Abstract
The skin of anuran species is a protective barrier against predators and pathogens, showing also chemical defense by substances that represent a potential source for bioactive substances. This review describes the current chemical and biological knowledge from the skin secretions of Leptodactylidae species, one of the most diverse neotropical frog families. These skin secretions reveal a variety of substances such as amines (12), neuropeptides (16), and antimicrobial peptides (72). The amines include histamine and its methylated derivatives, tryptamine derivatives and quaternary amines. The peptides of Leptodactylidae species show molecular weight up to 3364 Da and ocellatins are the most reported. The peptides exhibit commonly glycine (G) or glycine-valine (GV) as C-terminal amino acids, and the most common N-terminal amino acids are glutamic acid (E), lysine (K), and valine (V). The substances from Leptodactylidae species have been evaluated against pathogenic microorganisms, particularly Escherichia coli and Staphylococcus aureus, and the most active peptides showed MIC of 1-15 µM. Furthermore, some compounds showed also pharmacological properties such as immunomodulation, treatment of degenerative diseases, anticancer, and antioxidant. Currently, only 9% of the species in this family have been properly studied, highlighting a large number of unstudied species such as an entire subfamily (Paratelmatobiinae). The ecological context, functions, and evolution of peptides and amines in this family are poorly understood and represent a large field for further exploration.
Keywords: Antimicrobial peptides, Peptides, Amines, Antibiotic resistance
Background
Amphibian skin has a wide range of physiological functions, including defense against predators and microorganisms through the secretion of chemical substances, gas exchange, and water balance [1, 2]. These animals have a great variety of predators, such as mammals, birds, snakes, and spiders, resulting in a diverse array of defensive substances [1]. Alkaloids from poison frogs and toads (e.g. Dendrobatidae and Bufonidae), for example, can be noxious to predators, while proteins from Bufonidae, Hylidae, Leptodactylidae, and Odontophynidae can reduce palatability [3-6]. Amphibians are exposed to diverse environmental conditions, and their skin must protect them from microorganisms found in water, soil, and air [7-9]. As a result, they rely on chemical defenses, which can be peptide-based and supplemented by other substances, such as alkaloids. [10-12].
The metabolites associated with chemical defense are generally stored in the epithelial glands [9]. The most common glands present in amphibian skin are mucus and granular glands, although some species carry specialized glands with particular functions [13, 14]. Mucus glands are specific for mechanical functions, such as lubrication in aquatic environments and humidification in terrestrial environments [15]. Mucus is primarily commonly related to mechanical functions, lubrication, and humidification, but it also plays a role in water balance and gas exchange, exhibiting antimicrobial properties occasionally [1, 15]. Granular glands, on the other hand, are more specialized in defense against predators and microbial infections, which accumulate peptides, alkaloids, and amines, exhibiting various biological properties, such as prevention of microbial infections [16-18].
Due to the natural exposure to pathogens and the species diversity of amphibians, the study of skin secretions represents a great potential to discover new bioactive molecules [1, 16, 19, 20]. This represents a great opportunity to counter public health issues, such as bacterial infections exacerbated by resistant strains and the ability of bacteria to evade therapeutic antibiotics through biofilm formation. Bacterial infections also carry high morbidity and mortality rates, estimating an increase in deaths that may surpass cancer deaths in 2050 [21]. This estimation has been exacerbated by the drug-resistant bacteria, in particular Staphylococcus aureus and their resistant strains to methicillin (MRSA), beta-lactams, and carbapenems [22-24]. This health problem was intensified by the COVID-19 pandemic due to the irrational use of antibiotics [25, 26], as well as bacterial biofilms with recurrent infections [27]. Although previous reviews have presented the chemical composition of the skin secretion of anurans [11, 12], topics related to antimicrobial activities and ecological functions have been overlooked. Besides, other anuran families, such as Bufonidae and Dendrobatidae, overshadow leptodactylids species. Here, we review the current knowledge about the skin secretion of Leptodactylidae species and their potential applications. We restricted our research to the current species of the Leptodactylidae following Frost [20]. As the family systematics and taxonomy have been continuously modified [28-31], we update data of the species name to avoid confusion about chemistry, systematics, and chemotaxonomy (Additional file 1). Species without information about collection locality or with uncertainty about species determination were updated using synonymy by Frost [20].
Therefore, this review was based on previous chemical and biological studies from Leptodactylidae (Anura) focused on skin peptides and other substances, especially against pathogenic microorganisms, such as the antimicrobial peptides (AMPs), in addition to the ecology and evolution of the explored substances. The antimicrobial peptides (AMPs) of anurans from skin secretions have been targeted in several studies. They have also shown antiviral properties against several types of viruses, such as dengue, influenza A (H1N1 and H5N1), human immunodeficiency virus (HIV), human papillomavirus (HPV), herpes simplex, Zika virus, and SARS-CoV-2. Their antiviral mechanism actions have been described by interaction or disruption of capsid virus, suppression of gene expression, modulation of the immune system, blocking of the virus entry into cells, and inhibition of viral replication or synthesis of proteins [32].
