Abstract
Quantum dots (QDs) rendered water soluble for biological applications are usually passivated by several inorganic and/or organic layers in order to increase fluorescence yield. However, these coatings greatly increase the size of the particle, making uptake by microorganisms impossible. We find that adenine- and AMP-conjugated QDs are able to label bacteria only if the particles are <5 nm in diameter. Labeling is dependent upon purine-processing mechanisms, as mutants lacking single enzymes demonstrate a qualitatively different signal than do wild-type strains. This is shown for two example species, one gram negative and one gram positive. Wild-type Bacillus subtilis incubated with QDs conjugated to adenine are strongly fluorescent; very weak signal is seen in mutant cells lacking either adenine deaminase or adenosine phosphoribosyltransferase. Conversely, QD-AMP conjugates label mutant strains more efficiently than the wild type. In Escherichia coli, QD conjugates are taken up most strongly by adenine auxotrophs and are extruded from the cells over a time course of hours. No fluorescent labeling is seen in killed bacteria or in the presence of EDTA or an excess of unlabeled adenine, AMP, or hypoxanthine. Spectroscopy and electron microscopy suggest that QDs of <5 nm can enter the cells whole, probably by means of oxidative damage to the cell membrane which is aided by light.
Much progress has recently been made in using semiconductor nanocrystals as fluorescent labels for biological samples. These “quantum dots” (QDs) have advantages over traditional fluorescent probes due to their broad absorption spectra, narrow emission spectra, and resistance to photobleaching. Labeling with QDs has been demonstrated for multiplexed optical coding (10), with mammalian cells (33), with slime molds (16), and with bacteria (17) and protozoa (34).
While mammalian cells take up QDs by endocytosis, bacteria do not endocytose, and internalization of QDs by prokaryotes has so far not been observed. Such internal labeling would be desirable in many applications. Green fluorescent protein does not work in many strains, especially in anaerobes, and hence, general fluorescence labeling techniques are needed (11). Because QDs are both fluorescent and electron dense, fluorescence and electron micrographs can be directly compared for high-resolution imaging of internal mechanisms in single bacteria.
Three possible mechanisms through which nanocrystals could pass through bacterial cell walls and membranes are nonspecific diffusion, nonspecific membrane damage, and specific uptake. Nonspecific diffusion is unlikely with QDs; the largest globular proteins that can pass through intact cell walls of Bacillus subtilis have radii of ∼2 nm (6), while the smallest high-quality QDs measure ∼3 to 4 nm (4) (representing green emission for CdSe and blue emission for CdS [30]).
Entry of other types of nanocrystals into bacteria via nonspecific membrane damage has been established, particularly with highly reactive halogenated nanoparticles (31). QDs are able to transfer electrons to redox-active molecules (including oxygen) upon light exposure (24); the smaller the QD, the greater its redox potential (12, 15). The dependence of particle entry upon light exposure would strongly suggest this mechanism.
Specific transport is the last possibility. The pore sizes for specific transport can be somewhat larger than those for nonspecific diffusion; the largest pores known, permeases for excreting proteins from bacteria, have been shown to have pore openings of up to 6 nm (32). Most pore sizes are unknown, however, as the genes for the permeases have not been cloned.
We predicted that adenine compounds were likely candidates for specific processing by bacteria, because tags on the primary amino group of adenine usually do not inhibit recognition and uptake (as seen with purine analogues such as 6-benzylaminopurine) (5, 27). A theoretical study using the LiSSA (ligand search superposition and analysis) program found that adenine-containing compounds may bind to proteins in many different orientations and that substrate recognition is not dependent upon any particular free hydrogen residues (21). In a particular example, the enzyme adenine phosphoribosyltransferase was shown to be able to recognize adenine from either end of the purine ring (7).
The three questions to be answered were as follows: (i) are bacteria able to import adenine-conjugated QDs despite the large size, (ii) does this occur in a metabolism-specific manner, and (iii) is it dependent upon light exposure?
In this paper, we show that adenine-conjugated QDs are able to label wild-type Bacillus subtilis only when the size of the QDs is <5 nm. B. subtilis, a gram-positive bacterium, was chosen because its purine metabolism is well understood and strains with single mutations in adenine-processing enzymes are available. Elimination of the function of either adenine phosphoribosyltransferase (the apt mutant) or adenine deaminase (the ade mutant) (22) is sufficient to suppress labeling with QD-adenine. However, the mutants but not the wild type are labeled with QD-AMP.
Because gram-negative and gram-positive bacteria possess different cell walls and uptake mechanisms, the experiments were duplicated with a gram-negative bacterium, Escherichia coli. Metabolism-dependent labeling is again seen, but with qualitative differences in fluorescence time course and toxicity.
Evidence for specific uptake is thus seen, but dependence upon light is marked, especially with bare-core CdSe nanocrystals. The implications for the use of QDs as fluorescent labels and for toxicity are discussed in this context.
MATERIALS AND METHODS
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich.
QD synthesis and conjugation.
Six independent preparations of QDs were used, representing three colors each of bare CdSe QDs and of CdSe/ZnS core shell QDs. Size distributions were determined from fluorescence emission spectra and high-resolution transmission electron microscopy (TEM) (see below for details). Bare CdSe QDs were synthesized, characterized, solubilized with mercaptoacetic acid (MAA), and conjugated to primary-amine-containing molecules using the activator 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) as described previously (17). Concentrations of conjugate and QDs were adjusted to prevent aggregation: final concentrations used were 10 mM conjugate and ∼0.2 μM QDs. The pH of all solutions was adjusted to neutral (pH 6.5 to 7.5). Core shell QDs were produced after synthesis of CdSe QDs by capping with ZnS as described previously (4). Excessive aggregation and precipitation were seen when ZnS-capped QDs were solubilized with MAA, and thus, all experiments with core shell particles were performed after solubilization with dihydrolipoic acid (DHLA) as described previously (19). EDC coupling to DHLA conjugates was performed with lower concentrations of adenine or AMP (2 to 5 mM) and lower concentrations of QDs (0.05 to 0.1 μM). After conjugation, QDs were purified from residual EDC and unbound conjugate by dialysis against sterile distilled water (dH2O). Using the concentrations specified, little aggregation was seen after dialysis; the purified conjugates were spun for 2 min at 14,000 rpm in a microcentrifuge to remove any sedimented material.
Conjugates were prepared to the following molecules: adenine; AMP; wheat germ agglutinin (WGA), a lectin, as described previously (17); and adenine and AMP in a 1:1 ratio with polyethylene glycol with a molecular weight of 5,000 (PEG-5000). Amide bond formation was confirmed using Fourier transform infrared spectroscopy. For purine compounds, conjugation efficiency was estimated by measuring absorbance at 260 nm. Controls in which adenine and AMP were added to QDs without EDC were also prepared.
The quantum yield of the solubilized particles was measured and compared to the fluorescein dianion (∼92% in 0.1N NaOH) with 450-nm excitation.
Incubation with bacteria.
