Abstract
DNA microarray analysis showed that yfiD, yggB, and yggE genes were up-regulated when superoxide dismutase (SOD)-deficient Escherichia coli IM303 (I4) was cultivated under the oxidative stress generated by photoexcited TiO2, and pYFD, pYGB, and pYGE were constructed by inserting the respective genes into a pUC 19 vector. The content of reactive oxygen species (ROS) in IM303 (I4) cells carrying pYGE was reduced to 31% of ROS content in the control cells with pUC 19. In the culture of wild-type strain, E. coli MM294, in the medium with paraquat (10 μmol/l), maximum specific growth rate of the cells with pYGE was about five times higher than that of the control cells, with a decreased ROS content in the former cells. The introduction of pYGE also suppressed the occurrence of the cells with altered amino acid requirement in the culture of MM294 cells with paraquat.
Aerobic organisms have high reducing potential of molecular oxygen and preferentially utilize oxygen as a terminal electron acceptor in respiration for effectively generating biochemical energy for their proliferation (9). As a consequence of the aerobiosis, reactive oxygen species (ROS) such as superoxide anion ( · O2−), hydrogen peroxide (H2O2), and hydroxyl radical ( · OH) are often formed and can oxidize various biomaterials owing to their high reactivity, which causes DNA breakage, enzyme inactivation, membrane lipid peroxidation, and so on (3, 10, 20).
Several antioxidant systems exist in living cells to avoid cellular damage caused by oxidative stresses (17). Sox regulon is an example of an ROS-scavenging system, and it includes some gene products, for instance superoxide dismutase (SOD), that are induced under oxidative conditions via a SoxRS regulatory mechanism (4, 7, 15). SOD is found in almost all aerobic organisms and catalyzes the conversion of · O2− to H2O2. Thus, this enzyme is a key component in cellular defense against oxidative stress through reducing the intracellular concentration of · O2− to maintain cellular viability (1, 13). It was demonstrated that Escherichia coli mutants lacking in SOD displayed several defects in phenotypic features, such as auxotrophy for amino acids and high frequencies of spontaneous mutagenesis, when the cells were exposed to an aerobic condition (5, 16, 18).
In our previous work (12), it was reported that the population of SOD-deficient mutant of E. coli IM303 contained a trace of spontaneously derived variant cells which prevailed during the culture under the oxidative stress generated by photoexcited titanium dioxide (TiO2). This result suggested that the variant cells possibly acquired a certain defense system against the oxidative stress besides the SOD mechanism. In the present study, DNA microarray analysis was performed to find out up-regulated genes in the SOD-deficient mutant of E. coli exposed to the oxidative stress produced by photoexcited TiO2. The SOD-deficient and wild-type strains of E. coli were transformed with plasmids carrying the selected genes showing up-regulated expression under the oxidative condition. The transformed E. coli cells were cultivated under varied stress conditions to understand biological functions of the genes in terms of cell growth, ROS content, and amino acid requirement.
A mutant of E. coli, SOD-deficient strain IM303 (I4), and its wild-type strain MM294 were used throughout the experiments. Strain IM303 (I4) was one of isolates obtained from an original population of the mutant (12). A modified M9 medium containing 8 g/liter glucose and 5 mg/liter thiamine was used with supplementation of amino acid mixture (denoted as 2068 mixture, refer to the catalogue from American Type Culture Collection for an ATCC 2068 synthetic medium; 0.25 g Met, 0.25 g Arg, 0.30 g Tyr, 0.60 g Lys, 0.80 g Ile, 0.40 g Phe, 1.5 g Val, 0.50 g Asp, 2.0 g Thr, 0.80 g Glu, and 3.75 g Ser per liter). IM303 (I4) cells were grown at 37°C in an L-type test tube containing 10 ml of the medium with or without 10 mg/liter TiO2 particles (Degussa P25; Nippon Aerosil Co., Tokyo, Japan) under a shaking condition of 45 strokes per min. When necessary, the tube was irradiated at an incident light intensity of I = 12.5 W/m2 with 20-W black light fluorescent lamps (Type FL-20S BL-B; Matsushita Electric Industrial Co., Osaka, Japan). These stress conditions were confirmed to be sublethal for the growth of SOD-deficient E. coli (see the supplemental material). In the case of MM294 culture, the cells were grown in the test tube containing 10 ml of the modified M9 medium with 10 μmol/liter paraquat (PQ) under the same shaking condition in the absence of TiO2 and light. Growth of the cells was monitored by measuring an optical density at 660 nm (OD660). Maximum specific growth rate, μm, and lag time, tL, were estimated by fitting the obtained growth-curve data to the modified Gompertz equation (21).