Leptodactylid frogs
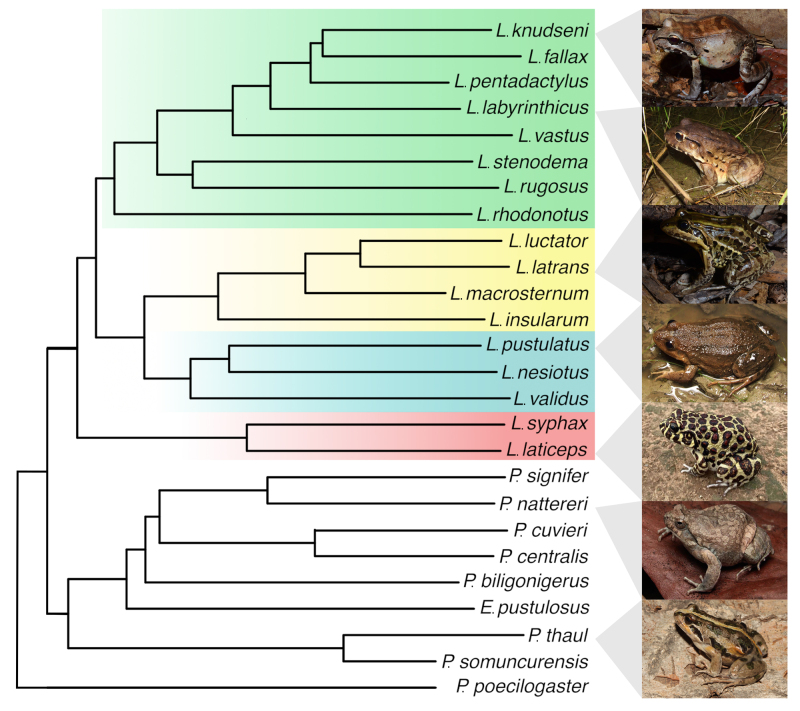
Leptodactylidae Werner, 1896 is one of the most diverse and widely distributed frog families in the neotropical region [20], and it presents large potential to research new bioactive compounds. Frogs in this family can be found from Mexico (Sonora) throughout Central and South America to Argentina and Brazil, including northern Antilles [20]. Leptodactylidae comprises more than 230 species (Figure 1), distributed in three monophyletic subfamilies: Leiuperinae, Leptodactylinae, and Paratelmatobiinae [33]. Leiuperine has 101 species distributed in five genera (Edalorhina, Engystomops, Physalaemus, Pleurodema, and Pseudopaludicola). Leptodactylinae shows 118 species distributed in four genera (Adenomera, Hydrolaetare, Leptodactylus, and Lithodytes), while Paratelmatobiinae represents 15 species in four genera (Crossodactylodes, Cochran, Paratelmatobius, Rupirana and Scythrophrys) [20, 33]. Leptodactylus, the most diverse genus in the family, includes 84 species arranged in four species groups (L. fuscus, L. latrans, L. melanonotus, and L. pentadactylus), according to molecular phylogeny, reproductive modes, anatomy, and additional behavioral characteristics [28, 34].
Figure 1. Phylogenetic tree of Leptodactylidae species with studies of skin secretion. Leptodactylus knudseni (Photo by Diego Santana), L. fallax, L. pentadactylus, L. labyrinthicus (Photo by Diego Santana), L. vastus, L. stenodema, L. rugosus, L. rhodonotus, L. luctator, L. latrans (Photo by Diego Santana), L. macrosternum, L. insularum, L. pustulatus (Photo by Diego Santana), L. nesiotus, L. validus, L. syphax, L. laticeps (Photo by Hugo Cabral), Physalaemus signifier, P. nattereri (Photo by Diego Santana), P. cuvieri, P. centralis, P. biligonigerus, Engystomops pustulosus, Pleurodema thaul (Photo by Diego Baldo), P. somuncurensis, Paratelmatobius poecilogaster (outgroup). Colours represent species groups of Leptodactylus: L. pentadactylus group (green), L. latrans group (yellow), L. melanonotus (blue), and L. fuscus group (red).
Most species of Leptodactylidae are terrestrial, can be found in open formations in forested areas, and feed in leaf litter or close to temporary ponds [28]. Although these species can commonly habit lowland ecosystems, several of them can reach high mountainous areas over 1200 meters above sea levels (m.a.s.l.), such as Leptodactylus fragilis, L. fuscus, L. savagei, and L. ventrimaculatus [35]. Further, L. colombiensis can reach 2800 m.a.s.l. in the Colombian Cordillera Oriental [35]. Additionally, several endemic species are from high-altitude ecosystems (e.g. Leptodactylus oreomantis and Physalaemus rupestris) [36, 37].
Representative species (Figure 1) for the study of skin metabolites from Leptodactylidae showed extensive distributions such as Leptodactylus knudseni (Bolivia, Brazil, Colombia, Ecuador, French Guiana, Guyana, Peru, Suriname, and Venezuela), L. fallax (Jamaica and Puerto Rico), L. pentadactylus (Brazil, Colombia, Ecuador, French Guiana, Guyana, Peru, Suriname, and Venezuela), L. labyrinthicus (Argentina, Brazil and Paraguay), L. vastus (Bolivia and Brazil), L. stenodema (Brazil, Colombia, Ecuador, French Guiana, Guyana, Peru, and Suriname), L. rugosus (Brazil, Guyana, and Venezuela), L. rhodonotus (Bolivia, Brazil, Colombia, and Peru), L. fallax (Jamaica and Puerto Rico), L. luctator (Argentina, Bolivia, Brazil, and Uruguay), L. latrans (Brazil), L. macrosternum (Argentina, Bolivia, Brazil, Colombia, French Guiana, Guyana, Paraguay, Peru, Suriname, Trinidad and Tobago, Uruguay, and Venezuela), L. insularum (Colombia, Costa Rica, Panama, Trinidad and Tobago and Venezuela), L. pustulatus (Brazil), L. nesiotus (French Guiana, Guyana, Suriname, Trinidad and Tobago), L. validus (Brazil, Colombia, Guyana, Suriname, Trinidad and Tobago, and Venezuela), L. syphax (Bolivia, Brazil, and Paraguay), L. laticeps (Argentina, Bolivia, and Paraguay), Physalaemus nattereri (Bolivia, Brazil, and Paraguay), P. cuvieri (Argentina, Bolivia, Brazil, Guyana, Paraguay, Uruguay, and Venezuela), P. centralis (Bolivia, Brazil, and Paraguay), P. bibigonigerus (Argentina, Bolivia, Brazil, Paraguay, and Uruguay), and Engystomops pustulatus (Ecuador and Peru) [20]. However, there are species with restricted distributions, such as Physalaemus. signifier (Brazil), Pleuroderma thaul (Argentina), and P. sumoncurensis (Argentina) [20].