Fifty to 200 μl of conjugate was added to 250 μl bacteria (logarithmic growth phase) and ∼0.8 ml of nutrient medium (final QD concentrations, ∼10 to 40 nM for bare CdSe and 2 to 10 nM for core shell CdSe/ZnS). Control cultures received an identical volume of H2O or a solution of unlabeled adenine or AMP for a final concentration of 2 mM. E. coli strains were purchased from the American Type Culture Collection (ATCC): wild-type ATCC 25922 and the adenine auxotroph ATCC 23804. B. subtilis strains were a gift from Per Nygaard, Institute of Molecular Biology, University of Copenhagen. Bacteria were grown as clonal cultures. Growth medium was Luria-Bertani (LB) medium unless stated otherwise; minimal medium consisted of 0.2% (wt/vol) ammonium sulfate, 1.4% dipotassium phosphate, 0.6% monopotassium phosphate, 0.1% sodium citrate, 0.02% magnesium sulfate, and 0.5% glucose (29). Unless otherwise stated, bacteria were grown in clear tubes in a clear plastic incubator under ambient room light. For cultures specified to be grown under “dark” conditions, the tube was wrapped in aluminum foil.
Two types of killed control cultures were included in all experiments: heat-killed bacteria (produced by placing tubes containing mid-log-phase cultures into boiling water for 10 min; no spore formation was seen with B. subtilis) and bacteria subjected to three rounds of freeze-thaw (−80°C to +37°C). Both controls were incubated with QDs in the same manner as the live cells. Other controls incubated with QDs were metabolically inhibited cultures (containing 1 mM EDTA) and competitive controls containing 2 mM unlabeled adenine or hypoxanthine. These latter controls were repeated three to five times but not included with all experiments.
Quantum dot controls were included with all experiments and consisted of the same concentration of QDs as in the bacterial cultures diluted into dH2O. These controls were placed into the incubator with the bacterial cultures and subjected to the same conditions of light, temperature, and shaking.
After incubation, bacterial cultures were pelleted at 14,000 rpm in a microcentrifuge and washed once in 1% saline to eliminate medium fluorescence and unbound QDs. The resulting suspension was subjected to spectrofluorimetry and fluorescence microscopy.
Spectroscopy.
Emission spectra of bacterial cultures were taken to correlate with the fluorescence images and to locate potential fluorescence emission in the UV or near-infrared spectroscopy. Emission spectra were measured with a Gemini EM plate reader in epifluorescence mode to minimize effects of solution turbidity. Absorbance spectra were recorded with a Hewlett Packard 8453 UV-vis spectrophotometer. Spectra were normalized to bacterial density by matching absorbance at 600 nm and emission at a wavelength significantly distant from the fluorescence of QDs being used (700 nm for green and yellow QDs and 460 nm for red QDs).
Light microscopy.
Light microscopy allowed for the identification of the success or failure of fluorescent labeling and a crude estimate of the cellular localization of the fluorescence. QDs and bacteria were examined using epifluorescence microscopy on a Nikon Eclipse E600 upright microscope with a Nikon PlanFluor 1005 lens (numerical aperture, 1.30) and a modified UV-DAPI (4′-6′-diamidino-2-phenylindole) filter cube set (excitation, 330 to 380 nm; dichroic mirror reflecting, 400 nm; longpass emission filter, beginning at 435 nm). Bacteria were flame fixed and examined unstained or examined live under a coverslip to eliminate possible effects of flame fixing.
Incubation times and growth curves.
Growth curves were obtained to measure QD toxicity in liquid medium and to determine the optimal times for incubation. Curves were obtained by incubating bacteria under culture conditions identical to those used in QD labeling experiments and measuring the optical density at 600 nm at each time point from 15 min to 5 h. In the case of B. subtilis, microscopic examination was used to determine whether sporulation had occurred. After the curves had been obtained, optimal times were chosen to be 1 h for E. coli and 3 h for B. subtilis, and all non-time-dependent data presented use these incubation times.
Long-term storage.
For long-term storage, washed cultures were repelleted and resuspended in fresh LB medium and then stored at 4°C in the dark.
Protoplasts.
Protoplasts were prepared to remove cell walls and any associated QDs from the cultures in order to test for effects on fluorescence and to recover internalized material from the bacterial cytoplasmic fraction. Protoplasts were prepared by adding lysozyme (0.5 to 1 mg/ml) to cell suspensions in protoplast buffer (30 mM Tris, pH 7.5, 5 mM EDTA, 50 mM NaCl, 30% sucrose [13]). After digestion for 30 min at 4°C, protoplasts were sedimented by centrifugation (3,000 × g), and the supernatant was discarded. Protoplasts were examined by epifluorescence microscopy with or without the presence of a 1 μM concentration of the amino naphthylethenyl pyridinium dye di-4-ANEPPS (Molecular Probes) for membrane visualization. TEM was used to confirm the absence of external particles and the presence of internal Cd and Se before rupture.
Recovery of the cytoplasmic fraction was accomplished by rupture of protoplasts and fractionation as described previously (13). Briefly, protoplasts were pelleted by centrifugation and resuspended in protoplast buffer without sucrose. The resulting solution was checked by microscopy to ensure that most of the protoplasts had lysed and was then centrifuged at 14,000 × g for 15 min. The supernatant was retained for high-resolution microscopy and fluorescence analysis, and the pellet (membrane fraction) was discarded.
Transmission electron microscopy.
To confirm nanocrystal uptake, bacteria were examined by TEM and energy-dispersive X-ray spectroscopy (EDS) using whole mounts and thin sections. An Akashi EM-002B microscope operating at 100 kV was used for low-resolution TEM and EDS; the area sampled by this instrument was approximately 8.8 nm. Acquisition rates were maintained at 10 to 20% dead time with 60 s of live time at 83 kV. The electron beam was defocused at the condenser lens to maintain counting rates below 1 kHz and live time efficiency of >95%. The selected area diffraction patterns were recorded with an INCA 3.04 Microanalysis Suite and digitized for analysis.
Absolute weight percent (wt%) and atomic percent (atomic%) of 10 different elements were measured in all EDS experiments: Mg, Al, Si, P, S, Cl, K, Ca, Fe, Se, and Cd. A weight percent of >2 was taken as significant. All measurements of P were taken outside cells because of high background levels of P within the cells.
Thin sections.
Thin sections for high- and low-resolution TEM were prepared by fixing freshly incubated bacteria in culture medium with 2.5% (vol/vol) glutaraldehyde. After 12 to 16 h of fixation, cells were washed three to five times in dH2O with centrifugation, and the final pellet was dispersed in Noble agar worms of desired length. Specimens were then dehydrated in ethanol and acetone before embedding in Epon resin. Sample resin blocks were trimmed and sectioned (50 to 60 nm) on an MT-X Ultramicrotome with a 45° Diatome diamond knife. Ultrathin sections were placed on 200-mesh formvar/carbon-coated copper grids. Control thin sections containing QD conjugates only or QD conjugates after exposure to culture medium only (no bacteria) were also prepared. These were exposed to glutaraldehyde and embedded in the same manner as the bacterial cultures.
High-resolution TEM.
High-resolution images were used to quantify nanocrystal size before and after ligand conjugation and after exposure to bacterial cells. Solubilized QDs, conjugated QDs, ruptured protoplasts, or thin sections were placed onto 200-mesh holey carbon copper grids and imaged using a Philips EM430 analytical transmission electron microscope operating at 300 kV. Particle sizes were measured and analyzed using the public domain software NIH Image (http://rsb.info.nih.gov/nih-image/).
RESULTS
Effects of adenine and AMP on QD emission spectra and photosensitivity.
