The threat of terrorist or criminal use of pathogenic organisms and their toxins remains a great concern in the United States. The anthrax letter attack of 2001 (12) raised the awareness of our vulnerability. It also demonstrated the need to perform microbial forensic analyses for attribution purposes in a bioterrorism event. As part of the effort to deter biological terrorism and strengthen the law enforcement response to such an act, the United States recently established a microbial forensic laboratory known as the National Bioforensics Analysis Center (NBFAC), which is part of the Department of Homeland Security and operates in partnership with the Federal Bureau of Investigation (FBI) (3). The NBFAC provides a central facility to conduct analysis of evidentiary material (20). Although the NBFAC's infrastructure and capabilities draw on the best scientific resources available in the United States and on some resources internationally, the practitioners of the nascent field of microbial forensics recognize that there remain significant gaps in both science and operations that must be filled to establish a more readily responsive and effective system.
A number of approaches to consider for the microbial forensic panoply that will help develop the field and provide greater capacity have been described previously (14). To build on those approaches and to identify the more pressing gaps, scientists from diverse disciplines convened a meeting from 19 to 21 April 2004 at the Banbury Center of Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. This meeting was sponsored by the Department of Homeland Security. The specific gaps identified were presented subsequently on 23 June 2004 at the White House Conference Center to a key group of scientists, stakeholders, and policy makers. In addition, the gaps requiring research investments have been identified by the FBI's Scientific Working Group on Microbial Genetics and Forensics (3). The discussions of the Banbury Microbial Forensics Group included how to coordinate and effectively focus resources that several agencies (both within the United States and internationally) bring to bear and how to address the need to engage the broader research community in the most important issues in this field. This report summarizes the major findings of both meetings. The belief is that the best policy for investments in microbial forensics can be established when there is input and guidance from the greater scientific community. Therefore, this document is intended to serve as a vehicle to elicit response from the community. Those who have constructive input should contact the authors.
ADDRESSABLE GAPS
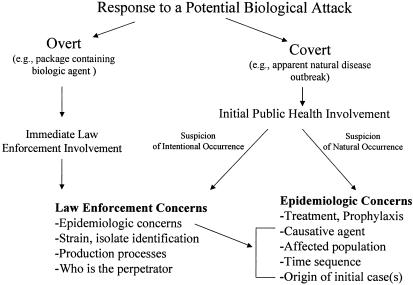
The variety of possible attack scenarios mandates the need to configure and prioritize the various research and development, validation, and operational elements of a microbial forensics program. The response to an overt or covert biological attack is currently governed by a national plan that coordinates public health and law enforcement responsibilities and leadership (Fig. 1). Several recent outbreaks of new or unusual diseases and acts of bioterrorism and biocrimes have occurred (1, 2, 7, 10, 11, 13, 18, 19). An end-to-end analysis of these events and the responses would help determine which agency interfaces, procedures, methods, and technologies work well and should be adopted as standard practices. It would also show where the investigation process could be improved. Microbial forensic analysis must encompass sample handling, collection, preservation, method selection, casework analysis, interpretation of results, validation, and quality assurance. Lessons learned from public health outbreak investigations and participation by forensics specialists in future investigations (e.g., food-borne disease outbreaks) could help to develop and advance the science and practice of microbial forensics. Such analyses may provide guidance for differentiating expeditiously between natural and intentional occurrences, which is an extremely important need for response.
FIG. 1.
General public health and law enforcement responsibilities regarding a potential biological attack.
Identification, collection, handling, and preservation of samples prior to arrival at the laboratory are crucial to avoid compromising subsequent assays. The challenge is to preserve signatures in the sample when it is removed from the crime scene. The sample collection and storage methods used for recent anthrax and ricin investigations may not be appropriate for other types of agents and scenarios. There are no standardized microbial evidence collection kits analogous to a sexual assault kit designed to collect evidence from a rape victim. Evidence collection procedures need to be developed with the intent, if possible, of preserving traditional forensic evidence, such as hair, fibers, fingerprints, and human DNA, as well as providing adequate material for microbial forensic analyses.