Skin metabolites of Leptodactylidae
The main substances described in the skin secretion of Leptodactylidae species are amines and peptides (Tables 1 and 2). These compounds were 12 amines from 15 species of one genus and 88 peptides classified as neuroactive peptides (16) and antimicrobial peptides (72) from 25 species of four genera. Leptodactylus is the genus with a higher number of peptides described.
Table 1. Amines from the skin secretion of the Leptodactylidae species.
| Amine | Species | Chromatographic analysis | Reference |
|---|---|---|---|
| 5-hydroxytryptamine (5-HT) (C10H12N2O, MW 176.2) | Leptodactylus labrosus | ACC | [40] |
| Leptodactylus labyrinthicus | PC | [41,42] | |
| Leptodactylus labyrinthicus | ACC | [40] | |
| Leptodactylus laticeps | PC | [40-42] | |
| Leptodactylus melanonotus | PC | [41,42] | |
| Leptodactylus pentadactylus | PC | [40-42] | |
| Leptodactylus petersii | PC | [41,42] | |
| Leptodactylus podicipinus | PC | [41,42] | |
| Leptodactylus rhodonotus | PC | [40,41] | |
| Leptodactylus vilarsi | ACC | [40] | |
| Leptodactylus stenodema | ACC | [40] | |
| 6-Methylspinaceamine (C7H11N3, MW 137.2) | Leptodactylus labyrinthicus | PC | [41,42] |
| Bufotenidine (C13H18N2O, MW 218.3) | Leptodactylus labrosus | ACC | [40] |
| Leptodactylus melanonotus | PC | [41,42] | |
| Leptodactylus pentadactylus | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus petersii | PC | [41,42] | |
| Leptodactylus podicipinus | PC | [41,42] | |
| Leptodactylus rhodonotus | PC | [41] | |
| ACC | [40] | ||
| Leptodactylus stenodema | ACC | [43] | |
| Leptodactylus vilarsi | ACC | [40] | |
| Candicine (C11H18NO+, MW180.3) | Leptodactylus pentadactylus | PC | [41,42] |
| Dehydrobufotenine (C12H15N2O+, MW 203.3) | Leptodactylus stenodema | ACC | [43] |
| Histamine (C5H9N3, MW 111.1) | Leptodactylus labyrinthicus | PC | [41,42] |
| ACC | [40] | ||
| Leptodactylus laticeps | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus pentadactylus | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus stenodema | ACC | [43] | |
| Leptodactylus vilarsi | ACC | [40] | |
| Leptodactyline (C11H18NO+, MW 180.3) | Leptodactylus bolivianus | PC | [41,42] |
| Leptodactylus bufonius | PC | [41] | |
| Leptodactylus labrosus | ACC | [40] | |
| Leptodactylus labyrinthicus | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus laticeps | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus latinasus | PC | [38] | |
| [41,42] | |||
| Leptodactylus macrosternum | PC | [41,42] | |
| Leptodactylus melanonotus | PC | [41,42] | |
| Leptodactylus pentadactylus | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus petersii | PC | [41,42] | |
| Leptodactylus podicipinus | PC | [41,42] | |
| Leptodactylus rhodonotus | PC | [41,42] | |
| ACC | [40] | ||
| Leptodactylus stenodema | ACC | [38] | |
| Leptodactylus vilarsi | ACC | [40] | |
| N,N-Dimethylhistamine (C7H13N3, MW 139.20) | Leptodactylus labyrinthicus | PC | [41,42] |
| N-Methyl-5-hydroxytryptamine (C11H14N2O, MW 190.2) | Leptodactylus pentadactylus | ACC | [40] |
| Leptodactylus stenodema | ACC | [38] | |
| Leptodactylus vilarsi | ACC | [40] | |
| Leptodactylus melanonotus | PC | [41] | |
| Leptodactylus petersii | PC | [42] | |
| N-Methylhistamine (C6H11N3, MW 125.2) | Leptodactylus labyrinthicus | PC | [41,42] |
| Spinaceamine (C6H9N3, MW 123.2) | Leptodactylus labyrinthicus | PC | [41,42] |
| Leptodactylus laticeps | PC | [41,42] | |
| Tyramine (C8H11NO, MW 137.2) | Leptodactylus pentadactylus | PC | [41,42] |
MW: Molecular weight. PC: Paper Chromatography. ACC: Alumina Chromatography column.
Table 2. Peptides from the skin secretion of the Leptodactylidae family.
