Abstract
Filamentous fungi have a high capacity for producing large amounts of secreted proteins, a property that has been exploited for commercial production of recombinant proteins. However, the secretory pathway, which is key to the production of extracellular proteins, is rather poorly characterized in filamentous fungi compared to yeast. We report the effects of recombinant protein secretion on gene expression levels in Aspergillus nidulans by directly comparing a bovine chymosin-producing strain with its parental wild-type strain in continuous culture by using expressed sequence tag microarrays. This approach demonstrated more subtle and specific changes in gene expression than those observed when mimicking the effects of protein overproduction by using a secretion blocker. The impact of overexpressing a secreted recombinant protein more closely resembles the unfolded-protein response in vivo.
Filamentous fungi have a high capacity for producing large amounts of extracellular proteins. A direct result of their saprophytic lifestyle is their ability to secrete a broad range of enzymes that enable them to metabolize a wide variety of extracellular polymeric substrates. Developments in molecular techniques have given rise to the potential use of filamentous fungi for production of a wide range of heterologous products (35). Production of specific nascent proteins in some industrial strains can exceed 30 g/liter (51). However, the secretion of recombinant protein to date has been far lower. Analysis of possible limiting factors point to bottlenecks at the posttranslational level (1, 17, 35), most likely along the secretory pathway. Most of our current knowledge of protein secretion comes from the study of temperature-sensitive mutants of the yeast Saccharomyces cerevisiae (5). Identification of a number of homologous proteins involved in secretion in other eukaryotes, including Aspergillus niger, indicates that the processes are highly conserved (1, 14). Despite the reasonable assumption that protein secretion in filamentous fungi will closely resemble that in yeast, the morphological and genetic differences between these organisms suggest that additional proteins or slightly different mechanisms may be involved (34).
Overexpression of recombinant proteins causes their intracellular accumulation (57), suggesting a breakdown or overloading of the secretory pathway. Some improvements in recombinant protein production have been seen from overexpressing chaperones and foldases in yeast (18, 19). However, the overexpression of equivalent proteins in aspergilli has had variable effects: some constructs showed increased intracellular recombinant protein production but no increase in extracellular levels, while others showed no improvement at all (27, 29, 36). The conflicting evidence surrounding overexpression of secretion-related genes emphasizes the complexities of protein-folding interactions, the specific effects of different products, and the possible need for a coordinated increase in expression to optimal levels (27). Accumulation of unfolded or inactive recombinant proteins puts pressure on the secretory pathway and activates the unfolded-protein response (UPR) (28, 39). The UPR triggers enhanced expression of genes involved with protein folding, secretion, and degradation (25, 48).
Previous attempts to characterize the genes associated with recombinant protein secretion in filamentous fungi have often relied on simulating the effects by inducing the UPR using secretion blockers such as dithiothreitol (DTT), brefeldin A, and tunicamycin. These methods have identified genes involved with protein folding, such as tigA (52) and cypB (7). Secretion blockers have also been used in combination with green fluorescent protein fusions to visualize unfolded protein and locate potential bottlenecks (15).
Bovine chymosin is an aspartyl protease present in the abomasum (fourth stomach) of unweaned calves and is used as a clotting agent in cheese manufacture (55). Early attempts to use the model organisms Escherichia coli (10, 31) and S. cerevisiae (13, 26) as hosts for chymosin production suffered from the problem of intracellular accumulation. Most work has since focused on various strains of Aspergillus (6, 9, 49, 55). Using translational fusions (57), strain improvement by mutagenesis and multicopy transformants (9, 56) has resulted in chymosin secretion up to commercial levels of over 1 g/liter.
Functional genomics reverses the normal course of genetics research by starting with the genes and attempting to define their function (32). Genomic approaches have been used previously to look at the effect of DTT on yeast growth (11) and its capacity to induce the UPR (48), providing valuable information on the yeast genes involved with secretion and the coordination between the UPR and endoplasmic reticulum (ER)-associated protein degradation. Most transcriptome analysis studies are performed in batch culture, where the growth rate and environment are continually changing. However, the experimental condition being studied may have an impact on the growth rate of cells. Changes in growth rate affect the pattern of gene expression; therefore, interpretation of experimental results is inherently problematic, and the use of chemostat cultures to reduce the number of conflicting variables that affect the analysis can greatly aid in data interpretation (60). Transcript profiling can be particularly beneficial to provide an overview of a pathway not previously characterized in a species with limited genome annotation and provide evidence towards functional assignments (42). Expressed sequence tag (EST) microarrays for Aspergillus nidulans have been constructed and thoroughly validated in our laboratory (43). The whole-genome assembly for A. nidulans was released in 2003 (http://www.broad.mit.edu), and the automated genome annotation predicted 9,541 putative open reading frames (ORFs). However, the majority of these ORFs are currently described as hypothetical genes of unknown function.
We describe the first attempt to analyze the influence of heterologous protein production and secretion by comparing the transcriptome of A. nidulans recombinant chymosin-producing and parental strains directly in continuous culture. In addition, we compare the effects on gene expression of the production of chymosin in chemostat culture with the effects of chymosin expression in batch culture in shake flasks. Finally, we evaluated how well the commonly used secretion blocker DTT was able to mimic the response to chymosin expression. By combining transcriptome data and comparative sequence analysis along with prior knowledge of the protein secretion pathway from yeast, we attempt to characterize the holistic response to recombinant protein secretion in aspergilli.
MATERIALS AND METHODS
Transcriptome analysis in shake flask (batch) and chemostat culture.
A pyrG mutant, single-copy recombinant chymosin-producing A. nidulans strain (6) was compared to its parental strain transformed with an empty vector. Shake flask cultures were grown in 500 ml of SCM medium (57) in 2l flasks, and all chemostat cultures were grown in carbon-limiting 5% SCM (2.5 g/liter maltose) medium. Fermentations were performed in a temperature-, pH-, and agitation-controlled Braun Biostat M fermenter at 30°C, pH 5.5, and 1,000 rpm with a working volume of 2.1 liters and on-line carbon dioxide monitoring as previously described (43). Dry weight samples from the vessel and overflow were taken to confirm steady-state growth (with a dilution rate of 0.1 h−1). Chymosin activity was measured using a simple microtiter plate method based on the method described by Emtage et al. (10).
Inducing the UPR with dithiothreitol.
The secretion blocker DTT was added to the parental strain to a final concentration of 20 mM in continuous culture at steady state, and samples were removed at various time intervals over 8 h. RNA extraction, labeling, and hybridization were performed using methods described previously (21, 43). Differential gene expression was calculated after global geometric normalization in MaxDView (http://www.bioinf.man.ac.uk/microarray/). All microarray hybridizations were performed with technical (dye flips) and biological (repeated cultures or fermentations) replicates. A protocol for determining significant changes in gene expression was followed as validated previously (43). The hybridizations for the shake flask experiment were performed on a first-generation (1G) microarray containing 4,092 ESTs from a conidial library and additional A. nidulans sequences deposited in GenBank (43). The chemostat and DTT-induced UPR hybridizations were performed on a larger microarray with an additional 1,722 ESTs derived from negative-subtraction hybridization (37). This second-generation (2G) microarray also included additional PCR products representing 20 known secretion-related genes. These were amplified using primers designed from the A. nidulans genome sequence and ClustalW (46) sequence alignments to the appropriate A. niger genes (42).