Extraction and purification of trace nucleic acid, protein, or other signatures from samples are significant challenges. While many tools and procedures exist, from swabs to high-efficiency particulate air (HEPA) vacuum filters, few have been rigorously validated, especially with regard to extraction efficiency. Moreover, existing methods are not necessarily known to all agencies that have a responsibility to the investigation. It will be essential to link the NBFAC with other organizations that routinely develop such protocols (e.g., the National Institute for Occupational Safety and Health, Food and Drug Administration, Centers for Disease Control and Prevention, United States Department of Agriculture, Environmental Protection Agency, and others) so that these procedures can be quantitatively compared and validated and can be improved when needed.
Molecular genetics, genomics, and informatics will be central to identification, virulence determination, pathogenicity characterization, and source attribution. The ultimate goal of source attribution is to be able to individualize a sample such that it can be traced to a unique source. That is unlikely with current capabilities. Consider the above-mentioned anthrax letter attack, where a multilocus variable number of tandem repeats technique for Bacillus anthracis strain identification was used for analysis (15). The data were qualitatively interpreted as the Ames strain and focused the investigation towards laboratory sources. Yet, no further attribution was possible. “Grand leaps” in sequencing technology to increase speed, to reduce cost, and to maximize efficiency for forensic analysis are needed. Accumulation of the existing genetic information of pathogens and near neighbors into accessible databases is essential. Methods for selecting valid nucleic acid signature sets and approaches for interpretation of evidentiary results need to be improved. Microbial diversity and the genomic features, forces, and mechanisms that affect diversity and evolutionary processes require greater understanding. The same holds true for the interrelationships between a specific microbe and those of proximate communities that might impact identification and characterization. It is also important to better understand how environment impacts variability.
Molecular typing has been used for some years in epidemiological studies, such as with cases of human immunodeficiency virus transmission between a donor and an unknowing recipient or victim (17). Unfortunately, the discriminatory power of the molecular-typing procedure is not well defined. There is a need for a more-systematic rational design of typing systems that exploit the growing knowledge base on genome plasticity and the informatics tools for determining sets of informative sites (markers). Validation studies should include determining the degree of confidence and the limits of analysis and interpretation. Examples of useful informatics tools are listed in Table 1. Comparing sequence data generally will require the development of effective lineage-based approaches for reconstructing evolutionary history, accommodating gene conversion/recombination, and considering the possible heterogeneous rates and patterns of mutations across a microbe's genome. Optimized and validated sequence alignments and phylogenetic algorithms that incorporate various features, motifs, and population factors are needed. These population factors include incorporation of clonal inheritance, inheritance by sexual reproduction, gene conversion, recombination, and horizontal gene transfer.
TABLE 1.
Useful bioinformatic tools for microbial forensics
| Bioinformatic tool(s) |
|---|
| Robust algorithm(s) for sequence alignmenta |
| Phylogenetic algorithm(s) based on different types of markers/loci/ sequence datab |
| Identification of informative sites (markers), not necessarily a single set, but possibly a set of alternatives, with the power to answer specific questions |
| Methods for quantitatively interpreting results from a case analysis, such as estimation of the age/time of the most recent common ancestor and the nearest set of neighbors with associated confidence boundsc |
| More-effective algorithm(s) for detecting genomic signatures of pathogenicity/virulence and antibiotic resistance of microbial agents |
| Capabilities for relating diversity to function |
| Comprehensive comparative and functional genomics capabilities |
| User-friendly interfaces for interpretation and visualization of data from a genomic analysis |
Robust against rate and pattern heterogeneity of site-specific mutations/substitutions, the presence of repeat elements (of varied lengths), recombination, and gene conversion.
Cooptimized with a robust algorithm(s) and incorporating clonal and sexually inherited markers, recombination, gene conversion, and horizontal gene transfer.
Taking into account factors listed in footnote a and incorporating population-related factors.
Nucleic acid-based methods, although extremely important, are unlikely to pinpoint a unique source. Thus, chemical and physical analyses of evidence almost certainly will be needed to attempt to obtain additional information. However, there is still a lack of understanding about how the chemical compositions of microorganisms and contaminants reflect the chemical compositions of the environments in which they were grown. For example, isotopic analyses relating to microbial diversity and growth conditions are needed. Matching of chemical and physical properties can help establish the relatedness of disparate incidents, and mismatches could have exclusionary power or signify a more complex event.