RNA extraction was conducted as follows, using IM303 (I4) cultures with and without the oxidative stress generated by light-irradiated TiO2. Cell suspension (18 ml) was withdrawn from the cultures at OD660 = 0.15 and was quickly mixed with an equal volume of cold ethanol containing 10% phenol to prevent degradation of intracellular RNA. The cells were collected by centrifugation for 5 min at 4°C and 5,000 × g and then were washed with the cold phenol-ethanol solution. Total RNA was extracted from the whole cells by employing an RNeasy Mini Kit together with an RNase-free DNase Set (QIAGEN Inc., Valencia, CA) according to the supplier's instruction. Purified RNA was recovered by ethanol precipitation after phenol-chloroform treatment.
Labeled cDNAs were prepared using an RNA Fluorescence Labeling Core Kit (Takara Bio Inc., Otsu, Japan) with dUTP-Cy3 or dUTP-Cy5 fluorescent nucleotide (Amersham Biosciences Ltd., Buckinghamshire, United Kingdom) according to the manufacturer's instruction. Assay of DNA microarray was conducted with an IntelliGene E. coli CHIP (Takara Bio Inc.) that contains immobilized cDNAs of 3,437 genes, corresponding to about 80% of predicted open reading frames in E. coli K-12. Hybridization was conducted according to the instruction, and fluorescent intensity on the DNA microarray plate was measured by a GMS 418 Array Scanner (Genetic MicroSystems Inc., Woburn, MA), followed by image analysis using an ImaGene (BioDiscovery Inc., Los Angeles, CA). The microarray profiling data were compared between RNA samples from IM303 (I4) cultures with and without the oxidative stress generated by photoexcited TiO2. For each gene set, the ratio of fluorescent intensities was recorded as a measure of difference in gene expression on the basis of those values from the cells cultivated in the absence of the oxidative stress. In the present study, the genes with the ratios of more than 3 and of less than 0.3 were respectively defined as up-regulated and down-regulated ones in the cells cultivated under the oxidative stress.
The constitutive regions of the selected genes were amplified by PCR using KOD-Dash DNA polymerase (Toyobo Co., Osaka, Japan) from the chromosomal DNA of MM294 cells. A pUC 19 vector was digested with appropriate restriction enzymes, ligated with the amplified DNA fragments with a Ligation-Convenience Kit (Nippon Gene Co., Tokyo, Japan), and then applied for transforming E. coli DH5α. The plasmid DNAs multiplied in the cells were extracted and purified by Wizard Plus SV Minipreps DNA Purification Systems (Promega Corp., Madison, WI).
IM303 (I4) and MM294 cells carrying pUC 19 vector or plasmid with a selected gene insertion were cultivated under indicated conditions, and an aliquot of culture broth (0.5 ml) was withdrawn from the test tube to determine the intracellular ROS content in a middle exponential growth phase (OD660 = 0.5). ROS content in the cells was quantified with 5-(and-6)-chloromethyl-2′,7′-dichlorofluorescin diacetate (C-6827; Molecular Probes Inc., Eugene, OR) and was expressed on an H2O2 equivalent basis, as described elsewhere (12).
To examine the alternation in amino acid requirement, the transformed MM294 cells were grown up to OD660 = 0.2 in the L-tube containing the modified M9 medium with 10 μmol/liter PQ at 37°C. After washing the cells with physiological saline by centrifugation (at 4°C and 10,000 × g for 5 min), a series of diluted cell suspensions was plated on the modified M9 solid media containing 2068 amino acid mixture and the mixture of 20 kinds of amino acids (denoted as 1904 mixture, refer to the catalogue from American Type Culture Collection for ATCC 1904 synthetic medium). The plates were then incubated for 48 h at 37°C. The alteration degree of amino acid requirement, Ad, for the transformed MM294 cells was estimated by numerating developed colonies by the equation Ad = (Na20 − Na11)/Na20, where Na11 and Na20 are the numbers of colonies on the modified M9 plates with 2068 mixture and with 1904 mixture, respectively.
