, D., F. Baumer, and H. Dellweg. 1978. Purification and properties of an extracellular β-glucosidase from Lenzites trabea. Eur. J. Appl. Microbiol. Biotechnol. 5:29-36. [Google Scholar]
- 15.Herr, D., F. Baumer, and H. Dellweg. 1978. Purification and properties of an extracellular endo-1,4-β-glucanase from Lenzites trabea. Arch. Microbiol. 117:287-292. [Google Scholar]
- 16.Irwin, D. C., M. Spezio, L. P. Walker, and D. B. Wilson. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, K. A., Jr., C. J. Houtman, Z. C. Ryan, and K. E. Hammel. 2001. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 67:2705-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, E. D., G. F. Lao, D. Irwin, B. K. Barr, A. Benjamin, and D. B. Wilson. 1993. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl. Environ. Microbiol. 59:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerem, Z., W. Bao, and K. E. Hammel. 1998. Rapid polyether cleavage via extracellular one-electron oxidation by a brown-rot basidiomycete. Proc. Natl. Acad. Sci. USA 95:10373-10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerem, Z., K. A. Jensen, and K. E. Hammel. 1999. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven Fenton reaction. FEBS Lett. 446:49-54. [DOI] [PubMed] [Google Scholar]
- 21.Kleman-Leyer, K., E. Agosin, A. H. Conner, and T. K. Kirk. 1992. Changes in molecular size distribution of cellulose during attack by white rot and brown rot fungi. Appl. Environ. Microbiol. 58:1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleman-Leyer, K. M., and T. K. Kirk. 1994. Three native cellulose-depolymerizing endoglucanases from solid-substrate cultures of the brown rot fungus Meruliporia (Serpula) incrassata. Appl. Environ. Microbiol. 60:2839-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, K. H., S. G. Wi, A. P. Singh, and Y. S. Kim. 2004. Micromorphological characteristics of decayed wood and laccase produced by the brown rot fungus Coniophora puteana. J. Wood Sci. 50:281-284. [Google Scholar]
- 24.Machuca, A., and A. Ferraz. 2001. Hydrolytic and oxidative enzymes produced by white- and brown-rot fungi during Eucalyptus grandis decay in solid medium. Enzyme Microb. Technol. 29:386-391. [Google Scholar]
- 25.Mansfield, S. D., J. N. Saddler, and G. M. Gubitz. 1998. Characterization of endoglucanases from the brown rot fungi Gloeophyllum sepiarium and Gloeophyllum trabeum. Enzyme Microb. Technol. 23:133-140. [Google Scholar]
- 26.Newman, R. H. 1994. Crystalline forms of cellulose in softwoods and hardwoods. J. Wood Chem. Technol. 14:451-466. [Google Scholar]
- 27.Paszczynski, A., R. Crawford, D. Funk, and B. Goodell. 1999. De novo synthesis of 4,5-dimethoxycatechol and 2,5-dimethoxyhydroquinone by the brown rot fungus Gloeophyllum trabeum. Appl. Environ. Microbiol. 65:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell, J. C., and M. Lever. 1972. New automated procedure for colorimetric determination of glucose. Biochem. Med. 6:543-547. [DOI] [PubMed] [Google Scholar]
- 29.Rättö, M., A.-C. Ritschkoff, and L. Viikari. 1997. The effect of oxidative pretreatment on cellulose degradation by Poria placenta and Trichoderma reesei cellulases. Appl. Microbiol. Biotechnol. 48:53-57. [Google Scholar]
- 30.Reverbel-Leroy, C., S. Pages, A. Belaich, J. P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritschkoff, A.-C., J. Buchert, and L. Viikari. 1994. Purification and characterization of a thermophilic xylanase from the brown rot fungus Gloeophyllum trabeum. J. Biotechnol. 32:67-74. [Google Scholar]
- 32.Sadana, J. C., and R. V. Patil. 1988. 1,4-β-d-Glucan cellobiohydrolase from Sclerotium rolfsii. Methods Enzymol. 160:307-314. [Google Scholar]
- 33.Schmidhalter, D. R., and G. Canevascini. 1993. Purification and characterization of two exocellobiohydrolases from the brown rot fungus Coniophora puteana (Schum ex Fr.) Karst. Arch. Biochem. Biophys. 300:551-558. [DOI] [PubMed] [Google Scholar]
- 34.Tomme, P., E. Kwan, N. R. Gilkes, D. G. Kilburn, and R. A. J. Warren. 1996. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. J. Bacteriol. 178:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varela, E., T. Mester, and M. Tien. 2003. Culture conditions affecting biodegradation components of the brown rot fungus Gloeophyllum trabeum. Arch. Microbiol. 180:251-256. [DOI] [PubMed] [Google Scholar]
- 36.Wood, T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19-25. [Google Scholar]
- 37.Zabel, R. A., and J. J. Morell. 1992. Wood microbiology: decay and its prevention. Academic Press, San Diego, Calif.