Instrumental analysis also can be useful by providing inferences regarding the processes used to produce and “weaponize” an agent preparation. Such information may indicate the sophistication of the perpetrator, identify how recently the materials were made, provide evidence regarding whether or not certain unique or unusual materials were used, and suggest whether the process lends itself to producing large quantities of the agent. There are significant challenges associated with developing a robust capability that will reliably permit such inferential uses of chemical and physical data. The knowledge base from which such deductions may be drawn is sparse. Furthermore, the existing information is not readily accessible or may be of questionable reliability.
Other challenges include the following. (i) With the use of surrogates for study and validation, the transferability of conclusions drawn from studies of surrogates to the actual agent may not be straightforward. It will be necessary to better understand the limitations of surrogates and how to choose the best one for a given study. (ii) The use of decontamination protocols prior to handling and analysis must be examined. The effect of such treatments on the relevant properties of the agent and on traditional forensic assays needs to be better understood. (iii) The best sample collection and preservation strategies must be established. As with nucleic acids mentioned above, it will be important to establish collection and storage methods that do not inadvertently distort or contaminate subtle structural or chemical signatures and that minimize their degradation. (iv) Decision tree algorithms that select and prioritize analytical methods must be developed. A systematic procedure for choosing the most-informative analytical tests becomes crucial, particularly when only limited or trace quantities of samples are available. It will also be important to formalize such considerations to expedite the development of the best analytical plan when an incident occurs. (v) A reference library of materials and data that can be used to make comparisons and to calculate statistical probabilities must be established.
Another area that has not been sufficiently exploited in identifying a perpetrator is the host response. Either by exposure or by prophylaxis, long-term versus primary immune response could “rule in or rule out” suspects. One of the most stable and specific host responses is the antibody response to pathogen or toxin antigens. In general, the presence of immunoglobulin M indicates an early response and immunoglobulin G a later response. Additionally, the pharmacokinetics of an antibiotic may be informative. Knowledge of how long an antibiotic persists in the body, how long its metabolites persist, and in which tissues they are deposited and for how long could be useful. This information may indicate if the perpetrator took protective measures, especially when he/she denies any contact or handling of a pathogenic agent. Such information needs to be developed further and collated in a unified database. This is a difficult challenge since there is little baseline information on antibiotic use among the general population or levels of detectable antibodies to the threat agents among the general population. Perhaps expression arrays may enhance capabilities.
An area that has not been given adequate attention is a nation's animal and plant agricultural economy and its associated food production and distribution system. These are particularly vulnerable and inviting targets for economic damage and, potentially, political destabilization. The U.S. food and fiber industry generates nearly $1 trillion in revenue annually (16), is a significant part of the U.S. economy, and is a tempting target. Agriculture today for many countries is a global enterprise with animals, plants, and products entering and leaving the country on a daily basis. Processing, packaging, and distribution occur at multiple sites. Many aspects of the agricultural system, including entry, production, processing, packaging, and distribution, are vulnerable to attack.
Although the spread of a number of agricultural or food-borne microorganisms or toxins could have a catastrophic impact on humans, far less attention has been focused on agents that could directly affect animal and plant populations or our food supply, except in instances where an animal pathogen is zoonotic. In contrast to direct attacks on human populations, potentially less technical complexity, sophistication, and infrastructure are required for the acquisition, production, and introduction of a bioagent targeting agriculture or food. Operational risks to would-be perpetrators planning and executing such operations are considerably low because many of the agents do not cause disease in humans. Any attack on our animal or food supply, if sequential or multifocal, could undermine the people's trust in their government, particularly in the absence of a quick and orchestrated response. To complicate attribution, there are, for example, 300,000 hospitalizations and 5,000 deaths per year due to naturally occurring food-borne illness in the United States (5); thus, the background noise from naturally occurring outbreaks may confound recognition of an unnatural, covert event (6, 9). Food-borne pathogens, such as Escherichia coli O157:H7, Salmonella spp., Shigella spp., and Vibrio cholerae, are readily available and have been used many times as weapons in biocrimes (4). Distinguishing natural from intentional animal and plant microbial outbreaks, and attributing outbreaks to the proper sources, will require substantial microbial forensic development. Therefore, investing in a robust microbial forensics and scientific investigative capacity for attribution of agricultural and food-borne pathogens and toxins makes sense as part of the nation's “tool kit” for biosecurity.