| Species | Type | Peptide | Extraction | Technique* | Sequence | MW | Tmass (Esmass) | Reference |
|---|---|---|---|---|---|---|---|---|
| Engystomops pustulosus | NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH₂ | 1284.6 | - | [54] |
| AMP | Tigerinin-1EP | NI | HPLC | GCKTYLIEPPVCT | 1424.7 | 1421.7 | [55] | |
| AMP | pustulosin-1 | NI | HPLC | FWKADVKEIGKKLAAKLAEELAKKLGEQ | 3141.6 | 3141.8 | [55] | |
| AMP | pustulosin-2 | NI | HPLC | FWKADVKEIGKKLAAKLAEELAKKLGEE | 3142.6 | 3142.8 | [55] | |
| AMP | pustulosin-3 | NI | HPLC | DWKETAKELLKKIGAKVAQVISDKLNPAPQ | 3318.7 | 3318.9 | [55] | |
| AMP | pustulosin-4 | NI | HPLC | DWKADAKDILKKIGAKIAQVISDKLNPAPQ | 3274.6 | 3274.8 | [55] | |
| Leptodactylus fallax | - | LASP | NI | HPLC | GLWDDLKAAAKKVVSSLASAAIEKL-NH | 2583.5 | 2513.9 | [56] |
| AMP | Ocellatin-F1/Fallaxin | NI | HPLC | GVVDILKGAAKDIAGHLASKVMNKL-NH₂ | 2547.5 | 2549 | [57] | |
| Leptodactylus insularum | AMP | Ocellatin-1I | NI | HPLC | GLLDLLKGAGKGLLTHLASQIa | 2117.3 | 2117.3 (2117.3) | [58] |
| AMP | Ocellatin-1I (1-16) | NI | HPLC | GLLDLLKGAGKGLLTH | 1605.0 | 1606.0 (1606.0) | [58] | |
| AMP | Ocellatin-2I | NI | HPLC | GLLDFFKGAGKELLTHLASQIa | 2257.2 | 2257.2 (2257.3) | [58] | |
| AMP | Ocellatin-2I (1-16) | NI | HPLC | GLLDFFKGAGKELLTH | 1745.0 | 1746.0 (1746.0) | [58] | |
| AMP | Ocellatin-3I | NI | HPLC | GVIDILKSLGKNILTNLASKLSDNTA | 2697.5 | 2698.5 (2698.6) | [58] | |
| Leptodactylus knudseni | AMP | Ocellatin-K1 | MS | HPLC | GVVDILKGAAKDLAGHLASKVMNKL | 2547.5 | 2547.65 | [59] |
| Leptodactylus labrosus | NP | Caerulein-like peptide | SOE | ACC | - | - | - | [40] |
| Leptodactylus labyrinthicus | AMP | Ocellatin-F1/Fallaxin | SS | HPLC | GVVDILKGAAKDIAGHLASKVMNKL-NH₂ | 2547.5 | 2545.4 (2546.5) | [60] |
| AMP | Ocellatin-LB1 | SS | HPLC | GVVDILKGAAKDIAGHLASKVM-NH₂ | 2192.2 | 2191.2 (2191.1) | [60] | |
| AMP | Ocellatin-LB2 | SS | HPLC | GVVDILKGAAKDIAGHLASKVMN-NH₂ | 2306.3 | 2305.0 (2304.9) | [60] | |
| NP | Caerulein | SOE | ACC | EQDY (HSO3) TGWMDF-NH2 | - | - | [54,61] | |
| NP | Caerulein-like peptide | SOE | ACC | - | - | - | [40] | |
| Leptodactylus laticeps | AMP | Ocellatin-L1 | NI | HPLC | GVVDILKGAAKDLAGHLATKVMNKL-NH₂ | 25614.7 | 2206.3 (2206.3) | [62] |
| AMP | Ocellatin-L2 | NI | HPLC | GVVDILKGAAKDLAGHLATKVMDKL-NH₂ | 25624.6 | 2564 | [63] | |
| AMP | Plasticin-L1 | NI | HPLC | GLVNGLLSSVLGGGQGGGGLLGGIL | 21642.2 | 2165.5 | [63] | |
| NP | Caerulein | SOE | ACC | EQDY (HSO3) TGWMDF-NH₂ | - | - | [54] | |
| NP | Caerulein-like peptide | SOE | ACC | - | - | - | [40] | |
| Leptodactylus latrans | AMP | Ocellatin-1.1 | EST | HPLC | GVVDILKGAGKDLLAH--------- | 16049.2 | - | [64] |
| AMP | Ocellatin-2.1 | EST | HPLC | GVLDIFKDAAKQLIA---------- | 16009.2 | - | [64] | |
| AMP | Ocellatin-3.1 | EST | HPLC | GVLDILKNAAKNILA---------- | 15519.3 | - | [64] | |
| AMP | Ocellatin-5 | EST | HPLC | AVLDILKDVGKGLLSHFMEKV-NH₂ | 23113.0 | 2312.8 | [64] | |
| AMP | Ocellatin-5.1 | EST | HPLC | AVLDILKDVGKGLL----------- | 14528.9 | - | [64] | |
| AMP | Ocellatin-6 | EST | HPLC | AVLDFIKAAGKGLVTNIMEKVG-NH₂ | 22732.8 | 2274.7 | [64] | |
| AMP | Ocellatin-6.1 | EST | HPLC | AVLDFIKAAGKGLVTNIM------- | 18600.5 | - | [64] | |
| Leptodactylus luctator | AMP | Ocellatin-1 | EST | HPLC | GVVDILKGAGKDLLAHLVGKISEKV-NH₂ | 25585.2 | 2559.1 (2560.0) | [65] |
| AMP | Ocellatin-10 | - | - | GLLDFLKAAGKGLVSNLIEKVG | 2241.3 | 2184.8 | [66] | |
| AMP | Ocellatin-11 | - | - | GVLDIFKDAAKQILAHAAEKIG | 2307.3 | 2250.8 | [66] | |
| AMP | Ocellatin-2 | EST | HPLC | GVLDIFKDAAKQILAHAAEKQI-NH₂ | 23783.3 | 2250.3 (2251.6) | [65] | |