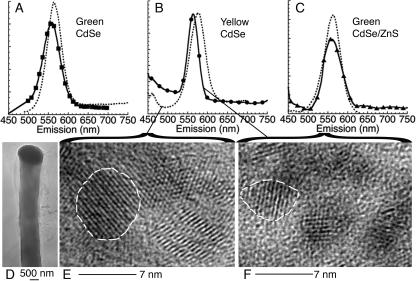
CdSe and CdSe/ZnS QDs were synthesized and solubilized, and the size distributions were measured by TEM before and after conjugation to adenine and AMP. Tested were three colors of bare CdSe QDs (green [emission peak, 575 nm; mean diameter ± standard deviation, 3.9 ± 0.4 nm], yellow [emission peak, 587 nm; mean diameter, 4.6 ± 0.4 nm], and red [emission peak, 615 nm; mean diameter, 5.4 ± 0.4 nm]) and three colors of core shell CdSe/ZnS QDs (green [emission peak, 565 nm; mean diameter, 3.6 ± 0.3 nm], yellow [emission peak, 590 nm; mean diameter, 5.5 ± 0.5 nm], and red [emission peak, 620 nm; mean diameter, 6.5 ± 0.5 nm]). The quantum yields of these particles were much lower than those of commercial polymer-capped QDs: ∼3% for CdSe QDs and ∼10% for CdSe/ZnS QDs before solubilization. After solubilization by MAA, quantum yields of bare QDs were ∼0.01%. CdSe/ZnS QDs solubilized by DHLA retained quantum yields of >1% (J. A. Kloepfer et al., unpublished data). Aggregation of adenine and AMP conjugates was minimal with bare CdSe and minor with CdSe/ZnS solubilized with DHLA.
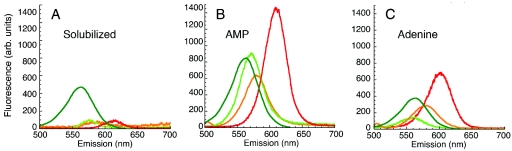
Adenine and AMP increased the photosensitivity of bare CdSe QDs over that of MAA-solubilized QDs in a manner that was dependent upon the size (color) of the nanocrystal. After 3 h of exposure to ambient room light at 37°C, MAA-solubilized QDs retained their initial weak fluorescence intensity (Fig. 1A). All colors of QD-AMP demonstrated fluorescence enhancement without aggregation. Green QD-AMP was enhanced 8-fold ± 2-fold, yellow QD AMP was enhanced 8-fold ± 0.5-fold, and red QD-AMP was enhanced 18-fold ± 4-fold (n = 4 independent preparations for each color; a representative preparation is shown in Fig. 1B). All colors showed a blue shift of 5.0 ± 0.2 nm with no change in peak width (n = 4 preparations).
FIG. 1.
Photoluminescence and photosensitivity of QD-only controls before and after conjugation to purine compounds. The concentrations of QDs in these samples were 40 nM for CdSe QDs and 2 nM for CdSe/ZnS QDs. All preparations were incubated at 37°C for 3 h under ambient room light. (A) Four distinct colors of QDs after solubilization with MAA, green-emitting CdSe/ZnS (dark green line; peak, 565 nm), green-emitting CdSe (light green line; peak, 575 nm), yellow-emitting CdSe (orange line; peak, 587 nm), and red-emitting CdSe (red line; peak, 615 nm). These spectra are equivalent to those seen before incubation (not shown; n > 4 independent preparations). (B) The same preparations and quantities of QDs after conjugation to AMP. (C) The same preparations and quantities of QDs after conjugation to adenine.
Red and yellow QD-adenine conjugates showed similar but smaller enhancement: 3.0-fold ± 0.8-fold for yellow, and 7-fold ± 1-fold for red. Green QD-adenine was not significantly enhanced and was occasionally quenched (1.0-fold ± 0.4-fold change) (n = 4 independent preparations for each color; a representative preparation is shown in Fig. 1C). All colors showed a blue shift of 11 ± 2 nm without change in peak width (n = 4).
The same effect was not seen with CdSe/ZnS core shell QDs. No peak shifts or changes in width were seen with any colors (n = 4 independent preparations), and enhancement was <2-fold for all conditions. Shown in Fig. 1A to C are green CdSe/ZnS QDs, which showed a 1.8-fold ± 0.6-fold enhancement with AMP and a slight reduction in luminescence with adenine, to 0.6-fold ± 0.4-fold intensity.
When incubation of QD-adenine and QD-AMP conjugates was performed in the dark, emission spectra remained indistinguishable from those of solubilized QDs in Fig. 1A (data not shown). The emission of such particles was too weak to permit reliable quantitative measurements of any peak shifts or intensity changes.
Size dependence of QD labeling and effects of light.
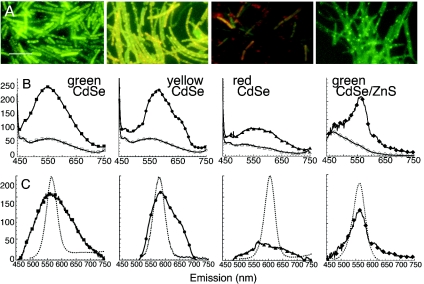
Unconjugated QDs, or QDs incubated with adenine but not EDC, showed no association with B. subtilis cells and no fluorescence of washed samples (data not shown; n > 10 samples; see also reference 17). With QD-adenine conjugates, the three smallest preparations led to strong fluorescent labeling: green CdSe, yellow CdSe, and green CdSe/ZnS (relative emission peak intensities normalized to bacterial density of 1.0 ± 0.1, 1.1 ± 1.1, and 0.72 ± 0.5, respectively; n = 3 preparations each). Epifluorescence micrographs and emission spectra of all of these preparations are shown in Fig. 2. The three largest preparations, yellow CdSe/ZnS, red CdSe, and red CdSe/ZnS, led to weak, blue-shifted fluorescence (emission peak intensities relative to green CdSe of <0.3; n > 3 preparations each). The case of red CdSe, which was representative, appears in Fig. 2.
FIG. 2.
Fluorescent labeling with four distinct colors of QD-adenine conjugates. All spectra are averages of at least three independent experiments, with error bars smaller than the symbols. All epifluorescence images were taken with the same gain and exposure time. Scale bar, 10 μm. (A) Epifluorescence images of green CdSe, yellow CdSe, red CdSe, and green CdSe/ZnS QD-adenine conjugates after 3 h of incubation with wild-type B. subtilis in LB culture medium. Labeling with green and yellow CdSe QDs is very strong and is broadened from the original QD color; labeling is strongest at the cell poles but is not restricted there. Labeling with red CdSe QDs is nearly absent. Labeling with CdSe/ZnS QDs shows no fluorescence broadening and a greater restriction to the cell poles. (B) Raw (not normalized or background-subtracted) emission spectra of the cultures in A (solid symbols) shown with autofluorescence spectra (open symbols). Autofluorescence was not significantly different between killed bacteria exposed to QDs and live bacteria without QDs; the size of the symbols represents the standard error of the observed variation. (C) Spectra after background subtraction. The spectra of the QD-only controls is included for comparison (▪▪▪▪▪). Green CdSe-adenine-B. subtilis (▪) yields a spectrum centered around the peak of the original QD-adenine (dashed line) but with a full width at half maximum (FWHM) broadened from 44 to 100 nm (Gaussian fit, R > 0.999). The spectrum of yellow CdSe-adenine-B. subtilis (•) also retains the original peak (dashed line) and is broadened slightly less than that of green (FWHM,80 nm; Gaussian fit, R > 0.99) but with a significant red tail. The spectrum of red CdSe-adenine-B. subtilis (▴) shows no real peak. The spectrum of green CdSe/ZnS (♦) is not significantly broadened and overlaps the QD-control spectrum almost perfectly.