Annotation of the microarray.
BLAST was used to identify ESTs representing predicted cDNA (ORF) sequences from the A. nidulans genome sequence (http://www.broad.mit.edu). The SignalP program (30) was used to look for predicted prepro signal sequences. All raw data are available at the Consortium for the Functional Genomics of Microbial Eukaryotes website (http://www.cogeme.man.ac.uk). All EST and cDNA sequences are available from Pipeonline (http://bioinfo.okstate.edu/pipeonline/) and the Oklahoma EST and cDNA database (http://www.genome.ou.edu/fungal.html). Sequences were also compared for sequence similarity to GenBank and the comprehensive yeast genome database (http://mips.gsf.de/genre/proj/yeast/index.jsp).
RESULTS
Transcriptome analysis in shake flask batch culture.
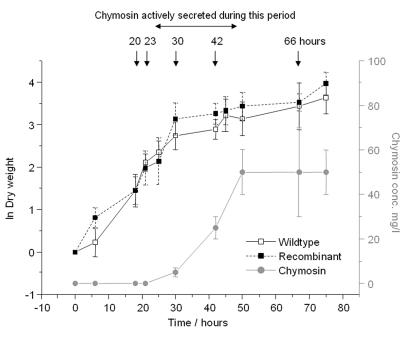
The effects of recombinant protein secretion were first analyzed by comparing the gene expression profiles of chymosin-producing and parental strains in shake flasks. Chymosin activity from the recombinant strain in the growth medium was first detected 20 h after inoculation and reached a level of approximately 50 mg liter−1 after 50 h (Fig. 1). RNA was prepared from samples taken from both strains between 20 and 66 h after inoculation. A total of 292 sequences on the 1G microarray were found to have increased expression after 30 h compared to samples taken at other time points. Of these, 179 sequences had a possible identity to genes in GenBank (using BLASTx), but only 24 ESTs or PCR products corresponded to 20 previously cloned Aspergillus genes (Fig. 2), demonstrating the limitations of our current view of the Aspergillus genome sequence. Expression profiles of non-Aspergillus species suggested by BLAST matches may be unreliable (43). Two of these genes, the ER chaperone bipA and protein disulfide isomerase pdiA, have been previously shown to be up-regulated during recombinant protein secretion (29, 36, 52). The unknown genes that were up-regulated included some with BLAST hits to secretion-related genes including chaperones, heat shock proteins, and GTP-binding proteins from other species.
FIG. 1.
Growth and chymosin production by recombinant and parental strains in batch shake flask culture. Samples were taken from both strains at times shown for comparative transcriptome analysis. ln (dry weight) and chymosin concentration are calculated from the average of three cultures, and error bars represent standard deviations.
FIG. 2.
Aspergillus genes in GenBank thought to be involved in recombinant protein secretion in batch shake flask culture. Red and green represent up- and down-regulation relative to white, which is equal to no change.
The genes that show increased expression in the chymosin-producing strain compared to its parent mostly fall into two distinct groups: those involved with the secretory pathway and those involved in metabolism. The high proportion of genes involved with metabolism is thought to be related to the fact that the increase in chymosin production coincides with the exponential growth phase. However, continual changes in cell density and medium composition during the exponential growth phase result in changes in gene expression, and it is therefore difficult to show which changes are due to recombinant protein expression and which are due to changes in other variables.
Transcriptome analysis in chemostat culture.
Both parental and chymosin-producing strains were grown in carbon-limited chemostat culture at a dilution rate (equal to the specific growth rate) of 0.1 h−1. The growth rate in a continuous culture is constant; therefore, samples taken at different intervals during steady state were physiologically equivalent. By comparing any samples from the two strains and calculating the mean changes, the number of possible variables can be minimized by having many biological replicates. Transcriptome analysis of both batch and continuous fermentations in A. nidulans has shown a large reduction in noise between samples taken during steady state compared with those take during replicate batch experiments (M. E. Gent et al., unpublished observation).
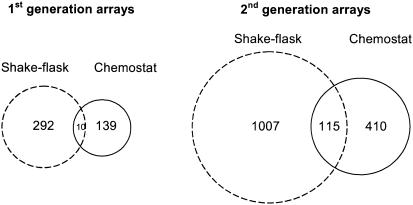
Only 139 sequences present on the 1G array showed significantly increased gene expression levels in the chymosin-producing strain relative to the parental strain grown in continuous culture. This is less than half the number of transcripts compared to those with higher levels in the batch culture analysis (292) after 30 h (Fig. 3). However, only 10 sequences showed significantly increased levels with both batch and continuous culture methods (Fig. 3); two of these ESTs and PCR products represent the ER chaperone, bipA. An additional 271 (total, 410) sequences in continuous culture and over 700 additional (total, 1,007) sequences in batch culture (after 30 h) had increased expression levels in the recombinant strain compared to those in the parental strain, using the 2G array. There was a much greater overlap (115 spots [>25% of the continuous culture sequences]) of transcripts with elevated levels in both culture methods using the 2G array (Fig. 3). Analysis of the putative identities (BLAST [GenBank]) of ESTs generated by negative-subtraction hybridization appears to suggest that they represent more genes involved with secretion and the unfolded-protein response than those from the conidial library. The negative-subtraction hybridization technique actively selects for low-abundance transcripts, so many genes involved with the UPR could be expected to exhibit low levels of expression during a glucose-grown conidiating culture.
FIG. 3.
Venn diagrams showing the number of sequences with increased expression when the parental and recombinant protein-producing strains in shake flask and chemostat cultures are compared. The circle area is proportional to the number of sequences. ESTs and PCR products from the first-generation arrays are also included on the second-generation arrays. Gene lists are available at http://www.cogeme.man.ac.uk.
Inducing the UPR with dithiothreitol.
The addition of DTT caused a significant and immediate reduction in the growth rate of the chemostat culture. However, a transcriptional response consistent with blocking secretion was not seen until 1 h after the addition of DTT, and this response was sustained at 2 and 4 h. This is slower than the response seen in similar transcriptome studies with S. cerevisiae. In one such study, the induction of target gene expression was essentially complete after 15 min (48), while in another study, it occurred within 30 min of DTT exposure (11). However, in both of these studies, DTT was added to the yeast cells during the mid-exponential phase, with a higher specific growth rate than that used in this study with A. nidulans; this would result in a quicker response. Transcription of genes encoding ribosomal proteins is directly correlated with growth rate (20, 24), and this was demonstrated 1 h after the addition of DTT to A. nidulans, when 34 out of 42 sequences (81%) thought to represent ribosomal genes were down-regulated.
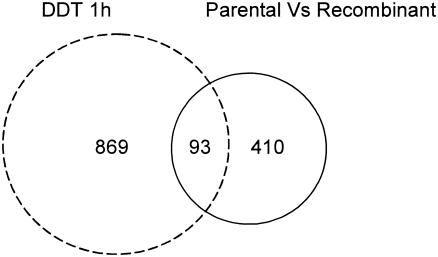
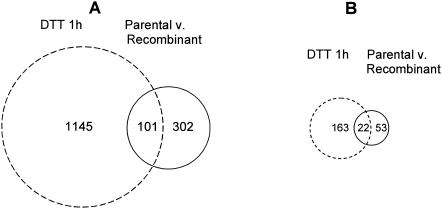
Our transcriptome data suggest that only ∼11% of the genes up-regulated in response to the addition of DTT also have increased expression levels in a chymosin-producing strain compared with its parental strain (Fig. 4). However, a greater proportion (23%) of genes with higher transcript levels in the recombinant strain also had increased expression levels in the DTT-treated culture.