| AMP | Ocellatin-3 | EST | HPLC | GVLDILKNAAKNILAHAAEQI-NH₂ | 22012.5 | 2200.8 (2202.5) | [65] | |
| AMP | Ocellatin-4 | EST | HPLC | GLLDFVTGVGKDIFAQLIKQI-NH₂ | 22743.0 | 2274.3 (2 274.2) | [67] | |
| AMP | Ocellatin-7 | - | - | GVVDILKDTGKKLLSHLMEKIG | 2393.4 | 2336.8 | [66] | |
| AMP | Ocellatin-8 | - | - | GVVDILKDTGKKLLSHLMEKVG | 2379.4 | 2322.8 | [66] | |
| AMP | Ocellatin-9 | - | - | GVLDIFKDTGKKLLSHLMEKVG | 2427.4 | 2370.8 | [66] | |
| AMP | P1-Ll-1577 | EST | LC | DEMKLDGFNMHLE-NH₂ | 15776.9 | - | [68] | |
| AMP | P2-Ll-1298 | EST | LC | AAGKGLVSNLLEK-NH₂ | 12987.6 | - | [68] | |
| AMP | P3-Ll-2085 | EST | LC | GLLDFLKAAGKGLVSNLLEK-NH₂ | 20852.2 | - | [68] | |
| Leptodactylus macrosternum | AMP | Ocellatin-C1 | MS | HPLC | GILDFFKGPVKNALAE | 1717.9 | 1718.2 | [59] |
| AMP | Ocellatin-C2 | MS | HPLC | GLLGKGGLLAKVLA | 13088.5 | 1310 | [59] | |
| Leptodactylus nesiotus | AMP | Ocellatin-1N | NI | HPLC | GAVVDILKGAGKNLLSLALNKLSEKV | 2649.6 | 2649.3 (2649.6) | [58] |
| AMP | Ocellatin-2N | NI | HPLC | GAVVDILKDTGKNLLSLALNKLSEKV | 2737.6 | 2737.3 (2737.6) | [58] | |
| AMP | Ocellatin-3N | NI | HPLC | GIFDVLKNLAKGVITSLASa | 1945.1 | 1945.1 (1945.3) | [58] | |
| AMP | Ocellatin-4N | NI | HPLC | GLFDVLKNLAKGVITSLASa | 1945.1 | 1945.1 (1945.3) | [58] | |
| Leptodactylus pentadactylus | AMP | Ocellatin-F1/Fallaxin | - | - | GVVDILKGAAKDIAGHLASKVMNKL-NH₂ | 25474.6 | - | [69] |
| AMP | Ocellatin-P1/ Pentadactylin | NI | HPLC | GLLDTLKGAAKNVVGSLASKVMELK-NH₂ | 25414.6 | 2540.5 (2540.5) | [70] | |
| NP | Caerulein | SOE | ACC | EQDY (HSO3) TGWMDF-NH₂ | - | - | [54] | |
| NP | Caerulein-like peptide | SOE | ACC | - | - | - | [40] | |
| AMP | Ocellatin-PT1 | EST | HPLC | GVFDIIKDAGKQLVAHAMGKIAEKV-NH₂ | 26374.7 | 2639.1 | [18] | |
| AMP | Ocellatin-PT2 | EST | HPLC | GVFDIIKDAGKQLVAHATGKIAEKV-NH₂ | 26074.7 | 2609 | [18] | |
| AMP | Ocellatin-PT3 | EST | HPLC | GVIDIIKGAGKDLIAHAIGKLAEKV-NH2 | 25285.1 | 2530 | [18] | |
| AMP | Ocellatin-PT4 | EST | HPLC | GVFDIIKGAGKQLIAHAMGKIAEKV-NH₂ | 2593.5 | 2595.1 | [18] | |
| AMP | Ocellatin-PT5 | EST | HPLC | GVFDIIKDAGRQLVAHAMGKIAEKV-NH₂ | 2665.5 | 2667.1 | [18] | |
| AMP | Ocellatin-PT6 | EST | HPLC | GVFDIIKGAGKQLIAHAMEKIAEKVGLNKDGN | 3363.8 | 3365.9 | [18] | |
| AMP | Ocellatin-PT7 | EST | HPLC | GVFDIIKGAGKQLIAHAMGKIAEKVGLNKDGN | 3291.8 | 3293.8 | [18] | |
| AMP | Ocellatin-PT8 | EST | HPLC | GVFDIIKGAGKQLIARAMGKIAEKVGLNKDGN | 3310.9 | 3312.9 | [18] | |
| Leptodactylus rhodonotus | NP | Caerulein-like peptide | SOE | ACC | - | - | - | [40] |
| NP | Caerulein | SOE | ACC | EQDY (HSO3) TGWMDF-NH₂ | - | - | [54] | |
| Leptodactylus rugosus | NP | Caerulein | SOE | ACC | EQDY (HSO3) TGWMDF-NH₂ | - | - | [54] |
| Leptodactylus stenodema | NP | Caerulein | SOE | ACC | EQDY (SO3) TGWMDF-NH2 | - | - | [54] |
| NP | Caerulein-like peptide | SOE | ACC | - | - | - | [43] | |
| NP | Caerulein-like peptide | SOE | ACC | - | - | [40] | ||
| Leptodactylus syphax | AMP | Ocellatin-S1/ Syphaxin | EST | HPLC | GVLDILKGAAKDLAGHVATKVINKI | 2543.5 | - | [71] |
| Leptodactylus validus | AMP | Ocellatin-V1 | NI | HPLC | GVVDILKGAGKDLLAHALSKLSEKV-NH₂ | 2560.5 | 2559.5 (2559.5) | [72] |
| AMP | Ocellatin-V2 | NI | HPLC | GVLDILKGAGKDLLAHALSKISEKV-NH₂ | 2574.5 | 2573.6 (2573.5) | [72] | |
| AMP | Ocellatin-V3 | NI | HPLC | GVLDILTGAGKDLLAHALSKLSEKV-NH₂ | 2547.5 | 2546.5 (2546.5) | [72] | |
| Leptodactylus vastus | AMP | Leptoglycin | EST | HPLC | GLLGGLLGPLLGGGGGGGGGLL | 1761.0 | 1762 | [73] |
| AMP | Ocellatin-K1 (1-21) | EST | HPLC | GVVDILKGAAKDLAGHLASKV | 2061.2 | 2062,44 | [74] | |