The autofluorescence from control cultures was minimal, and all controls were similar. Live cells without QDs showed the same spectra both in ordinary medium and in medium supplemented with 2 mM adenine. The spectra from killed controls with QDs overlapped those of the controls that contained live cells but no QDs (Fig. 2B). Subtraction of the autofluorescence spectrum led to an emission peak that was broadened in the case of the bare CdSe QDs; the core shell QDs retained their initial spectrum (Fig. 2C).
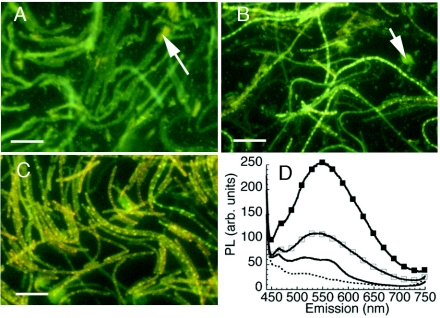
Corresponding to what was seen with QDs alone, the fluorescence spectrum and intensity of bacterial labeling with bare CdSe conjugates changed qualitatively with light exposure. When B. subtilis was exposed to QD-adenine in the dark, very little cell-associated fluorescence was seen. Aggregates of QDs were seen in the background (Fig. 3A). When the QD-adenine conjugates were preexposed to light and then incubated with B. subtilis in the dark, some cell-associated fluorescence was seen, but it was limited and blue shifted (Fig. 3B). Only when the conjugates were exposed to bacteria under room light was the typical pattern of QD fluorescence obtained (Fig. 3C and D).
FIG. 3.
Effects of light exposure before and after addition of bare CdSe QD-adenine conjugates to B. subtilis. Scale bar, 10 μm. (A) Live bacteria exposed to yellow QD-adenine for 3 h in the dark. Very little labeling is apparent; aggregates of QDs showing the initial QD color are found throughout the culture (arrow). (B) Live bacteria exposed to yellow QD-adenine for 3 h in the dark; the QD conjugate was preexposed to room light for 3 h at 37°C. The aggregates in the background are now blue shifted (arrow), but there is still limited cellular labeling. (C) Live bacteria exposed to yellow QD-adenine for 3 h under room light. Note the absence of free QDs in the solution. (D) Emission spectra of A to C. ▪, incubated under room light; □, preexposed and incubated in the dark; ▪▪▪▪▪, not preexposed, incubated in the dark; dashed line, control (heat-killed B. subtilis exposed to QD under room light). PL, photoluminescence; arb., arbitrary.
The lack of fluorescent labeling under dark conditions could represent either a lack of particle uptake or uptake of nonfluorescent particles. However, the former case could be readily established. When incubation was performed in the dark, the QDs were sedimented by centrifugation with the bacterial pellet, leading to a precipitate the color of the QDs; pellets incubated under room light were white (not shown).
Metabolism-specific labeling of B. subtilis.
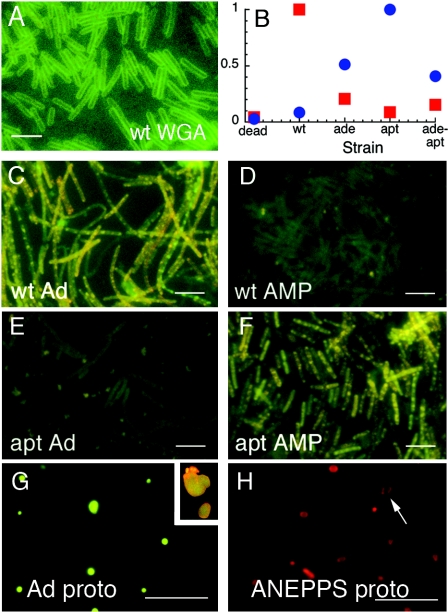
The size dependence of the fluorescent labeling, and the appearance of fluorescence throughout the cells, suggested that QD-adenine conjugates were being taken up by B. subtilis (for contrast, an image of B. subtilis labeled with the noninternalized QD-wheat germ agglutinin conjugate is given in Fig. 4A). We thus incubated QD-adenine conjugates with adenine metabolism mutants of B. subtilis known to show reduced or absent adenine uptake: a mutant lacking adenine deaminase (the ade mutant), one lacking adenosine phosphoribosyltransferase (the apt mutant), and a double mutant lacking both (the ade-apt mutant). Since these mutants, especially the apt mutant, are predicted to take up adenosine or AMP at increased rates (3, 20), we also incubated all mutants and the wild type with QD-AMP conjugates.
FIG. 4.
Metabolism-specific fluorescent labeling of B. subtilis with QD-adenine and QD-AMP. QDs are yellow emitting, corresponding to the second column in Fig. 1. All points are averages of n = 3 (error bars smaller than symbols); scale bars, 5 μm. (A) B. subtilis incubated with QD-WGA conjugates shows a typical pattern of external labeling for contrast with appearance of internal labeling. wt, wild type. (B) Relative fluorescence peaks of various strains exposed to QD-adenine (red squares) and QD-AMP (blue circles). Peaks were normalized to the brightest strain, which controlled for differences in quantum yield between QD-adenine and QD-AMP. (C) Wild type with QD-adenine. (D) Wild type with QD-AMP. (E) The apt mutant with QD-adenine (Ad). (F) The apt mutant with QD-AMP. The ade mutant and ade-apt double mutant are similar but not quite as bright (not shown). (G) Protoplasts (proto) of the wild type after incubation with QD-adenine. Insert, upper right, QD-adenine protoplasts after staining with the membrane dye di-4-ANEPPS. (H) Control protoplasts cannot be visualized unstained, but staining with d-4-ANEPPS shows the appearance of membrane labeling in the absence of cell wall (arrow).
Results showed extreme differences between the strains. For QD-adenine, a strong fluorescence signal was evident only in the wild type. With QD-AMP, all mutants yielded some signal, while the wild type's signal was very weak; the strongest signal was seen with the apt mutant. The wild type and the apt mutant showed the greatest differences between adenine and AMP: the wild type showed a 7.3-fold- ± 0.2-fold-stronger signal with adenine than with AMP (n = 4 independent experiments, measured at the peak of the emission spectrum which corresponded with the peak of the original QD conjugate), whereas the apt mutant was the opposite (18.3-fold ± 0.3-fold stronger with AMP than with adenine; n = 3). The other mutants were less extreme: for the ade mutant, labeling was 4.0-fold ± 0.2-fold stronger for AMP than for adenine (n = 3), but the AMP signal was only 0.5 ± 0.1 times as strong as in the apt mutant. For the double mutant, a 4.3-fold- ± 0.2-fold-stronger signal was seen with AMP than with adenine (n = 4), with the AMP signal 0.4 ± 0.2 times as strong as for the apt mutant. These results are summarized graphically in Fig. 4B, and epifluorescence images of the wild type and ade mutants labeled with yellow CdSe QDs are given in Fig. 4C to F. Results were similar for all colors and consistent with the size selection described in Fig. 2 (data not shown).