FIG. 4.
Venn diagrams showing the number of up-regulated spots when different approaches for induction of UPR are compared using the G2 array. The circle area is proportional to the number of spots. Gene lists are available at http://www.cogeme.man.ac.uk.
Annotation of the microarray.
Determining the identities of the genes that the ESTs represent is an ongoing, continually evolving process in tandem with work on the genome sequence. A major breakthrough was the release of the automated gene calls, which enabled the ESTs to be linked directly to the ORFs they represent. The estimated number of EST sequences on the array that represent ORFs (Table 1) is rather low; theoretically, every EST should represent an ORF. In reality, however, some of the ESTs may represent genes whose ORFs have not been predicted due to incorrect gene calling, small ORF size (<100 amino acids), or gaps in the genome sequence. Alternatively, poor sequencing of the ESTs themselves may mean that they fail to correspond to any part of the genome sequence. Even if all the ESTs could be matched to putative ORFs, perhaps only half of the estimated total genes in the genome would be represented on the microarray. However, not all of the predicted ORFs may result in transcribed and translated proteins.
TABLE 1.
Comparison of the annotation of the microarrays
| Characteristic | No. (%) of sequences detected by microarray
|
|
|---|---|---|
| 1G | 2G | |
| Total no. of spots | 4,352 | 6,272 |
| No. ESTs | 3,752 | 5,579 |
| No. PCR products | 340 | 363 |
| Empty/DMSOa | 260 | 330 |
| Duplicated sequences | 520 (13) | 622 (10) |
| Unique sequences | 3,572 (87) | 5,320 (90) |
| Sequences representing ORFs | 3,304 (81) | 4,597 (77) |
| Unique A. nidulans ORFs represented (of predicted 9,541) | 2,080 (22) | 2,790 (29) |
DMSO, dimethyl sulfoxide.
Comparing induction of the UPR using different approaches.
Transcripts of the UPR transcription factor (HacA) were significantly higher in both the chymosin-producing and DTT-treated strains, (3.2-fold and 1.7-fold, respectively). This is in accordance with the observation that, during the UPR, HacA up-regulates its own transcription (28) and demonstrates that the addition of DTT and expression of bovine chymosin both activated the UPR. Sequences representing many known A. nidulans GTP-binding proteins, cyclophilins, and heat shock proteins (SrgA, SrgB, SrgE, VpsB, CypA, PinA, and HscA) had increased transcript levels with both approaches (Table 2 and Fig. 5). The protein disulfide isomerase genes PdiA (AN0248.2), PrpA (AN0075.2), and TigA (AN3592.2) all had significantly elevated transcript levels in response to chymosin expression, whereas only PdiA was up-regulated in response to DTT addition. Calnexin was not seen to be up-regulated with either approach despite having a putative UPR element in its promoter sequence and a previous report of elevated mRNA levels in a strain expressing bovine prochymosin fused to the catalytic domain of glucoamylase (53). Only a few putative orthologs of genes that encode proteins in the ER-associated degradation pathway (ERAD) were represented on the array. However, transcript levels of a putative ortholog of HRD3 were significantly increased in both the DTT-induced and chymosin-producing strains, confirming previous observations that the UPR and ERAD are intimately linked (2, 48).
TABLE 2.
Changes (n fold) in transcript levels of A. nidulans secretion-related genes and putative yeast orthologs due to recombinant protein secretion and/or induction of the UPR with DTTh
| Function | Yeast gene | A. nidulans ORF | ID or source | Change (fold)e
|
|
|---|---|---|---|---|---|
| Chemostat | DTT (1h) | ||||
| Protein folding/UPR | |||||
| ER chaperone bipA | KAR2 | AN2062.2 | 2000Sep131300_3520a | 1.31e | 3.90e |
| 2000Sep131300_240a | 1.21e | 2.64e | |||
| Protein disulfide isomerase pdiAh | PDI1 | AN7436.2 | PCR | 3.24e | 1.33e |
| 2000Sep131300_1629a | −1.01 | 1.30e | |||
| Protein disulfide isomerase prpA | −j | AN0248.2 | PCR | 2.57e | 1.06 |
| Protein disulfide isomerase tigA | −j | AN0075.2 | PCR | 1.49e | −1.24e |
| Calnexin—chaperone, folding clxA | CNE1 | AN3592.2 | 2000Sep131300_3254a | 1.00 | −1.02 |
| Chaperone of the ER lumen | CER1 | AN0847.2 | −j | −j | −j |
| Cyclophilin | CPR1 | AN8605.2 | 2000Sep131300_1548a | 1.02 | 1.79e |
| Cyclophilin cypA | CPR3 | AN3814.2 | pncs916aG10.SP6a | 1.21e | 1.27e |
| Cyclophilin (cis-trans) | CPR8 | AN4467.2 | cypB (AF107254)d | −j | −j |
| Peptidyl-prolyl cis-trans isomerase D | CPR5 | ||||
| Peptidyl-prolyl cis-trans isomerase | CPR2 | ||||
| Member of the cyclophilin family | CPR6 | AN4583.2 | pncs905aE03.SP6a | 1.37e | 4.71e |
| Member of the cyclophilin family | CPR7 | N12F7 SP6a | 1.23e | 1.58e | |
| Peptidyl-prolyl cis-trans isomerase precursor | CPR4 | Contig 111b | −j | −j | |
| PPlase, FK506-binding protein | FPR1 | AN3598.2 | 2000Sep131300_1483a | −1.02 | −1.38e |
| FK506/rapamycin-binding protein of the ER | FPR2 | AN8343.2 | −j | −j | −j |
| Proline cis-trans isomerase | FPR3 | AN3908.2 | k0f12a1.f1b | −j | −j |
| Nucleolar peptidyl-prolyl cis-trans isomerase | FPR4 | ||||
| Mitotic peptidyl-prolyl cis-trans isomerase pinA | PIN1 | AN6145.2 | PCR | 1.68e | 1.99e |
| Stress response transcription factor srrA | SKN7 | AN3688.2 | 2000Sep131300_325a | 1.46e | −j |
| Heat shock protein, 70 kDa | SSA1 | AN5129.2 | N8G4.SP6a | 1.36e | 1.52e |
| SSA2 | SETIIpL.3-B10a | 1.21e | 2.03e | ||
| SSA3 | 10_2_C5.SP6a | 1.27e | 1.30e | ||
| SSA4 | N5G5.SP6a | 1.17 | 1.23e | ||
| Heat shock protein, 70 kDa | SSB1 | AN4616.2 | Pncs916aH11a | −j | −j |
| Heat shock protein, 70 kDa, hscA | SSB2 | X98931c | PCR | 5.96e | 2.07e |
| Heat shock protein, 70 kDa | ECM10 | AN6010.2 | Pncs920aB09a | 1.37e | 1.14e |
| Heat shock protein, 60 kDa | HSP60 | AN6089.2 | N15GE11.SP6a | 1.34e | 2.09e |
| N13C11.SP6a | 1.37e | 2.00e | |||
| N5G7.SP6a | 1.21e | 1.86e | |||
| SETIPL2G2.SP6a | 1.69e | 1.72e | |||