| AMP | Ocellatin-K1(1-16) | EST | HPLC | GVVDILKGAAKDLAGH | 1562.9 | 1563,82 | [74] | |
| Physalaemus biligonigerus | NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH₂ | 1284.6 | - | [54,75,76] |
| NP | Tachykinins | - | - | - | - | - | [61] | |
| Physalaemus centralis | AMP | PEP1 _N4 | EST | HPLC | GLKEFMKGLAKTALEHIAGALA | 2268.3 | 2268.2 (2268.0) | [77] |
| AMP | PEP2_N5 | EST | HPLC | GLKEFMKGLAKTALEKIAGALA | 2259.3 | 2259.3 (2259.1) | [77] | |
| AMP | PEP4_N6 | EST | HPLC | GLKEFIKGLAKTALEKIAGALA | 2241.3 | 2241.3 (2241.3) | [77] | |
| AMP | PEP5_N7 | EST | HPLC | GLKEFMKDLAKTVVEKIAGALA | 2331.3 | 2331.3 (2331.2) | [77] | |
| NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH2 | 1284.6 | - | [54] | |
| NP | Tachykinins | - | - | - | - | - | [61] | |
| Physalaemus cuvieri | NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH2 | 1284.6 | - | [54] |
| Physalaemus nattereri | AMP | Nattererin-1 | EST | HPLC | QPQPSFKNIVAGAIKVAAEKALNKIMDKLG-NH₂ | 3178.8 | - | [78] |
| AMP | Nattererin-2 | EST | HPLC | QPQPSFRNIVAGAIKVAAEKALNKIMDKLG-NH₂ | 3206.8 | - | [78] | |
| AMP | Ocellatin-1 | EST | HPLC | GVVDILKGAGKDLLAHLVGKISEKV-NH₂ | 2558.5 | - | [78] | |
| AMP | Ocellatin-3 | EST | HPLC | GVLDILKNAAKNILAHAAEQI-NH₂ | 2201.3 | - | [78] | |
| AMP | Ocellatin-5 | EST | HPLC | AVLDILKDVGKGLLSHFMEKV-NH₂ | 2311.3 | - | [78] | |
| NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH2 | 1284.6 | - | [54] | |
| AMP | Antioxidin-I | EST | HPLC | TWYFITPYIPDK | 1542.8 | 1543.69 | [2] | |
| AMP | Nattererin-1 | EST | HPLC | QPQPSFKNIVAGAIKVAAEKALNKIMDKLG-NH₂ | 3178.8 | - | [79] | |
| AMP | Nattererin-2 | EST | HPLC | QPQPSFRNIVAGAIKVAAEKALNKIMDKLG-NH₂ | 3206.8 | - | [79] | |
| NP | (des-Arg9)-Bradykinin | EST | HPLC | RPPGFSPF | 904.4 ( 904.5) | [79] | ||
| NP | (Hyp3)-Bradykinin | EST | HPLC | RPHypGFSPFR | 1076.5 (1076.6) | [79] | ||
| NP | (Hyp3)-Bradykinin-VD | EST | HPLC | RPHypGFSPFRVD | 1290.6 (1290.7) | [79] | ||
| NP | (Hyp3, Thr6)-Bradykinin | EST | HPLC | RPHypGFTPFR | 1090.5 (1090.6) | [79] | ||
| NP | (Hyp3, Thr6)-Bradykinin | EST | HPLC | RPHypGFTPFRIY | 1366.73 (1366.8) | [79] | ||
| NP | (Thr6)-Bradykinin | EST | HPLC | RPPGFTPFR | 1074.5 (1074.6) | [79] | ||
| NP | (Thr6)-Phyllokinins | EST | HPLC | RPPGFTPFRIY | 1350.73 (1350.8) | [79] | ||
| NP | (Thr6, des-Arg9)-Bradykinin | EST | HPLC | RPPGFTPF | 918.4 (918.54) | [79] | ||
| NP | (Val1, Thr6)-Bradykinin | EST | HPLC | VPPGFTPFR | 1017.5 (1017.6) | [79] | ||
| NP | (Val1, Thr6)-Bradykinin-SPA | EST | HPLC | VPPGFTPFRSPA | 1272.6 (1272.7) | [79] | ||
| NP | (Val1, Thr6)-Bradykinin-VD | EST | HPLC | VPPGFTPFRVD | 1231.6 (1231.7) | [79] | ||
| NP | (Val1, Thr6, des-Arg9)-Bradykinin | EST | HPLC | VPPGFTPF | 861.4 (861.5 ) | [79] | ||
| NP | Bradykinin | EST | HPLC | RPPGFSPFR | 1060.5 (1060.6) | [79] | ||
| NP | SO (Hyp3, Thr6)-Phyllokinins | EST | HPLC | RPHypGFTPFRIY(SO3H) | 1446.6 (1446.7) | [79] | ||
| NP | SO (Thr6)-Phyllokinins | EST | HPLC | RPPGFTPFRIY(SO3H) | 1430.6 (1430.8) | [79] | ||
| Physalaemus signifer | NP | Physalaemin | SOE | ACC | EADPDKFYGLM-NH₂ | 1284.6 | - | [54] |
| Pleurodema somuncurense | AMP | somuncurin-1 | EST | HPLC | FIIWPLRYRK-NH₂ | 1390.8 | 1390.8 | [80] |
| AMP | somuncurin-2 | EST | HPLC | FILKRSYPQYY-NH₂ | 1476.8 | 1476.8 | [80] | |
| AMP | somuncurin-3 | EST | HPLC | DDGEEEAESEEANPEENTEGEKKKKCRRRKGSKLLRRCRGVKI-NH₂ | 4986.5 | 4986.5 | [80] | |
| AMP | somuncurin-4.1 | EST | HPLC | TIYPLRSAE-NH₂ | 1048.6 | 1048.6 | [80] | |