Results were highly reproducible when QDs were added to cultures in mid- to late log phase. Addition of particles to bacteria in stationary or declining phase rarely led to fluorescent labeling (n > 20 for all phases tested; 19/20 were successful in log phase, and 2/20 showed some labeling in stationary phase). When QDs were added in early log phase, bacteria often died or sporulated, as determined by microscopic inspection showing broken cells and endospores. Use of minimal growth medium did not affect results.
Labeling of any strain was inhibited 80 to 100% with the addition of EDTA or an excess of unlabeled adenine or hypoxanthine (n > 5 independent experiments).
Digestion of cell walls with lysozyme resulted in protoplasts that were highly fluorescent, indicating that the QDs had penetrated at least through the cell wall to the plasma membrane (Fig. 4G and H). While not definitive in itself, this allowed for removal of the cell wall-associated QDs and for the recovery of the cytoplasmic fraction for electron microscopy.
Labeling of E. coli.
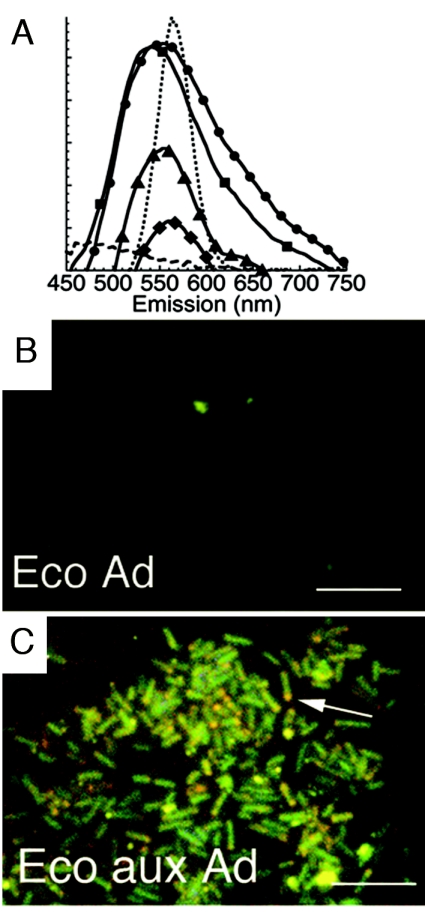
The metabolism-specific labeling seen with B. subtilis suggested possible involvement of specific import mechanisms. Since these mechanisms are often very different in gram-positive and gram-negative bacteria, we repeated the experiments using a gram-negative bacterium (E. coli) for which an adenine auxotrophic mutant was available (ATCC 23804). No visually apparent labeling of wild-type E. coli (ATCC 25922) incubated with adenine- or AMP-conjugated QDs was seen. Similarly, labeling was not seen with the adenine auxotrophic mutant as long as it was incubated in adenine-rich LB medium. However, when incubated in minimal medium, the auxotroph showed strong fluorescence with both adenine and AMP conjugates. Spectra and appearance did not differ significantly from what was seen in B. subtilis except for the presence of a greater red tail in the broadened emission when bare CdSe QDs were used. Results using green CdSe QDs are shown in Fig. 5A to C.
FIG. 5.
Fluorescent labeling of E. coli with QD-adenine and QD-AMP. Scale bar, 5 μm. (A) Fluorescence spectra (arbitrary units) after 1 h of incubation of the adenine auxotrophic mutant with QD-adenine (•) and QD-AMP (▪), the wild type with QD-adenine (▴) and QD-AMP (♦), and the heat-killed auxotroph with QD-adenine () (n > 3; error bars smaller than symbols). The spectrum of the original QDs is included for comparison (dotted line). (B) Wild type incubated with QD-adenine (Ad) for 30 min in minimal growth medium. (C) Adenine auxotroph (aux Ad) under the same conditions; note red shifts at cell poles (arrow).
Growth inhibition by conjugated QDs.
Toxicity of the QD conjugates was investigated by examining growth curves and fluorescence time course. Growth of B. subtilis was significantly inhibited but not arrested by the presence of QD-AMP and QD-adenine conjugates (Fig. 6A). Results were indistinguishable for bare CdSe and CdSe/ZnS QDs (n = 3 experiments each). However, B. subtilis cultures exposed to CdSe conjugates also often displayed extremely elongated cells (refer again to Fig. 2A and 4C, where cells >10 μm long are common). This was not seen with CdSe/ZnS QDs (Fig. 2A).
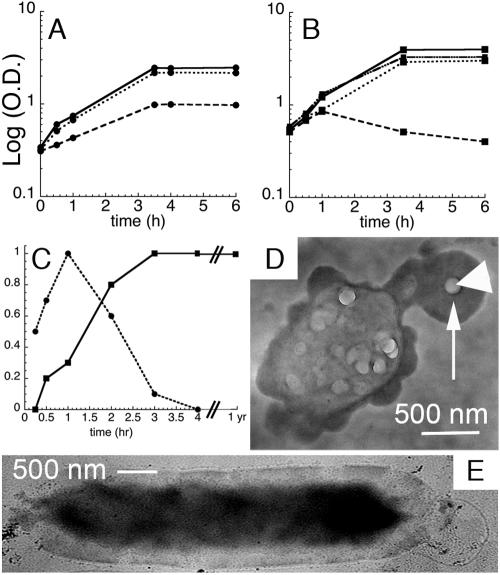
FIG. 6.
Effects of QD labeling on growth and survival (n > 3 for all data points; error bars smaller than symbols). (A) Growth curves for the B. subtilis apt mutant at 37°C, showing a log of optical density at 600 nm versus time in h for incubations with no QDs (———), QDs that show strong cell-associated fluorescence (QD-AMP (), and QDs that do not show significant cell fluorescence (QD-AMP with PEG-5000 [•••••]). (B) Growth curves for adenine auxotrophic E. coli at 37°C in LB medium with no QDs (———), in minimal medium with no QDs (−/B), in minimal medium with QD-AMP (•••••), and in minimal medium with QD-AMP-PEG (—•••-). Growth curves were begun at mid-log phase to reflect optimal time for QD addition. (C) Relative fluorescence intensity at peak compared to incubation time for B. subtilis (▪) and E. coli (•). (D) Whole-mount TEM of an E. coli cell after 4 h of incubation with QD-adenine. EDS values within the dark area (arrow) were 14 atomic% ± 4 atomic% Cd and 9% ± 2% Se (n = 100); the large particle (arrowhead) contained negligible Cd and Se. (E) Whole-mount TEM of a QD-adenine-labeled B. subtilis cell after 1 year at 4°C, showing internal electron-dense material (14 atomic% ± 2 atomic% Cd, 8% ± 1% Se; n = 60) and an absence of sporulation. (Unlabeled controls are nearly invisible [not shown].) (a Cd/Se ratio between 1.2 and 2.4 is consistent with whole QDs [Fig. 8]). Note the presence of many QD aggregates after this long period of storage; this is typical for QDs stored in the presence of water.
Only those cultures that displayed fluorescence that appeared internalized showed slowed growth or cell elongation. Growth curves were indistinguishable from those of controls for QD conjugates that labeled only the cell perimeters, whether the QDs were CdSe or CdSe/ZnS (tested were the wild type with QD-AMP, the apt mutant with QD-adenine, and B. subtilis with QD-WGA, QD-PEG-5000, and QD-AMP-polyethylene glycol [at a 1:1 ratio]). The size of the pellet could be used as a reliable visual marker of whether or not labeling had occurred. A growth curve seen with cell wall-targeted particles is given in Fig. 6A.