| Heat shock protein, 30 kDa | −j | AN5781.2 | N14GA2.SP6a | 1.32e | 2.42e |
| 10_1_C8a | 1.30e | 1.21e | |||
| UPR transcription factor hacA | HAC1 | AN9397.2 | PCR | 3.24e | 1.73e |
| Serine/threonine protein kinase UPR | IRE1 | AN0235.2 | 2000Sep131300_4522a | −1.04 | −1.16 |
| Translocation | |||||
| Translocon (alpha subunit) | SEC61 | AN7721.2 | Contig 212b | −j | −j |
| Translocon (beta subunit) | SSS1 | AN4589.2 | −j | −j | −j |
| Translocon (gamma subunit) | SBH1 | AN0417.2 | −j | −j | −j |
| ER protein-translocation complex subunit | SEC62 | AN6269.2 | −j | −j | −j |
| ER protein-translocation complex subunit | SEC63 | AN0834.2 | 2000Sep131300_564a | 1.04 | 5.47e |
| ER protein-translocation complex subunit | SEC71 | AN1442.2 | c8a09a1.f1b | −j | −j |
| ER protein-translocation complex subunit | SEC72 | AN7623.2 | −j | −j | −j |
| Signal sequence processing protein | SEC11 | AN3126.2 | −j | −j | −j |
| 19-kDa signal recognition particle | SEC65 | AN0643.2 | −j | −j | −j |
| Signal sequence receptor alpha subunit | SRP101 | AN6627.2 | −j | −j | −j |
| Signal recognition particle protein srpA | SRP54 | AN8246.2 | Contig 1155b | −j | −j |
| Vacuolar protein sorting | |||||
| Sec1-like protein (Golgi to endosome) vpsA | VPS45 | AN6531.2 | f5f09a1.r1b | −j | −j |
| VPS plasma membrane to endosome sagA | END3 | AN1023.2 | PCR | −1.01 | 1.20e |
| VPS (Golgi to plasma membrane) | SEC1 | AN4724.2 | −j | −j | −j |
| VPS (endosome to vacuole) | VPS4 | AN3061.2 | −j | −j | −j |
| VPS (Golgi to vacuole) | VPS41 | AN4876.2 | 2000Sep131300_2938a | 1.06 | 1.32e |
| VPS (vacuolar sorting protein) | VPS10 | AN6118.2 | −j | −j | −j |
| Probable VPS,gS. pombe (T50328)i | −f | AN0854.2 | SETIPL3G1.SP6a | 1.23e | 1.27e |
| Sorting nexin, retrieval from endosomes | SNX41 | AN6351.2 | pncs902aB07.SP6a | 3.23e | 1.24e |
| Probable vacuolar sorting protein hbrA | VPS33 | AN2418.2 | PCR | 1.20e | −j |
| Protein degradation | |||||
| ER degradation | DER1 | AN8852.2 | y4h09a1.f1b | −j | −j |
| Ubiquitin-conjugating enzyme (UBC7 ortholog) | UBC7 | AN5351.2 | −j | −j | −j |
| Ubiquitin-conjugating enzyme (UBC7 paralog) | −j | AN8258.2 | Contig 1112b | −j | −j |
| Zinc finger for ER degradation E3 | HRD1 | AN1488.2 | Contig 794b | −j | −j |
| HMG-CoAg reductase degradation | HRD3 | AN0810.2 | pncs911aB01.SP6a | 1.49e | 1.53e |
| Ubiquitin-specific protease | DOA4 | AN2072.2 | −j | −j | −j |
| E2 ubiquitin-conjugating enzyme—peroxin | PEX4 | AN8258.2 | −j | −j | −j |
| Ubiquitin-activating enzyme | UBA1 | AN2174.2 | 2000Sep131300_1506a | 1.02 | 1.87e |
| 26S proteasome regulatory subunit | RPT3 | AN2904.2 | 2000Sep131300_1182a | 1.05 | 1.31e |
| E2 ubiquitin-conjugating enzyme | UBC12 | AN5344.2 | 2000Sep131300_790a | 1.07 | 1.01 |
| Cellular export and secretion | |||||
| Reticulocyte binding protein | SEC7 | AN6709.2 | −j | −j | −j |
| Secretory component protein, putative | SHR3 | AN1845.2 | 2000Sep131300_3678a | 1.00 | 1.27e |
| Secretory pathway GDP-dissociation inhibitor | GDI1 | AN5895.2 | −j | −j | −j |
| Putative myo-inositol-1-phosphate synthase | INO1 | AN7625.2 | 2000Sep131300_3013a | 1.22e | −1.29e |
| N1C4.SP6a | 1.22e | −1.29e | |||
| Cytosolic factor phosphatidylinositol transfer | SEC14 | AN0243.2 | −j | −j | −j |
| Putative secretory component nsfA | SEC18 | AN3098.2 | pncs902aE06.SP6a | 1.20e | 1.24e |
| Involved in nonclassical protein export pathway | NCE102 | AN7683.2 | 2000Sep131300_1208a | 8.90e | −j |
| Secretory pathway Ca2+-ATPase pmrA | PMR1 | AN7464.2 | −j | −j | −j |
| Glycosylation/modification | |||||
| Oligosaccharyltransferase beta subunit | WBP1 | AN4683.2 | −j | −j | −j |
| Oligosaccharyltransferase delta subunit | SWP1 | AN1683.2 | −j | −j | −j |
| Oligosaccharyltransferase epsilon subunit | OST2 | AN4031.2 | 2000Sep131300_883a | −1.02 | −1.25 |
| Required for asparagine-linked glycosylation | ALG11 | AN5725.2 | aD02.b1.SP6a | 1.06 | −1.26 |
| Mannosyltransferase | ALG2 | AN6874.2 | 2000Sep131300_4144a | 1.01 | −5.64e |
| Regulates mannosylphosphorylation | MNN4 | AN4030.2 | 2000Sep131300_2411a | 1.01 | 1.02 |
| Mannose-1-phosphate guanyltransferase | PSA1 | AN1911.2 | contig 627b | 1.22e | −j |
| Vesicle trafficking/transport | |||||
| GTPase (ER to Golgi) sarA | SAR1 | AN0411.2 | 2000Sep131300_2639a | 1.02 | 1.00 |
| GTPase (ER to golgi Rab2 homolog) srgD | rab2f | AN5106.2 | 2000Sep131300_2624a | 1.06 | 1.35e |
| GTPase (cis to medial Golgi) srgB | YPT1 | AN4281.2 | PCR | 1.22e | 1.43e |
| GTPase (Golgi to plasma membrane) srgA | SEC4 | AN6974.2 | PCR | 1.70e | 1.38e |
| GTPase (intra-Golgi) srgE | YPT31 | AN0347.2 | PCR | 1.49e | 2.27e |
| YPT32 | |||||
| GTPase (Golgi to vacuole) srgC | YPT6 | AN7602.2 | −j | −j | −j |
| GTPase (late endosome to vacuole) avaA/srgF | YPT7 | AN0089.2 | −j | −j | −j |
| GTPase (plasma membrane to endosome) srgG | YPT51 | AN4915.2 | 2000Sep131300_3122a | −1.05 | −1.22e |
| YPT53 | |||||
| GTPase (early to late endosome) srgH | YPT52 | AN3842.2 | N8G5.SP6a | 1.24e | −1.03 |
| GTPase (late endosome to plasma membrane) srgL | rab11f | AN0069.2 | −j | −j | −j |
| Protein transport protein | SEC13 | AN4317.2 | pncs912aF05.SP6a | 1.04 | 1.07 |
| Targeting and fusion of ER-Golgi vesicles | TRS120 | AN6533.2 | 2000Sep131300_717a | 1.24e | −1.29e |
| ER lumen protein-retaining receptor | ERD2 | AN9415.2 | pncs920aA07.SP6a | −1.02 | −1.29 |
| Coatomer complex beta chain of vesicles | SEC26 | AN1177.2 | 2000Sep131300_2823a | −1.03 | 1.05 |
| Coatomer complex beta chain of vesicles | SEC27 | AN5972.2 | pncs917aB12.SP6a | 12.82e | 1.46e |
| ADP-ribosylation factor-like protein, ras | ARL3 | AN1126.2 | 2000Sep131300_1470a | −1.06 | −1.26e |
| Putative syntaxin (T-SNARE) protein (ssoA?) | SSO1 | AN3416.2 | 2000Sep131300_1894a | −1.08 | −1.22e |
| SSO2 | |||||
| Lipid/inositol metabolism | |||||
| sn-1,2-Diacylglycerol ethanolamine | EPT1 | AN4778.2 | pncs906aE03.SP6a | 1.28e | −1.25e |
| Fatty acyltransferase | SLC1 | AN6139.2 | 2000Sep131300_2940a | −1.05 | 1.69e |