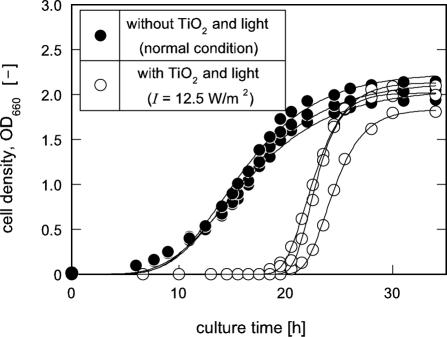
Previous work described that the original population of SOD null mutant of E. coli, IM303, contained a trace of spontaneously derived variant cells, and these variant cells were permitted to survive and prevail during the culture with the sublethal oxidative stress generated from light-irradiated TiO2 (12). As shown in Fig. 1, in the culture of a typically selected isolate, IM303 (I4), with TiO2 and light (I = 12.5 W/m2), maximum specific growth rate, μm, of the cells was found to be approximately two times higher than that of the cells cultivated in the absence of TiO2 and light irradiation (normal culture condition), although a prolonged lag time of tL ≈ 21 h was calculated in the culture with TiO2 receiving light.
FIG. 1.
Growth profiles of a typical isolate, IM303 (I4), from an SOD-deficient mutant of E. coli. The cells were grown in the modified M9 medium with 2068 amino acid mixture with and without TiO2 and light. The solid lines were drawn by fitting the data to the modified Gompertz equation (21).
We then searched the causative genes for physiological changes by means of comparing the difference of total gene expression levels in IM303 (I4) cells cultivated under the conditions with and without the oxidative stress from photoexcited TiO2. DNA microarray analysis indicated 25 up-regulated and 40 down-regulated genes under the oxidative stress (data not shown). Among the 25 up-regulated genes, yfiD, yggB, and yggE genes, which are classified to the genes encoding functionally unknown or hypothetical proteins in GenoBase (http://ecoli.aist-nara.ac.jp/GB5/search.jsp), were focused to be candidates exhibiting the potential to defend bacterial cells. Concerning yfiD gene, a few researchers reported that protein YfiD (14.3 kDa) was expressed mainly at low medium pH, and it was predicted that YfiD endows bacterial cells with tolerance to oxidative stresses (6, 19). The two genes yggB and yggE, which also showed comparatively high expression under the oxidative stress, are functionally still unclear, although Levina et al. (14) suggested that YggB protein, together with MscS, having a mechanosensitive channel activity, may be involved in the regulation of turgor pressure in bacterial cells. As yggE is closely located downstream of yggB, it is likely that these genes constitute a polycistronic transcription unit having protective functions against cellular stresses. In this context, it was expected from the DNA microarray analysis and several papers (6, 14, 19) that yfiD, yggB, and yggE gene products may function to protect bacterial cells or to recover cellular damage from the oxidative stress. These three genes were selected to be cloned into pUC 19 vector, and the resultant plasmids were named pYFD, pYGB, and pYGE, respectively.
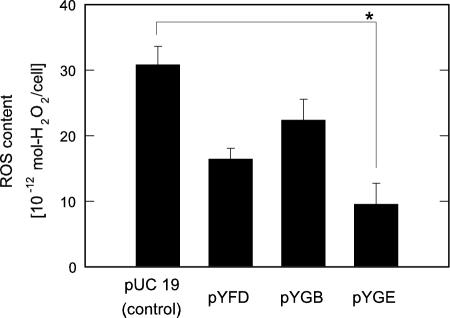
SOD deficiency frequently induces the deteriorated growth of an E. coli mutant even in a rich medium with amino acid supplementation and the failure in restoring vitality due to damage to some biosynthetic pathways of amino acids in the cells (1, 5). Thus, it is most likely that ROS such as · O2− are accumulated in excess to damage the SOD-deficient cells when cultivated with oxygen supply under an aerobic condition. We investigated the effect of the selected genes on the ROS contents in IM303 (I4) cells cultured under a normal aerobic condition. As shown in Fig. 2, the ROS level in IM303 (I4) cells carrying pYGE appreciably dropped to 31% of the control cells carrying pUC 19 (P < 0.001). In the cases of IM303 (I4) cells carrying pYFD and pYGB, on the other hand, the reduction in the ROS contents was not so significant as compared with that in the control cells (P > 0.002).
FIG. 2.
ROS contents in an SOD-deficient mutant of E. coli, IM303 (I4), carrying pUC 19, pYFD, pYGB, and pYGE. The cells were grown in the modified M9 medium with 2068 amino acid mixture as well as 50 mg/liter ampicillin and 10 μmol/liter isopropyl-β-D-thiogalactopyranoside in the absence of TiO2 and light. The asterisk indicates statistical significance (P < 0.001).