For the E. coli auxotrophic mutant, growth in minimal medium was limited, and cells began to die after 60 min. The presence of QD-adenine or QD-AMP conjugates restored growth to almost full wild-type levels. As with B. subtilis, cell wall-targeted conjugates did not affect growth (Fig. 6B).
The reduction in growth seen in E. coli was much less than that seen in B. subtilis (percentage of control at 2 h, 101% ± 5% for E. coli compared to 60% ± 2% for B. subtilis; percentage of control at 6 h, 76% ± 4% for E. coli compared to 39% ± 3% for B. subtilis; n = 3). However, the fluorescence of B. subtilis remained nearly constant after reaching its peak, whereas the fluorescence of labeled E. coli began to decline after 1 h and was undetectable after 4 to 5 h (Fig. 6C). Electron micrographic examination of cells suggested that E. coli cells were able to extrude QDs, whereas the particles remained in and/or on B. subtilis cells. The extrusion was of Cd and Se in ratios consistent with whole QDs, not of Cd alone (Fig. 6D and E). When kept in rich medium at 4°C, QD-labeled B. subtilis cells retained their fluorescence and electron density for at least 1 year (Fig. 6C and E).
There was no growth inhibition seen when bacteria were incubated with CdSe QDs in the dark; this was consistent with lack of uptake as seen in Fig. 3.
Properties of internalized QDs determined from thin sections.
Because bare CdSe QDs showed evidence of possible breakdown and because of concerns over the toxicity of Cd (8), the properties of internalized particles were studied by TEM and EDS in order to quantify their surface elemental composition. Thin sectioning resulted in images of the inside of labeled bacteria and internalized QDs.
Cells that displayed no fluorescence (e.g., wild-type B. subtilis with AMP or the mutants with adenine) did not display any noticeable electron-dense areas or any measurable Cd or Se content (<2% each for >100 spots tested per cell, with >50 cells tested). In preparations that displayed fluorescent labeling, crystals consistent with whole QDs were seen in the cell membrane and throughout the cytoplasm (Fig. 7A [for B. subtilis] and B [for E. coli]). Control cells were nearly invisible, without electron dense areas (not shown; n > 100 cells from n = 5 preparations).
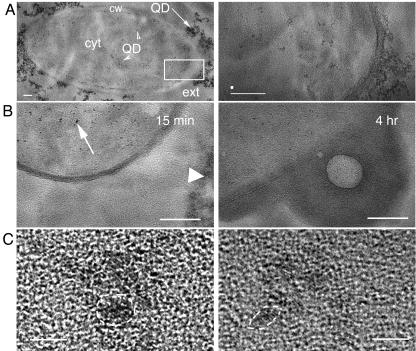
FIG. 7.
TEM images of QDs in embedded ultrathin sections. (A) Transverse thin section of the B. subtilis ade mutant incubated with yellow QD-AMP. Scale bar, 100 nm. Left, low resolution showing QDs inside and outside the cell. Visible are the cell wall (cw), cytoplasm (cyt), and cell exterior (ext). QDs are seen outside the cell (arrow) and inside the cytoplasm (arrowheads). The boxed area is shown in close-up on the right, where distinct particles are clearly visible inside the cytoplasm. EDS values from these particles are given in Results; the white spot above the scale bar shows the size of the EDS beam. (B) Transverse thin section of E. coli auxotroph incubated with QD-AMP. Scale bar, 100 nm. Left, cultures incubated for 15 min, showing many QDs inside the cell (arrow) as well as remaining outside (arrowhead). Right, cultures incubated with QD for 4 h, showing expulsion. (C) High resolution of QDs within thin sections of B. subtilis. Scale bar, 5 nm. Although the thickness of the thin sections does not permit sufficient resolution for quantitative analysis of particle size, the QD lattice structure can be visualized, confirming that the particles are QDs (examples encircled by dashed lines).
Particular areas of the thin sections were then imaged at high resolution, confirming the QD lattice structure within the cells (Fig. 7C). Because the sections were thicker than the QDs, quantitative size distributions could not be obtained by this method, but confirmation of sizes significantly greater than 2 nm was obtained.
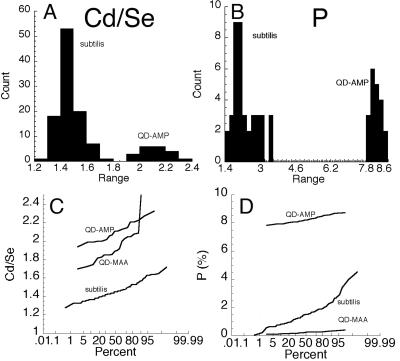
EDS results showed certain significant changes in the elemental composition of the surface of the QDs exposed to bacterial cells. The only elements affected were Cd, Se, and, in the case of AMP conjugates, P. Thus, this technique could be used to monitor the surface of the AMP-QDs during incubation by examination of the Cd/Se ratio and the absolute weight percent of P (the Cd/Se ratio could be followed inside the cells but P could not because of high intracellular P). For QDs without AMP, P made up <1 wt% of QDs. P levels of QD-AMP conjugates were significant (8.25 ± 0.06 wt%; n = 20) but diminished with exposure to live B. subtilis (reduced to 1.56 ± 0.57 wt% after 2 h; n = 100). At the same time, the Cd/Se ratio diminished slightly but significantly (Fig. 8).
FIG. 8.
Alterations in surface elemental composition of QD-AMP conjugates exposed to and/or taken up by B. subtilis cells. All values were taken after 4 h of incubation under room light. (A) Histogram of Cd/Se ratios of QD-AMP conjugates in H2O (QD-AMP) compared to those found inside and immediately outside live B. subtilis cells (subtilis) (no significant difference between the latter two groups was seen; P > 0.2). (B) Histogram of weight percents of P for conjugates in H2O (QD-AMP) compared to those exposed to (but found outside of) live cells (subtilis). (C) Probability plot of Cd/Se ratios for QD-AMP conjugates, solubilized QD-MAA, and QD-AMP inside live cells (subtilis). (D) Probability plot of wt% of P for QD-AMP, QD-MAA, and QD-AMP exposed to live cells (subtilis).
No Cd-rich inclusions were seen in these cells, unlike what was seen with certain types of QD-protein conjugates in our previous study (17).
Properties of internalized QDs determined from ruptured protoplasts.
The soluble, cell-free material recovered from ruptured protoplasts was used to obtain extremely high-resolution images of internalized material from all QD types which showed internalization. This cytoplasmic fraction showed measurable fluorescence. The resulting spectra from bare green QDs were nearly equivalent to those of the original conjugated QDs (Fig. 9A). The peak of core shell green QDs was also nearly identical to the original, which was not surprising given that the whole cells also showed a narrow peak (Fig. 9C). However, for the largest QDs that labeled successfully, the bare yellow QDs, a significant blue shift as well as spectral narrowing were observed in the ruptured protoplasts (Fig. 9B).
FIG. 9.