Many of the issues and dimensions discussed earlier in this paper apply equally well to analysis of bioweapons directed towards agricultural and food targets. A farm-to-table tracking system to facilitate trace-back is under development and will help significantly. However, fungal plant pathogens are biologically more complex than their bacterial and viral counterparts. Whole-genome sequencing of a fungal genome is far from being a trivial task and is even more challenging considering that there are at least 8,000 fungal species known to cause plant diseases (8). The Banbury Microbial Forensics Group believes that completely securing our agricultural and food production and distribution systems from attack is not possible. Therefore, a strong forensic capability is needed for attribution of animal, plant, and food-borne pathogens and toxins to provide the law enforcement, intelligence, agriculture, public health, and homeland security communities with information to assist in identifying perpetrators of biocrimes and bioterrorism and to serve as a deterrence factor. It is important to address basic and applied questions and issues regarding microbial forensics as it applies to agricultural and food threat agents and associated physical evidence with the same vigor as for human pathogens and toxins. We strongly recommend that a microbial forensics meeting, with diverse scientific representation, be convened as soon as possible to begin addressing attribution when agriculture or food is the target.
CONCLUSION
Scientists with diverse expertise are needed to provide the basis for a successful microbial forensics program. Input from the broad scientific community and a robust system of peer review are essential for developing a national microbial forensics system. Such input has already resulted in the establishment of quality assurance guidelines for laboratories performing this type of work (3). While some capabilities do exist to carry out a forensic investigation of a bioterrorism act or a biocrime, information contained within the evidence that may provide clues for attribution currently cannot be exploited fully. The most-pressing gaps described here (Table 2) can provide direction for the NBFAC and for the scientific community so that the nation's assets can be utilized effectively to enhance microbial forensics and to protect our country.
TABLE 2.
Summary of areas to address further for improving microbial forensics capabilities
| Area(s) recommended for further exploration |
|---|
| Effective approaches for distinguishing between an intentional and natural event |
| End-to-end evaluation of past cases (e.g., lessons learned from emerging disease outbreaks) |
| Sample collection, handling, and preservation protocols |
| Extraction protocols |
| Validation of analytical and interpretive protocols |
| Microbial diversity |
| Bioinformatics toolbox |
| Quantitative conclusions |
| Databases of genomes and signatures (of pathogens and near neighbors) and of methods (genetic, chemical, physical) |
| Unit process analysis of plausible recipes for production |
| Use of surrogates |
| Effects of decontamination on evidence |
| Systematic procedures for selecting and ordering analytical methods (decision trees) |
| Host immune responses and pharmacokinetics of prophylactic drugs |
Acknowledgments
This is publication no. 04-16 of the Laboratory Division of the FBI.
Names of commercial manufacturers are provided for identification only, and inclusion does not imply endorsement by the FBI.
REFERENCES
- 1.Azad, A. F. 2004. Prairie dog: cuddly pet or Trojan horse? Emerg. Infect. Dis. 10:542-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briese, T., X. Y. Jia, C. Huang, L. J. Grady, and W. I. Lipkin. 1999. Identification of a Kunjin/West Nile-like flavivirus in brains of patients with New York encephalitis. Lancet 354:1261-1262. [DOI] [PubMed] [Google Scholar]
- 3.Budowle, B., S. E. Schutzer, A. Einseln, L. C. Kelley, A. C. Walsh, J. A. Smith, B. L. Marrone, J. Robertson, and J. Campos. 2003. Public health. Building microbial forensics as a response to bioterrorism. Science 301:1852-1853. [DOI] [PubMed] [Google Scholar]
- 4.Carus, W. S. 2002. Bioterrorism and biocrimes: the illicit use of biological agents since 1900. Fredonia Books, Amsterdam, The Netherlands.