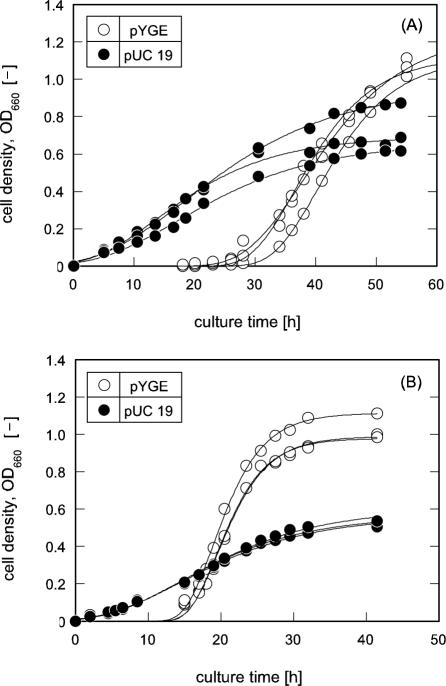
In general, cell growth rate can be linked to an intracellular ROS level, which may be fluctuated in response to oxidative stress against cells, and especially SOD null strains with attenuated DNA-repairing systems exhibit limited growth under oxidative stress in aerobic circumstances (11). As demonstrated in Fig. 3A, IM303 (I4) cells carrying pYGE were remarkably encouraged in their growth activity when compared with that of the control cells carrying pUC 19. The averaged μm value of IM303 (I4) cells with pYGE was 0.06 h−1, which was about three times higher than that of the counterpart.
FIG. 3.
Growth profiles of E. coli cells carrying pUC 19 and pYGE. (A) SOD-deficient mutant of E. coli, IM303 (I4). The cells were grown in the modified M9 medium with 2068 amino acid mixture as well as 50 mg/liter ampicillin and 10 μmol/liter isopropyl-β-D-thiogalactopyranoside (IPTG) in the absence of TiO2 and light. (B) Wild-type E. coli MM294 cells. The cells were grown in the modified M9 medium with 2068 amino acid mixture as well as 10 μmol/liter PQ, 50 mg/liter ampicillin, and 10 μmol/liter IPTG. The solid lines were drawn by fitting the data to the modified Gompertz equation (21).
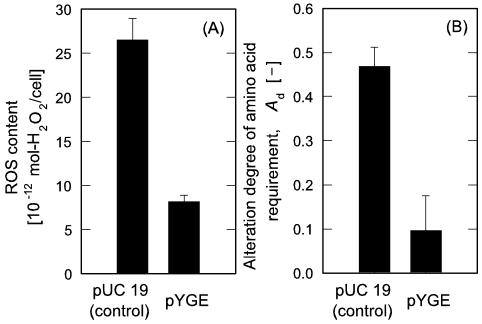
For the extended examination of yggE gene function to reduce intracellular ROS and recover cell growth, the wild-type strain, MM294 cells transformed by pYGE, was cultivated in the medium with 10 μmol/liter PQ, at which the growth rate of the cells was suppressed to about a quarter of that in PQ-free culture (data not shown). As shown in Fig. 3B, MM294 cells with pYGE displayed the profile of significantly recovered growth when compared with the suppressed proliferation of the control cells carrying pUC 19. The averaged μm value of MM294 cells influenced by the yggE gene was 0.1 h−1, which was about five times higher than that of the control cells. We also compared the intracellular ROS contents between the transformants of MM294 cells carrying pUC 19 and pYGE cultivated in the presence of PQ. As indicated in Fig. 4A, the ROS level in the cells with pYGE was approximately 31% of that in the control cells (P < 0.001). It is worth noting that the introduction of pYGE led to a decrease in the ROS content in the wild-type cells cultivated with PQ as a generator of oxidative stress, in accordance with the enhancement of the μm value. These results suggest that E. coli cells get relief from the oxidative stress at an intracellular level and that increased ROS level in the cells by exogenous stress generator can be moderated by yggE gene product.
FIG. 4.
Influence of the yggE gene on ROS content and alteration degree of amino acid requirement in wild-type E. coli MM294 cells. (A) ROS content. (B) Alteration degrees of amino acid requirement. The cells were grown in the modified M9 medium with 2068 amino acid mixture as well as 10 μmol/liter PQ, 50 mg/liter ampicillin, and 10 μmol/liter IPTG.