| 3-Hydroxy-3-methylglutaryl-coenzyme A reductase 2 | HMG2 | AN3817.2 | 2000Sep131300_2868a | 1.27e | −1.84e |
| Cell wall biogenesis | |||||
| Involved in cell wall biogenesis, architecture | ECM3 | AN0930.2 | N1H1.SP6a | 1.52e | 1.20e |
| P-type ATPase—unknown function | SPF1 | AN3146.2 | pncs919aG12.SP6a | 1.34e | 1.07 |
| Strong similarity to Chs6p FMP50 | FMP50 | AN3122.2 | pncs913aA03.SP6a | −1.01 | 3.39e |
Pipeonline (http://bioinfo.okstate.edu/pipeonline).
Oklahoma cDNA and EST sequencing database (http://www.genome.ou.edu/fungal.html).
Not predicted by automated annotation.
GenBank accession number.
Significant fold change (43).
No homolog in S. cerevisiae.
VPS, vacuolar protein sorting.
Boldface type indicates previously cloned Aspergillus genes.
Schizosaccharomyces pombe.
−, no data available.
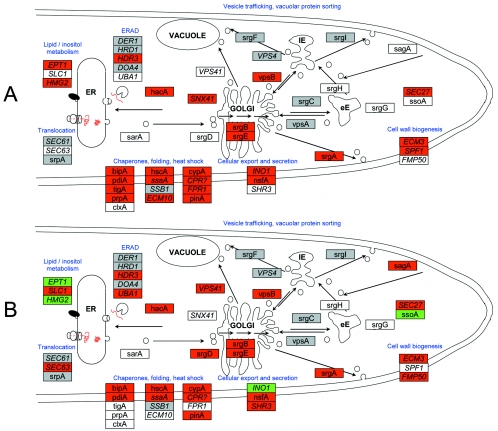
FIG. 5.
Model of the secretory pathway and UPR under different conditions. (A) Recombinant protein secretion induction of UPR. (B) Secretion blocker induction of UPR. Boxes represent known and putative secretion genes. Orthologs to yeast genes are in italics. Red, genes up-regulated; green, genes down-regulated; white, no significant change; grey, not present on the microarray. lE, late endosome, eE, early endosome.
Comparative genomic analysis of several Aspergillus species has demonstrated that the genes involved with targeting to the ER in aspergilli are highly conserved with those previously identified in yeast (40). Two routes for transporting of proteins across the ER membrane in S. cerevisiae have been described (44). Identification of A. niger homologs to S. cerevisiae Srp54p (47) and Kar2p/BipA (52) suggested that both signal recognition particle-dependent and -independent routes are also present in filamentous fungi. Conesa et al. (5) stated that “whether these two ER targeting mechanisms also operate in filamentous fungi remains to be determined.” However, identification of putative orthologs to all the major proteins involved with ER targeting following genome sequencing (Table 2) confirms that both mechanisms are conserved in aspergilli. Multiple sequence alignments of ORFs from several fungal species demonstrated that A. nidulans AN7623.1 represents two independent hypothetical proteins (40). The second ORF (beginning after 308 amino acids) represents a putative ortholog to the yeast translocation protein SEC67/72, while the first represents a hypothetical protein with a corresponding transcript sequence (contig 597) in the University of Oklahoma cosmid and cDNA sequencing database. Prediction of multiple genes as a single ORF in the A. nidulans genome has been reported (12). ORF AN6720.2 clearly represents three distinct genes, specifying a homolog of fructose-2,6-bisphosphatase, AvaB (EMBL accession number AB090888), and HbrB, a probable vacuolar sorting protein involved with hyperbranching mutants (12). It is thought that AN0643.2, a predicted 258-codon ORF, represents the ortholog of Sec65p. The automated annotation also predicted an overlapping 1,388-codon ORF (AN0642.2), with the first 236 residues identical to the secretion protein, which is most likely an error in the gene prediction algorithm. Unfortunately, only one of the putative genes involved with ER targeting, the ortholog to SEC63 (AN0834.2), appears to be present on the array. This gene was significantly up-regulated (5.47-fold) following the addition of DTT but was not differentially expressed in the comparison between the chymosin-producing and parental strains.
Additional known vesicle-trafficking genes (sagA and srgD) and sequences thought to represent orthologs of other secretion and ERAD-related genes (SHR3, SLC1, EPT1, FMP50, UBA1, and RPT3) had greater changes (severalfold) in the DTT-induced rather than recombinant protein secretion-induced UPR (Table 2 and Fig. 5). This suggests that the addition of DTT causes much greater and less-specific disruption of the secretory pathway than secretion of a recombinant protein. Secretion-related ORFs that showed particularly high (severalfold) changes in response to recombinant protein secretion were AN7683.2, a putative ortholog of NCE102, involved in a nonclassical protein export pathway, and AN5972.2, a putative ortholog of SEC27, the coatomer complex of β-chain vesicles. A putative A. nidulans ortholog to INO1 (AN7625.2), an inositol-1-phosphate synthase, was significantly down-regulated by the addition of DTT but had increased transcript levels in the recombinant protein-secreting strain, confirming previous observations that the UPR signal transduction pathway is not involved directly in transcriptional regulation of INO1 (3).
Repression of secretion under stress.
To fully understand the effects of recombinant protein production on cells, it is also highly valuable to look at the transcripts that are down-regulated, particularly those encoding secreted proteins, which have previously been shown to be significantly down-regulated following inhibition of secretion (25). This observation has given rise to a novel feedback mechanism, repression of secretion under stress (33). All sequences that had reduced transcript levels and were assigned to ORFs were searched for predicted signal sequences using SignalP (30). Of the arrayed sequences significantly down-regulated after the addition of DTT and during recombinant protein secretion, 22 out of 102 (22%) (Fig. 6) were predicted to encode secreted proteins. Most of these sequences represent unknown genes, although ORFs of known function included xylanase 2 and several putative secreted proteases. Predicted secreted proteins that had significantly reduced transcript levels in the recombinant strain in the chemostat comparison included xylanase 1, α-amylase, and cellobiose hydrolase, which were all previously reported to show reduced expression when protein secretion is inhibited in Trichoderma reesei (33).