- 5.Centers for Disease Control and Prevention. 2004. Diagnosis and management of foodborne illnesses: a primer for physicians and other health care professionals. Morb. Mortal. Wkly. Rep. Recomm. Rep. 53:1-33. [PubMed] [Google Scholar]
- 6.Check, E. 2004. Food panel calls for beefed up response to mad cow disease. Nature 427:575. [DOI] [PubMed] [Google Scholar]
- 7.Childs, J. E. 2004. Zoonotic viruses of wildlife: hither from yon. Arch. Virol. 18(Suppl.):1-11. [DOI] [PubMed] [Google Scholar]
- 8.Cubeta, M. A., and G. A. Payne. Biology and detection of fungal pathogens of humans and plants. In R. G. Breeze, B. Budowle, and S. E. Schutzer (ed.), Microbial forensics, in press. Academic Press, San Diego, Calif.
- 9.Dalton, R., and E. Check. 2004. Beef blockade greets first mad cow in United States. Nature 427:5. [DOI] [PubMed] [Google Scholar]
- 10.Di Giulio, D. B., and P. B. Eckburg. 2004. Human monkeypox. Lancet Infect. Dis. 4:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filice, G. A. 2004. SARS, pneumothorax, and our response to epidemics. Chest 125:1982-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, V. P., S. L. Lukacs, T. Handzel, J. Hayslett, S. Harper, T. Hales, V. A. Semenova, S. Romero-Steiner, C. Elie, C. P. Quinn, R. Khabbaz, A. S. Khan, G. Martin, J. Eisold, A. Schuchat, and R. A. Hajjeh. 2002. Opening a Bacillus anthracis-containing envelope, Capitol Hill, Washington, D.C.: the public health response. Emerg. Infect. Dis. 8:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jernigan, D. B., P. L. Raghunathan, B. P. Bell, R. Brechner, E. A. Bresnitz, J. C. Butler, M. Cetron, M. Cohen, T. Doyle, M. Fischer, C. Greene, K. S. Griffith, J. Guarner, J. L. Hadler, J. A. Hayslett, R. Meyer, L. R. Petersen, M. Phillips, R. Pinner, T. Popovic, C. P. Quinn, J. Reefhuis, D. Reissman, N. Rosenstein, A. Schuchat, W.-J. Shieh, L. Siegal, D. L. Swerdlow, F. C. Tenover, M. Traeger, J. W. Ward, I. Weisfuse, S. Wiersma, K. Yeskey, S. Zaki, D. A. Ashford, B. A. Perkins, S. Ostroff, J. Hughes, D. Fleming, J. P. Koplan, J. L. Gerberding, and the National Anthrax Epidemiologic Investigation Team. 2002. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg. Infect. Dis. 8:1019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keim, P. 2003. Microbial forensics: a scientific assessment. American Academy of Microbiology, Washington, D.C.
- 15.Keim, P., and K. L. Smith. 2002. Bacillus anthracis evolution and epidemiology. Curr. Top. Microbiol. Immunol. 271:21-32. [DOI] [PubMed] [Google Scholar]
- 16.Lipton, K. L., W. Edmundson, and A. Manchester. 1998. The food and fiber system: contributing to the U.S. and world economies. Agriculture information bulletin 742. U.S. Department of Agriculture, Washington, D.C.
- 17.Metzker, M. L., D. P. Mindell, X. M. Liu, R. G. Ptak, R. A. Gibbs, and D. M. Hillis. 2002. Molecular evidence of HIV-1 transmission in a criminal case. Proc. Natl. Acad. Sci. USA 99:14292-14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watts, J. 2004. SARS under control, but lab-safety questions remain. Lancet 363:1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein, R. A. 2004. Planning for epidemics—the lessons of SARS. N. Engl. J. Med. 350:2332-2334. [DOI] [PubMed] [Google Scholar]
- 20.White House Press Secretary. 2004. Biodefense for the 21st century. Presidential directive HSPD-10. White House, Washington, D.C.