Digestion with lysozyme followed by rupture of protoplasts and recovery of cytoplasmic fraction releases QDs with narrow spectra and restricted sizes. (A) Green CdSe QDs extracted from bacteria (•) show a slight blue shift from the QD-AMP controls (dashed line) (5 ± 2 nm) and no significant change in FWHM. (B) Bare yellow CdSe QDs (•) exhibit a greater blue shift (15 ± 5 nm) accompanied by a spectral narrowing (FWHM of 26 ± 3 nm for the ruptured protoplasts compared to the value for QD-AMP controls of 46 ± 1 nm [dashed line]). (C) Green core shell CdSe/ZnS QDs (▴) show no significant changes in spectrum relative to the QD-AMP controls (dashed line). (D) Whole-mount image of B. subtilis protoplast. Cd and Se are detectable throughout the cell, but large concentrations are found at the pole (dark area). (E) High-resolution TEM image of the largest individual particles observed in whole mounts of yellow QDs conjugated to AMP (corresponding to the spectrum of the dashed line in B). A dashed line is drawn around a single crystal for clarity; the lattice structure may vary from crystal to crystal. (F) High-resolution image of the largest particles observed from ruptured protoplasts of cells labeled with yellow QDs (corresponding to the ruptured protoplast curve (•) in B). A dashed line is drawn around a sample crystal for clarity.
Protoplasts were examined by TEM before rupture to confirm a lack of external QD material (Fig. 9D). After rupture, direct deposition of the fluorescent supernatant onto TEM grids revealed individual QDs with size distributions consistent with the spectra. For green QDs, bare and core shell, no significant size alterations were seen (not shown). However, for yellow CdSe QDs, the average particle size was reduced from 4.6 ± 0.4 nm to 3.8 ± 0.2 nm (n > 50 each) (Fig. 9E and F).
DISCUSSION
Limited success has been achieved with quantum dots as internal labels of living cells because of their inability to cross cell walls. This work demonstrates the ability of the smallest QDs (with or without ZnS shells) to enter bacteria based upon specific metabolic activities and thus opens the field for use of such QDs as fluorescent and/or electrodense probes of bacterial metabolism, at least in the case of ligands which possess flexible or “fuzzy” recognition templates that allow them to be recognized after substantial alterations (21).
Entry of adenine- and AMP-conjugated QDs into bacteria is seen to be highly dependent upon nanocrystal size, as expected for cells which do not endocytose. QDs >5 nm in diameter do not lead to any significant cell labeling. QDs 3.5 to 4.5 nm in diameter lead to strong fluorescent labeling with very little background. Since these particles are substantially too large to enter via nonspecific mechanisms, it is likely that specific uptake processes are involved in the internalization of these conjugates.
It is unlikely that the QDs are actually passing through purine permeases, which are likely to have pore sizes of only several angstroms (25). The observed dependence of labeling upon light exposure suggests another mechanism: the purine-conjugated QDs bind the cells in a metabolism-specific manner but cannot enter the cell. Upon light exposure, electron transfer between the QD and the surrounding water-based medium leads to reactive oxidizing species which cause transient membrane damage, allowing for particle entry. The smaller particles are more strongly oxidizing; the lack of labeling seen with red particles could result more from the lessened reactivity of these QDs rather than their size per se. This mechanism could be potentially be exploited for novel techniques of gene or drug delivery employing light activation (1). Lessened reactivity also explains the weaker labeling seen with CdSe(ZnS) core shell particles despite their higher quantum yield. Development of methods for improvement of the brightness and stability of bare-core particles, without compromising core accessibility, is thus desirable for the development of light-sensitive labels or antibiotics.
Similar mechanisms appear to work in both a gram-positive species (B. subtilis) and a gram-negative species (E. coli). The elimination of a single enzyme, adenosine phosphoribosyltransferase, from B. subtilis is sufficient to eliminate QD-adenine uptake and permit the uptake of QD-AMP, which does not label the wild type. Elimination of adenine deaminase has a qualitatively similar but less dramatic effect. In E. coli, strong uptake of both conjugates is seen only in adenine auxotrophs incubated in minimal medium.
Despite the release of Cd2+ ions upon light exposure, which is cytotoxic to many mammalian cells (1, 8), significant cell death or sporulation is not seen in bacterial cultures exposed to these conjugates. Slowed rates of growth are seen in internally labeled cultures, but this is likely to reflect nonspecific mechanisms more than heavy metal release, since nanocrystals made of less toxic materials such as Ag (28) and Si (23) also slow bacterial growth in a similar fashion. However, some suggestion of heavy metal damage was seen when B. subtilis cells were incubated with bare CdSe QD conjugates under room light, which was not seen with CdSe/ZnS QDs or in the dark. This was elongation of cells to >10 μm in many cases, reflecting decreased rates of cell division (2, 26). Very low concentrations of Cd, too low to be cytotoxic or detectable in solution, are known to inhibit DNA repair enzymes and may thus inhibit cell division (14).
The toxicity seen in E. coli was much less than that in B. subtilis, but recovery of cell growth was accompanied by loss of fluorescence and expulsion of electron-dense material consistent with whole QDs from the cells. Gram-negative bacteria are known to be able to extrude many materials through extrusion pumps; in one study, Pseudomonas aeruginosa was shown to expel Ag nanocrystals (18).
The loss of the external Cd shell of the QDs incubated with bacteria was confirmed by TEM with EDS analysis. The lattice structure of internalized QDs remains intact, but the outer surface displays Se enrichment. Signs of nanocrystal pulverization, such as large Cd inclusions inside the bacteria, are not seen.
A further interesting feature of labeling by bare CdSe QDs is that spectral broadening is seen when the particles are taken up by bacteria, with a significant red tail. A blue shift could be explained by particle breakdown, but a red shift must be due to other mechanisms. When the cytoplasmic fraction of the labeled bacteria is isolated, the spectrum of this soluble fraction is narrowed once again. This suggests that bare, adenine-conjugated QDs in the cell wall or cell membrane of labeled cells show spectral alterations.
The red shift seen was not red enough to represent emission from trap states created by photooxidation, which occur in the near-infrared range (∼750 nm) (data not shown). Interestingly, more red shift was seen with E. coli than with B. subtilis, implying that different cell wall compositions are important. A recent study reported a blue shift of QD-DNA conjugates upon binding to bacteria (9), confirming that energy or electron transfer mechanisms are likely to play a role in observed spectra. Understanding the mechanism of the broadening could lead to the design of environmentally sensitive probes that cannot be created with overpassivated capped nanocrystals.
It was interesting that a sufficient number of nanocrystals to yield a fluorescence spectrum could be isolated from the bacterial cytoplasmic fraction. These particles cannot be considered as representative of all of the fluorescent particles found in cultures of cells, since particles associated with the cell membrane and cell wall, as well as any particles in the cytoplasmic fraction that might have been pulled down by centrifugation, were removed. However, some conclusions may still be drawn from these experiments. The spectrum of the smallest (green-emitting) particles was the same in this fraction as in the original conjugated QDs; however, that of the yellow-emitting particles was blue shifted and narrowed. Correspondingly, electron micrographs showed a measurable reduction in size of only the yellow particles. This was not due to photooxidation, as the controls in the QD-only incubation were exposed to identical conditions of heat, light, and H2O. The narrowing of the spectrum, without a blue shoulder, suggests exclusion of largest particles rather than overall reduction in particle size. This is consistent with the size exclusion results obtained using different QD colors but allows for greater precision in the cutoff: the fluorescence showed a sharp cutoff at 590-nm emission or a QD size of ∼3.8 nm. If this method can be generalized to other import mechanisms, it will prove a useful tool for probing interactions between ligands, cell walls, membranes, and membrane proteins.
Acknowledgments
This material is based upon work performed at the California Institute of Technology, Jet Propulsion Laboratory, supported by a contract from the National Aeronautics and Space Administration.
We thank Ken Nealson for critical reading of the manuscript and Carol Garland for the high-resolution TEM images.