| AMP | somuncurin-4.2 | EST | HPLC | YYQVSEERRRDLASLARLYALAR-NH₂ | 2798.5 | 2798.5 | [80] | |
| AMP | somuncurin-4.2a | EST | HPLC | DLASLARLYALAR-NH₂ | 1431.8 | 1431.8 | [80] | |
| AMP | somuncurin-4.3 | EST | HPLC | NNEENELRRRVSFNRAVIHSLLG-NH₂ | 2722.4 | 2722.5 | [80] | |
| AMP | somuncurin-4.3a | EST | HPLC | VSFNRAVIHSLLG-NH₂ | 1411.8 | 1411.8 | [80] | |
| AMP | somuncurin-4.4 | EST | HPLC | GIVSYHPRSSD-NH₂ | 1216.6 | 1216.6 | [80] | |
| AMP | thaulin-3 | EST | HPLC | NLVGSLLGGILKK-NH₂ | 1310.8 | 1310.8 | [80] | |
| AMP | thaulin-Sl | EST | HPLC | DLLNGLLNPVLGIANGLTGGLVKK-NH₂ | 2388.4 | 2388.4 | [80] | |
| Pleurodema thaul | AMP | Gly-Thaulin-1 | SY | HPLC | GNGNLLGGLLRPVLGVVKGLTGGLGKK | 2586.6 | - | [81] |
| AMP | Thaulin-1 | SY | HPLC | NGNLLGGLLRPVLGVVKGLTGGLGKK | 2529.5 | 2531.08 | [81] | |
| AMP | Thaulin-2 | SY | HPLC | ELLGGLLDPVLGVANALTGGIIKK | 2360.4 | 2361.85 | [81] | |
| AMP | Thaulin-3 | SY | HPLC | NLVGSLLGGILKK | 1310.8 | 1311.63 | [81] | |
| AMP | Thaulin-4 | SY | HPLC | DDGEEAESEAANPEENTVGG | 2018.8 | 2019.92 | [81] |
*: chromatographic technique applied for separation or purification of constituents. MW: Molecular weight. NP: Neuropeptide. AMP: Antimicrobial Peptides. ES: Electrical Stimulation. NI: Norepinephrine Injection. SOE: Solvent Extraction. MS: Manual Stimulation. SS: Skin Scraping. ACC: Alumina chromatography column. HPLC: High performance liquid chromatography. Mass expressed in Daltons: Real mass (Theoretical mass). a: Denotes C-terminal -amidation.
Amines
Amines have been described from Leptodactylidae species as summarized in Table 1. The first isolated substance from the skin of a Leptodactylidae species was the biogenic amine leptodactyline, which was isolated in 1959 from Leptodactylus luctator under the name of Leptodactylus ocellatus [38]. Biogenic amines are nitrogenous organic molecules with low molecular weight yielded from the decarboxylation of amino acids or amination and transamination of aldehydes or ketones. These biogenic amines, which are associated with several biological activities, have also been identified in plants, animals, and microorganisms [39].
Erspamer (1971) classified the amines of amphibians into three groups: indole alkylamines, imidazole alkylamines, and hydroxyphenyl alkylamines, and all of them were registered from Leptodactylus spp. [1, 41, 44]. Leptodactyline (Figure 2) was the first m-hydroxyphenyl alkylamine described in animals, and among its functions are: the paralyzation of skeletal muscle, the induction of ganglion stimulation, and the nicotinic actions [45]. This amine has been registered in several other species of Leptodactylus (Table 1, Figure 2).
Figure 2. Chemical structures of the amines of Leptodactylidae species.
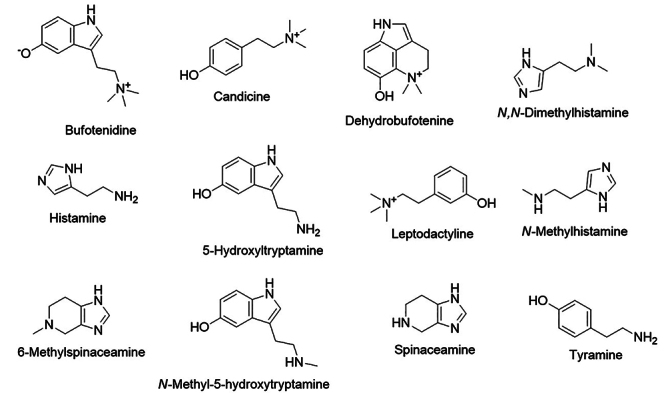
Candicine (Figure 2) is another hydroxyphenyl alkylamine that was first isolated from plants of the family Cactaceae, and it is also found in anuran Leptodactylus pentadactylus. This amine has similar effects observed for leptodactyline in mammals but with lower activity [44].
Indole alkylamines found in Leptodactylus are 5-hydroxytryptamine (5-HT) and its N-methylated derivatives (Table 1). These compounds are also reported from other species of families Ascaphidae, Ceratophryidae, Hylidae, Pelobatidae, Phyllomedusidae, Ranidae, and Rhinodermatidae [46]. Bufotenidine, an indole alkylamine, was initially isolated from European Bufo vulgaris, but it was also found in several species of Bufo and other families and genera [47], including species from Leptodactylidae (Table 1).