FIG. 6.
Venn diagrams showing the number of transcripts with reduced levels when induction of the UPR is compared using the 2G array. (A) All spots. (B) Spots representing secreted proteins. The circle area is proportional to the number of spots. Gene lists are available at http://www.cogeme.man.ac.uk.
DISCUSSION
A large degree of variation was seen in the gene expression profiling using different approaches. However, by combining multiple approaches and excluding growth rate effects, it was possible to obtain a clearer view of the transcriptional effects of secreting recombinant proteins in vivo. Less than half the number of genes showed increased gene expression in a recombinant protein-producing strain in continuous, rather than batch, culture conditions. A chemostat approach allows experimental growth conditions to be carefully controlled, thereby greatly reducing the complexity of the system and allowing a much more focused analysis of the factors affecting gene expression (20). An additional advantage of chemostat cultures for hybridization array experiments is that the turnover rate ensures that the average transcript profile will more closely represent the current conditions.
While many of the severalfold changes appear relatively low, these values have a high degree of statistical assurance, as discussed previously (43). In addition to producing data congruent with the activation of the UPR, we have also highlighted the subtle differences between the two strains that are discernible under more highly controlled chemostat conditions compared to the physiologically more variable conditions in shake flasks or with nonselective secretion blockers such as DTT.
A large number of secretion-related genes previously characterized in S. cerevisiae appear to be highly conserved in A. nidulans. If predicted ORFs have significant sequence similarity and correlated expression profiles when subjected to the same conditions as those for previously characterized genes from other species, then it can be assumed that these genes represent conserved orthologs allowing putative functional assignments. Analysis of EST sequences can be highly valuable for identifying errors in the predicted coding sequences of the automated genome annotation (40). Transcriptome data can be used effectively to provide supporting evidence for functional assignments and can help to identify genes which are limited to the filamentous fungi (41, 42). For example, vesicle transport in aspergilli is thought to be more complex than that in yeast due to the greater abundance of secreted proteins, the filamentous morphology, and polarized apical tip growth. For example, a homolog to mammalian rab2 GTPase (SrgD) and to the mammalian annexin family (34) have been reported in aspergilli but are absent from the genome of S. cerevisiae. Many of the secretion-related genes identified for A. nidulans (Table 2) have also been characterized in T. reesei by linking the BLAST hits to ESTs with gene ontology terms (8), thus demonstrating a high level of conservation.
Prior to whole-genome sequencing, a number of Aspergillus orthologs to secretion-related GTPases had been identified. There are 10 or 11 Ypt/Rab genes in yeast (23) and ∼50 secretion-related GTPases in mammalian cells (45). These genes are responsible for different steps in protein trafficking and were isolated from secretion-defective mutants which accumulated proteins at different stages of the secretory pathway (5). It was very straightforward to assign A. nidulans ORFs as orthologs to the Ypt/Rab genes previously identified in A. niger. Some of these GTPases, which are thought to have the same or similar function in S. cerevisiae, appear to have a single homolog in aspergilli. The GTPase srgE is an ortholog of Ypt31p and Ypt32p in yeast; these genes share an identical effector region, and single-gene disruptions are phenotypically neutral, while disruption of both is fatal (16). Two A. nidulans ORFs were identified as putative orthologs of two of the remaining yeast GTPases; one ORF, provisionally named SrgG (AN4915.1), appears to represent YPT51 and YPT53, which are thought to be remnants of the whole-genome duplication event in yeast (59), and the other, SrgH (AN3842.1), is thought to represent YPT52. Fragments of cDNA that represent these ORFs were predicted to encode GTPases as described previously by Punt et al. (34) and have now been assembled to full-length cDNAs. (contigs 1227 and 1665, respectively, in the Oklahoma cDNA sequencing database). Both sequences are also represented by EST sequences on the 2G microarray but were not significantly differentially expressed either during chymosin expression or following DTT treatment. SrgI is a putative homologue of Rab11A, which is found only in mammals and targets to the membrane from the recycling complex (38). The only GTPase showing differential expression in response to UPR induction in S. cerevisiae was Ypt10p (48), but this does not appear to be conserved in filamentous fungi. Both YPT10 and YTP11 have no mutant phenotypes and appear to be nonessential in yeast (23). Many of the previously identified GTPases had increased transcript levels upon UPR induction in this study, suggesting a greater role for vesicle trafficking in the Aspergillus UPR.
Peptidylpropyl isomerases catalyze the isomerization of cis and trans peptide bonds on the N-terminal side of proline residues (5) and are thought to be involved in secretion and the unfolded-protein response. In yeast, eight cyclophilins and four FK506-binding proteins have been identified. However, in the Aspergilli, putative orthologs of only four cyclophilins and three FK506-binding proteins could be found in the whole-genome sequences (Table 2). It is not clear whether these discrepancies are due to duplication of the yeast genes or errors in gene calling for aspergilli. The cyclophillin CypB has been previously isolated from both A. niger and A. nidulans (7, 22), but it appears to be homologous to three distinct genes in yeast. The CypB protein has a 23-amino-acid ER retention sequence, and the 5′-untranslated region includes three sequences resembling UPR elements. Expression of cypB is up-regulated in the presence of tunicamycin and DTT (7) and during the production of tissue plasminogen activator (58). However, no representative sequences were present on the microarray, and when cypB was overexpressed in a tissue plasminogen activator-producing strain, the level of the product was not increased. The heat shock protein HscA, a 70-kDa homolog to yeast Ssb2p (Table 2), is conserved in A. nidulans (GenBank accession no. X98931). However, it appears that the gene overlaps the far end of supercontig 1.22 and so has not been assigned an ORF by the Broad annotation. Only one ORF was identified to represent the yeast heat shock proteins Ssa1p to Ssa4p. This ORF (AN5129.2), putatively named SsaA, was represented by four different ESTs that all had increased transcript levels under both DTT and recombinant protein UPR induction.
Chymosin contains three disulfide bonds, one of which is indispensable for correct protein folding (4), which would explain why mRNAs for the protein disulfide isomerases PdiA, TigA, and PrpA were all significantly higher in the recombinant protein-producing strain than in the parental strain (Table 2). Disruption of prpA has been shown to reduce bovine chymosin production in A. niger, but overexpression had little effect (54). Perhaps coregulation is important, since constitutive expression of the entire UPR in a recombinant Aspergillus niger var. awamori strain resulted in a 2.8-fold increase in chymosin yield (50).
Our global view of transcript levels implicate many secretion-related genes being involved with the UPR response as a result of recombinant protein secretion. Previous studies have shown that secretion of a variety of recombinant proteins can induce the UPR (28, 50) and may indicate that there are both general and recombinant product-specific responses, which cannot be mimicked by using a secretion blocker.