It is known that SOD-deficient mutants are highly sensitive to oxidative stress and mutagenic DNA lesions are promoted in an aid-of-Fenton reaction in the cells, enhancing the frequency of oxygen-dependent mutagenesis (1, 2). Several researchers reported that the SOD deficiency imposed upon auxotrophies of E. coli, especially for some amino acids, when the cells suffered from oxidative stress under an aerobic condition (5, 8, 18). Therefore, we investigated the occurrence of altered amino acid requirement by estimating the Ad value in the culture of MM294 cells under an oxidative stress. As seen in Fig. 4B, the Ad value of MM294 cells with pYGE cultivated with 10 μmol/liter PQ was notably lowered at a level of Ad = 9.6%, the value of which corresponded to one-fifth reduction, as compared with the control cells with pUC 19 cultivated with 10 μmol/liter PQ (P < 0.001). Under the same culture condition, MM294 cells without any plasmid showed the value of Ad = 40%, which was comparable to the cells with pUC 19. These results demonstrated that the introduction of yggE gene effectively suppressed the emergence of the cells with strict amino acid requirement. It is considered that the biosynthetic pathways of amino acids in MM294 cells may be progressively deteriorated under an oxygenated condition, and some of the cells may lose their ability to proliferate under insufficient nutrient conditions. In this viewpoint, it is likely that the changes in amino acid requirement can be responsible for spontaneous occurrence of mutagenic alterations. The result shown in Fig. 4B can support the consideration that yggE gene product protects the genes concerning amino acid biosyntheses from oxidative damage through reducing intracellular ROS to a moderate harmless level.
With respect to yggE gene product, however, details of an antioxidant mechanism are still unclear. Further examinations will be undertaken to elucidate a working mechanism of yggE gene product and to understand a new strategy of living cells to fight against oxidative stress.
Supplementary Material
Acknowledgments
The present study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas (417) (no. 15033241) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farr, S. B., R. D'Ari, and D. Touati. 1986. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc. Natl. Acad. Sci. USA 83:8268-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel, T., and N. J. Holbrook. 2000. Oxidants, oxidative stress and the biology of ageing. Nature 408:239-247. [DOI] [PubMed] [Google Scholar]
- 4.Gaudu, P., N. Moon, and B. Weiss. 1997. Regulation of the soxRS oxidative stress regulon. J. Biol. Chem. 272:5082-5086. [DOI] [PubMed] [Google Scholar]
- 5.Gort, A. S., and J. A. Imlay. 1998. Balance between endogenous superoxide stress and antioxidant defense. J. Bacteriol. 180:1402-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, M. J., S. S. Yoon, and S. Y. Lee. 2001. Proteome analysis of metabolically engineered Escherichia coli producing poly(3-hydroxybutyrate). J. Bacteriol. 183:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidalgo, E., and B. Demple. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imlay, J. A., and I. Fridovich. 1992. Suppression of oxidative envelope damage by pseudoreversion of a superoxide dimutase-deficient mutant of Escherichia coli. J. Bacteriol. 174:953-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inaoka, T., Y. Matsumura, and T. Tsuchido. 1998. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J. Bacteriol. 180:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keyer, K., A. S. Gort, and J. A. Imlay. 1995. Superoxide and the production of oxidative DNA damage. J. Bacteriol. 177:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, S. Y., M. Nishioka, and M. Taya. 2004. Promoted proliferation of SOD-deficient mutant of Escherichia coli under oxidative stress induced by photoexcited TiO2. FEMS Microbiol. Lett. 236:109-114. [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y. C., C. D. Miller, and A. J. Anderson. 2000. Superoxide dismutase activity in Pseudomonas putida affects utilization of sugars and growth on root surfaces. Appl. Environ. Microbiol. 66:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levina, N., S. Tötemeyer, N. R. Stokes, P. Louis, M. A. Jones, and I. R. Booth. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18:1730-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liochev, S. I., L. Benov, D. Touati, and I. Fridovich. 1999. Induction of the soxRS regulon of Escherichia coli by superoxide. J. Biol. Chem. 274:9479-9481. [DOI] [PubMed] [Google Scholar]
- 16.Nunoshiba, T., F. Obata, A. C. Boss, S. Oikawa, T. Mori, S. Kawanishi, and K. Yamamoto. 1999. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 274:34832-34837. [DOI] [PubMed] [Google Scholar]
- 17.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 18.Touati, D. 2002. Investigating phenotypes resulting from a lack of superoxide dismutase in bacterial null mutants. Methods Enzymol. 349:145-154. [DOI] [PubMed] [Google Scholar]
- 19.Wagner, A. F. V., S. Schultz, J. Bomke, T. Pils, W. D. Lehmann, and J. Knappe. 2001. YfiD of Escherichia coli and Y06I of bacteriophage T4 as automonous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Bioph. Res. Commun. 285:456-462. [DOI] [PubMed] [Google Scholar]
- 20.Woodmansee, A. N., and J. A. Imlay. 2002. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electron to intracellular free iron. J. Biol. Chem. 277:34055-34066. [DOI] [PubMed] [Google Scholar]
- 21.Zwietering, M. H., L. Jongenburger, F. M. Rombouts, and K. van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.