REFERENCES
- 1.Bakalova, R., H. Ohba, Z. Zhelev, M. Ishikawa, and Y. Baba. 2004. Quantum dots as photosensitizers? Nat. Biotechnol. 22:1360-1361. [DOI] [PubMed] [Google Scholar]
- 2.Burdett, I. D., T. B. Kirkwood, and J. B. Whalley. 1986. Growth kinetics of individual Bacillus subtilis cells and correlation with nucleoid extension. J. Bacteriol. 167:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, S., D. R. Tomchick, D. Wolle, P. Hu, J. L. Smith, R. L. Switzer, and H. Zalkin. 1997. Mechanism of the synergistic end-product regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase by nucleotides. Biochemistry 36:10718-10726. [DOI] [PubMed] [Google Scholar]
- 4.Dabbousi, B. O., J. Rodriguez-Viejo, F. V. Mikulec, J. R. Heine, H. Mattoussi, R. Ober, K. F. Jensen, and M. G. Bawendi. 1997. (CdSe)ZnS core-shell quantum dots: synthesis and characterization of a size series of highly luminescent nanocrystallites. J. Physical Chem. B 101:9463-9475. [Google Scholar]
- 5.de Koning, H., and G. Diallinas. 2000. Nucleobase transporters (review). Mol. Membr. Biol. 17:75-94. [DOI] [PubMed] [Google Scholar]
- 6.Demchick, P., and A. L. Koch. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denessiouk, K. A., V. V. Rantanen, and M. S. Johnson. 2001. Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins 44:282-291. [DOI] [PubMed] [Google Scholar]
- 8.Derfus, A. M., W. C. W. Chan, and S. N. Bhatia. 2004. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 4:11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwarakanath, S., J. G. Bruno, A. Shastry, T. Phillips, A. John, A. Kumar, and L. D. Stephenson. 2004. Quantum dot-antibody and aptamer conjugates shift fluorescence upon binding bacteria. Biochem. Biophys. Res. Commun. 325:739-743. [DOI] [PubMed] [Google Scholar]
- 10.Han, M., X. Gao, J. Z. Su, and S. Nie. 2001. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 19:631-635. [DOI] [PubMed] [Google Scholar]
- 11.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 12.Haram, S. K., B. M. Quinn, and A. J. Bard. 2001. Electrochemistry of CdS nanoparticles: a correlation between optical and electrochemical band gaps. J. Am. Chem. Soc. 123:8860-8861. [DOI] [PubMed] [Google Scholar]
- 13.Harry, E. J., B. J. Stewart, and R. G. Wake. 1993. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol. Microbiol. 7:611-621. [DOI] [PubMed] [Google Scholar]
- 14.Hartwig, A., M. Asmuss, I. Ehleben, U. Herzer, D. Kostelac, A. Pelzer, T. Schwerdtle, and A. Burkle. 2002. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ. Health Perspect. 110(Suppl. 5):797-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyer, P., and H. Weller. 1994. Size-dependent redox potentials of quantized zinc-oxide measured with an optically transparent thin-layer electrode. Chem. Phys. Lett. 221:379-384. [Google Scholar]
- 16.Jaiswal, J. K., H. Mattoussi, J. M. Mauro, and S. M. Simon. 2003. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 21:47-51. [DOI] [PubMed] [Google Scholar]
- 17.Kloepfer, J. A., R. E. Mielke, M. S. Wong, K. H. Nealson, G. Stucky, and J. L. Nadeau. 2003. Quantum dots as strain- and metabolism-specific microbiological labels. Appl. Environ. Microbiol. 69:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyriacou, S. V., W. J. Brownlow, and X. H. Xu. 2004. Using nanoparticle optics assay for direct observation of the function of antimicrobial agents in single live bacterial cells. Biochemistry 43:140-147. [DOI] [PubMed] [Google Scholar]
- 19.Mattoussi, H., J. M. Mauro, E. R. Goldman, G. P. Anderson, V. C. Sundar, F. V. Mikulec, and M. G. Bawendi. 2000. Self-assembly of CdSe-ZnS quantum dot bioconjugates using an engineered recombinant protein. J. Am. Chem. Soc. 122:12142-12150. [Google Scholar]
- 20.Meyer, E., and R. L. Switzer. 1979. Regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase activity by end products. J. Biol. Chem. 254:5397-5402. [PubMed] [Google Scholar]
- 21.Moodie, S. L., J. B. Mitchell, and J. M. Thornton. 1996. Protein recognition of adenylate: an example of a fuzzy recognition template. J. Mol. Biol. 263:486-500. [DOI] [PubMed] [Google Scholar]
- 22.Nygaard, P., P. Duckert, and H. H. Saxild. 1996. Role of adenine deaminase in purine salvage and nitrogen metabolism and characterization of the ade gene in Bacillus subtilis. J. Bacteriol. 178:846-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Péreza, L., M. Floresa, J. Avalosa, L. San Miguel, O. Resto, and L. Fonseca. 2003. Comparative study of the growth curves of B. subtilis, K. pneumoniae, C. xerosis and E. coli bacteria in medium containing nanometric silicon particles. Mat. Res. Soc. Symp. Proc. 737:F3.6.1-F3.6.6. [Google Scholar]
- 24.Petruska, M. A., A. P. Bartko, and V. I. Klimov. 2004. An amphiphilic approach to nanocrystal quantum dot-titania nanocomposites. J. Am. Chem. Soc. 126:714-715. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez, M. A., R. Tryon, S. Pierce, G. Vasudevan, and S. M. Landfear. 2004. Functional expression and characterization of a purine nucleobase transporter gene from Leishmania major. Mol. Membr. Biol. 21:11-18. [DOI] [PubMed] [Google Scholar]
- 26.Sargent, M. G. 1975. Control of cell length in Bacillus subtilis. J. Bacteriol. 123:7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxild, H. H., and P. Nygaard. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 169:2977-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sondi, I., and B. Salopek-Sondi. 2004. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 275:177-182. [DOI] [PubMed] [Google Scholar]
- 29.Spizizen, J. 1958. Citation classic—transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steckel, J. S., J. P. Zimmer, S. Coe-Sullivan, N. E. Stott, V. Bulovic, and M. G. Bawendi. 2004. Blue luminescence from (CdS)ZnS core-shell nanocrystals. Angew. Chem. Int. Ed. Engl. 43:2154-2158. [DOI] [PubMed] [Google Scholar]
- 31.Stoimenov, P. K., R. L. Klinger, G. L. Marchin, and K. J. Klabunde. 2002. Metal oxide nanoparticles as bactericidal agents. Langmuir 18:6679-6686. [Google Scholar]
- 32.Wang, H. W., Y. Chen, H. Yang, X. Chen, M. X. Duan, P. C. Tai, and S. F. Sui. 2003. Ring-like pore structures of SecA: implication for bacterial protein-conducting channels. Proc. Natl. Acad. Sci. USA 100:4221-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, X., H. Liu, J. Liu, K. N. Haley, J. A. Treadway, J. P. Larson, N. Ge, F. Peale, and M. P. Bruchez. 2003. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat. Biotechnol. 21:41-46. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, L., S. Ang, and W. T. Liu. 2004. Quantum dots as a novel immunofluorescent detection system for Cryptosporidium parvum and Giardia lamblia. Appl. Environ. Microbiol. 70:597-598. [DOI] [PMC free article] [PubMed] [Google Scholar]