From our study and previous work involving yeast (11, 48), it seems clear that a large number of genes are involved in the UPR. Travers et al. (48) reported transcriptome analysis of the UPR induced by DTT and tunicamycin in S. cerevisiae. Their study identified 381 genes apparently involved in the UPR, with just over a quarter of these genes (103) known to be associated with the secretory pathway, UPR, or ERAD. They conceded that the high stringency of their data analysis criteria resulted in an underestimation of the full scope of the UPR (48). Indeed, transcript levels for KAR2, INO1, and HAC1 were not significantly elevated, despite the observations of other studies (11, 28). Our data lead us to question whether all of the genes identified in previous experiments are really associated with the UPR and suggest that many may be artifacts of the particular experimental approach employed.
From the data presented here, expression profiling of different strains, either those lacking secretion-related genes or those secreting alternative recombinant products, may represent the most reliable approaches for the future. Our data are consistent with the idea that the UPR has at least three effects: to improve protein folding and transport, to degrade unfolded proteins, and to allow fewer secretory proteins to enter the ER (25). We have demonstrated both similarities and differences in the genes induced by the UPR using different approaches and have highlighted the conservation of the secretory pathway in filamentous fungi. Our data stress the importance of minimizing the number of confounding variables in microarray experiments. While the addition of DTT does induce the UPR, it also retards growth of the mycelia, causing wide-ranging effects on the gene expression profile. A much smaller number of genes were significantly differentially expressed by inducing the UPR with the secretion of a recombinant protein than by the addition of a secretion blocker. A greater proportion of the genes identified represent known UPR or secretion-related genes, demonstrating the more subtle and specific effects of the UPR in vivo.
Acknowledgments
Genencor International Inc. is thanked for funding the fabrication of microarrays by M.E.G. and, in conjunction with the BBSRC, for a CASE studentship to A.H.S. K.L. was supported within the frame of the GAPSIA consortium, a part of the BBSRC “Exploiting Genomics” Initiative. This work was carried out at the Transcriptome Resource Facility of the COGEME consortium, which is part of the “Investigating Gene Function Initiative” of the BBSRC.
We thank Rolf Prade for the kind gift of the EST clones.
REFERENCES
- 1.Archer, D. B. 2000. Filamentous fungi as microbial cell factories for food use. Curr. Opin. Biotechnol. 11:478-483. [DOI] [PubMed] [Google Scholar]
- 2.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5:729-735. [DOI] [PubMed] [Google Scholar]
- 3.Chang, H. J., E. W. Jones, and S. A. Henry. 2002. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162:29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, H., G. Zhang, Y. Zhang, Y. Dong, and K. Yang. 2000. Functional implications of disulfide bond, Cys206-Cys210, in recombinant prochymosin (chymosin). Biochemistry 39:12140-12148. [DOI] [PubMed] [Google Scholar]
- 5.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, D., G. L. Gray, L. J. Wilson, K. J. Hayenga, M. H. Lamsa, M. W. Rey, S. Norton, and R. A. Berka. 1987. Controlled expression and secretion of bovine chymosin in Aspergillus nidulans. Bio/Technology 5:369-376. [Google Scholar]
- 7.Derkx, F., and S. M. Madrid. 2001. The foldase CYPB is a component of the secretory pathway of Aspergillus niger and contains the endoplasmic reticulum retention signal HEEL. Mol. Genet. Genomics 266:537-545. [DOI] [PubMed] [Google Scholar]
- 8.Diener, S. E., N. Dunn-Coleman, P. Foreman, T. D. Houfek, P. J. Teunissen, P. van Solingen, L. Dankmeyer, T. K. Mitchell, M. Ward, and R. A. Dean. 2004. Characterization of the protein processing and secretion pathways in a comprehensive set of expressed sequence tags from Trichoderma reesei. FEMS Microbiol. Lett. 230:275-282. [DOI] [PubMed] [Google Scholar]
- 9.Dunn-Coleman, N. S., P. Bloebaum, R. M. Berka, E. Bodie, N. Robinson, G. Armstrong, M. Ward, M. Przetak, G. L. Carter, and R. LaCost. 1991. Commercial levels of chymosin production by Aspergillus. Bio/Technology 9:976-981. [DOI] [PubMed] [Google Scholar]
- 10.Emtage, J. S., S. Angal, M. T. Doel, T. J. Harris, B. Jenkins, G. Lilley, and P. A. Lowe. 1983. Synthesis of calf prochymosin (prorennin) in Escherichia coli. Proc. Natl. Acad. Sci. USA 80:3671-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatherar, I. M., S. Pollerman, N. Dunn-Coleman, and G. Turner. 2004. Identification of a novel gene hbrB required for polarised growth in Aspergillus nidulans. Fungal Genet. Biol. 41:463-471. [DOI] [PubMed] [Google Scholar]
- 13.Goff, C. G., D. T. Moir, T. Kohno, T. C. Gravius, R. A. Smith, E. Yamasaki, and A. Taunton-Rigby. 1984. Expression of calf prochymosin in Saccharomyces cerevisiae. Gene 27:35-46. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, C. L., D. B. Archer, D. J. Jeenes, J. H. Doonan, B. Wells, A. J. P. Trinci, and G. D. Robson. 2000. A glucoamylase::GFP gene fusion to study protein secretion by individual hyphae of Aspergillus niger. J. Microbiol. Methods 42:39-48. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, C. L., V. Khalaj, A. F. Ram, D. B. Archer, J. L. Brookman, A. J. P. Trinci, D. J. Jeenes, J. H. Doonan, B. Wells, P. J. Punt, C. A. van den Hondel, and G. D. Robson. 2000. Glucoamylase::green fluorescent protein fusions to monitor protein secretion in Aspergillus niger. Microbiology 146:415-426. [DOI] [PubMed] [Google Scholar]
- 16.Goud, B., A. Salminen, N. C. Walworth, and P. J. Novick. 1988. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell 53:753-768. [DOI] [PubMed] [Google Scholar]
- 17.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Harmsen, M. M., M. I. Bruyne, H. A. Raue, and J. Maat. 1996. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl. Microbiol. Biotechnol. 46:365-370. [DOI] [PubMed] [Google Scholar]
- 19.Hayano, T., M. Hirose, and M. Kikuchi. 1995. Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett. 377:505-511. [DOI] [PubMed] [Google Scholar]
- 20.Hayes, A., N. Zhang, J. Wu, P. R. Butler, N. C. Hauser, J. D. Hoheisel, F. L. Lim, A. D. Sharrocks, and S. G. Oliver. 2002. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26:281-290. [DOI] [PubMed] [Google Scholar]
- 21.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 22.Joseph, J. D., J. Heitman, and A. R. Means. 1999. Molecular cloning and characterization of Aspergillus nidulans cyclophilin B. Fungal Genet. Biol. 27:55-66. [DOI] [PubMed] [Google Scholar]
- 23.Lazar, T., M. Gotte, and D. Gallwitz. 1997. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci. 22:468-472. [DOI] [PubMed] [Google Scholar]
- 24.Lim, F. L., A. Hayes, A. G. West, A. Pic-Taylor, Z. Darieva, B. A. Morgan, S. G. Oliver, and A. D. Sharrocks. 2003. Mcm1p-induced DNA bending regulates the formation of ternary transcription factor complexes. Mol. Cell. Biol. 23:450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, I. M., and M. J. Chrispeels. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15:561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellor, J., M. J. Dobson, N. A. Roberts, M. F. Tuite, J. S. Emtage, S. White, P. A. Lowe, T. Patel, A. J. Kingsman, and S. M. Kingsman. 1983. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene 24:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martin. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 28.Mulder, H. J., M. Saloheimo, M. Penttila, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genomics 271:130-140. [DOI] [PubMed] [Google Scholar]
- 29.Ngiam, C., D. J. Jeenes, and D. B. Archer. 1997. Isolation and characterisation of a gene encoding protein disulphide isomerase, pdiA, from Aspergillus niger. Curr. Genet. 31:133-138. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Nishimori, K., Y. Kawaguchi, M. Hidaka, T. Uozumi, and T. Beppu. 1982. Expression of cloned calf prochymosin gene sequence in Escherichia coli. Gene 19:337-344. [DOI] [PubMed] [Google Scholar]
- 32.Oliver, S. G. 1996. From DNA sequence to biological function. Nature 379:597-600. [DOI] [PubMed] [Google Scholar]
- 33.Pakula, T. M., M. Laxell, A. Huuskonen, J. Uusitalo, M. Saloheimo, and M. Penttila. 2003. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 278:45011-45020. [DOI] [PubMed] [Google Scholar]
- 34.Punt, P. J., B. Seiboth, X. O. Weenink, C. van Zeijl, M. Lenders, C. Konetschny, A. F. Ram, R. Montijn, C. P. Kubicek, and C. A. van den Hondel. 2001. Identification and characterization of a family of secretion-related small GTPase-encoding genes from the filamentous fungus Aspergillus niger: a putative SEC4 homologue is not essential for growth. Mol. Microbiol. 41:513-525. [DOI] [PubMed] [Google Scholar]
- 35.Punt, P. J., N. van Biezen, A. Conesa, A. Albers, J. Mangnus, and C. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 36.Punt, P. J., I. A. van Gemeren, J. Drint-Kuijvenhoven, J. G. Hessing, G. M. van Muijlwijk-Harteveld, A. Beijersbergen, C. T. Verrips, and C. A. van den Hondel. 1998. Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein in the secretion of homologous and heterologous proteins in black Aspergilli. Appl. Microbiol. Biotechnol. 50:447-454. [DOI] [PubMed] [Google Scholar]
- 37.Ray, A., S. Macwana, P. Ayoubi, L. T. Hall, R. Prade, and A. J. Mort. 2004. Negative subtraction hybridization: an efficient method to isolate large numbers of condition-specific cDNAs. BMC Genomics 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren, M., G. Xu, J. Zeng, C. Lemos-Chiarandini, M. Adesnik, and D. D. Sabatini. 1998. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saloheimo, M., M. Valkonen, and M. Penttila. 2003. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 40.Sims, A. H. 2004. Comparative and functional genomics of the Aspergilli and its relation to protein secretion. PhD thesis. The University of Manchester, Manchester, United Kingdom.
- 41.Sims, A. H., N. S. Dunn-Coleman, G. D. Robson, and S. G. Oliver. 2004. Glutamic protease distribution is limited to filamentous fungi. FEMS Microbiol. Lett. 239:95-101. [DOI] [PubMed] [Google Scholar]
- 42.Sims, A. H., M. E. Gent, G. D. Robson, N. Dunn-Coleman, and S. G. Oliver. 2004. Combining transcriptome data with genomic and cDNA sequence alignments to make confident functional assignments for Aspergillus nidulans genes. Mycol. Res. 108:853-857. [DOI] [PubMed] [Google Scholar]
- 43.Sims, A. H., G. D. Robson, D. C. Hoyle, S. G. Oliver, G. Turner, R. A. Prade, H. H. Russell, N. S. Dunn-Coleman, and M. E. Gent. 2004. Use of expressed sequence tag analysis and cDNA microarrays of the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 41:199-212. [DOI] [PubMed] [Google Scholar]
- 44.Stirling, C. J. 1999. Protein targeting to the endoplasmic reticulum in yeast. 1997 Fleming Lecture. Microbiology 145:991-998. [DOI] [PubMed] [Google Scholar]
- 45.Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153-208. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, S. A., E. J. Golightly, and D. S. Yaver. 1995. Nucleotide sequence of the Aspergillus niger srpA gene. Gene 167:337-338. [DOI] [PubMed] [Google Scholar]
- 48.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 49.Tsuchiya, K., K. Gomi, K. Kitamoto, C. Kumagai, and G. Tamura. 1993. Secretion of calf chymosin from the filamentous fungus Aspergillus oryzae. Appl. Microbiol. Biotechnol. 40:327-332. [DOI] [PubMed] [Google Scholar]
- 50.Valkonen, M., M. Ward, H. Wang, M. Penttilä, and M. Saloheimo. 2003. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 69:6979-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Hondel, C. A., P. J. Punt, and R. F. van Gorcom. 1992. Production of extracellular proteins by the filamentous fungus Aspergillus. Antonie Leeuwenhoek 61:153-160. [DOI] [PubMed] [Google Scholar]
- 52.van Gemeren, I. A., P. J. Punt, A. Drint-Kuyvenhoven, M. P. Broekhuijsen, A. van't Hoog, A. Beijersbergen, C. T. Verrips, and C. A. van den Hondel. 1997. The ER chaperone encoding bipA gene of black Aspergilli is induced by heat shock and unfolded proteins. Gene 198:43-52. [DOI] [PubMed] [Google Scholar]
- 53.Wang, H., J. Entwistle, E. Morlon, D. B. Archer, J. F. Peberdy, M. Ward, and D. J. Jeenes. 2003. Isolation and characterisation of a calnexin homologue, clxA, from Aspergillus niger. Mol. Genet. Genomics 268:684-691. [DOI] [PubMed] [Google Scholar]
- 54.Wang, H., and M. Ward. 2000. Molecular characterization of a PDI-related gene prpA in Aspergillus niger var. awamori. Curr. Genet. 37:57-64. [DOI] [PubMed] [Google Scholar]
- 55.Ward, M. 1990. Chymosin production in Aspergillus, p. 83-105. In S. A. Leong and R. A. Berka (ed.), Molecular industrial mycology: systems and applications. Marcel Dekker, New York, N.Y.
- 56.Ward, M., L. J. Wilson, and K. H. Kodama. 1993. Use of Aspergillus overproducing mutants, cured for integrated plasmid, to overproduce heterologous proteins. Appl. Microbiol. Biotechnol. 39:738-743. [DOI] [PubMed] [Google Scholar]
- 57.Ward, M., L. J. Wilson, K. H. Kodama, M. W. Rey, and R. M. Berka. 1990. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Bio/Technology. 8:435-440. [DOI] [PubMed] [Google Scholar]
- 58.Wiebe, M. G., A. Karandikar, G. D. Robson, A. J. P. Trinci, J. L. Candia, S. Trappe, G. Wallis, U. Rinas, P. M. Derkx, S. M. Madrid, H. Sisniega, I. Faus, R. Montijn, C. A. van den Hondel, and P. J. Punt. 2001. Production of tissue plasminogen activator (t-PA) in Aspergillus niger. Biotechnol. Bioeng. 76:164-174. [DOI] [PubMed] [Google Scholar]
- 59.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 60.Wu, J., N. Zhang, A. Hayes, K. Panoutsopoulou, and S. G. Oliver. 2004. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA 101:3148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]