The amines belonging to the imidazole alkylamines class described in Leptodactylidae include histamine and spinaceamine and their derivatives. Histamine and its derivates have been reported in Leptodactylus (Table 1), and induce cardiac stimulation and vasoconstriction, comparable to the stimulation effects of adrenaline in mammals [47,48]. Besides, some of these amines are also present in other anurans from Bufonidae, Hylidae, Telmatobiidade, Alsodidae, Odontophrynidae, Myobatrachidae, Microhylidae, Ranidae, Pipidae, Heleophrynidae, and Hyperoliidae [46, 49]. They are also reported only in the genus Leptodactylus of Leptodactylidae (Table 1). Spinaceamine is reported only from L. laticeps and L. labyrinthicus (Figure 1) [50]. Tyramine is a common amine described in both animals and plants [51], but it has been reported only for the Anura L. pentadactylus (Table 1).
Peptides
The peptides are also a large group of substances described in the skin secretions of Leptodactylidae, mainly antimicrobial peptides (AMPs). Peptides are long chains of amino acids linked by a peptide bold [52]. Generally, frog peptides are cationic, varying 8 to 48 amino acid residues with several hydrophobic amino acids and predominant conformation of amphipathic α-helix [53]. Currently, over 80 peptides have been described from Leptodactylidae species (Table 2). For example, somuncurin-3 (DDGEEEAESEEANPEENTEGEKKKKCRRRKGSKLLRRCRGVKI-NH₂) is the greater and (Val1, Thr6, des-Arg9)-Bradykinin (VPPGFTPF) is the smallest peptide, which was described from Pleurodema somuncurense and Physalaemus nattereri, respectively (Table 2).
The peptide constituents of the skin secretions of Leptodactylidae species have an α-helical form, usually reported with an NH2 terminal. The most abundant peptides reported are ocellatins (Table 2). In addition, the most common C-terminal amino acids are glycine (G) or glycine-valine (GV) sequence, while N-terminal amino acids are glutamic acid (E), lysine (K), and valine (V). Glycines (G) seem to be a recurrent amino acid in multiple positions, some peptides are mainly constituted by G (e.g. leptoglycine, plasticin-L1, and Gly-Thaulin-1). Leucines (L) and lysines (Lys) are frequently observed in multiple positions.
There are two major categories of peptides in amphibian skin secretion: the neuroactive peptides (NP) and the antimicrobial peptides (AMPs) [82, 83]. The neuroactive peptides from Leptodactylidae are physalaemin (tachykinin family), bradykinins and their derivatives, caeruleins, and the caeruleins-like peptides (Mean = 1167.3 SD = 47.7; Max = 1446.6; Min = 861.4; n = 16) (Table 2).
Physalaemin, an NP, was reported from several Physalaemus species (Table 2). It exhibits positive effects in stimulating the intestine, ileum, duodenum, bladder, pancreas, and stomach, displaying intense hypotensive activity in mammals. Additionally, it can also induce saliva production and lacrimal secretion in several mammals and some birds [40, 41, 75, 83].
Bradykinin peptides are found in many anuran species, but they were recorded only in Physalaemus from the Leptodactylidae family. Bradykinins contain C-terminal COOH residues, and they are considered the main peptides reported from skin secretions of anurans [83, 84]. Bradykinins exhibit effects on smooth muscles, showing gastrointestinal effects in mammals, and they are also involved in the pain response, and potent immunostimulatory effects [44, 82, 84]. Only Physalaemus nattereri shows bradykinins in the family (Table 2, Figure). Barbosa et al. [79] described bradykinins by sequencing granular and inguinal glands from P. nattereri and observed the genes related to bradykinins are expressed more in inguinal glands, which may be related to behavioral defenses.
The peptide caerulein is a neuropeptide among the most studied in anurans (Table 2). This peptide and caerulein-like polypeptides are described in several Leptodactylus species. They have shown a stimulant effect on gastric and pancreas secretions, resulting in acute pancreatitis, being able to stimulate the musculature of the gut, except in the duodenum. Other effects of caerulein include the reduction of blood pressure at very low doses and sedative effects [44, 82]. Caerulein has also been reported as having potent analgesic properties with an effect 2,000 times higher than morphine [85]. This peptide has been described from L. labyrinthicus, L. laticeps, L. pentadactylus, L. rhodonotus, L. rugosus, and L. stenodema (Table 2, Figure 1).
Although Leptodactylidae has several neuroactive peptides in their skin secretion, a great diversity of antimicrobial peptides (AMPs) has been also described, highlighting the interest in this family for research of antimicrobial molecules. The AMPs have variations in molecular weight (Mean = 7449.8 SD = 957.3; Max = 26374.6; Min = 1048.5; n = 84) and number of amino acids (Table 2).
AMPs are grouped according to their structure as ⍺-helice, ꞵ-sheet, cyclic, and extended peptides, and they constitute the innate immunity system of several organisms, including plants, microorganisms, invertebrates, and vertebrates [86, 87]. Generally, these peptides are amphipathic molecules, containing hydrophobic residues and cationic properties [86, 87]. Due to their properties, AMPs can interact with bacteria membranes and induce a disturbance on its surface, leading to a loss of integrity or developing channels to increase the membrane permeability [86, 88, 89]. Additionally, some AMPs seem to be able to penetrate the bacteria membranes and influence metabolic processes, such as the synthesis of DNA, RNA, and proteins [87].
Several frog peptides, such as plasticin-1 and ocellatin-F1, have been described as solvent-dependent conformations by circular dichroism (CD) and Nuclear Magnetic Resonance (NMR) studies [72, 90, 91]. Plasticin-1, for example, shows a random coil conformation in water, β-sheet in methanol, and α-helical in the solvent trifluoroethanol and water 1:1 (v/v) [92]. The antimicrobial activity of peptides has been related to the complex interactions of factors that include their conformation (α-helicity), hydrophobicity, charge, and amphipathicity [93-95]. Ocellatin-F1 exhibits a strong correlation between its antimicrobial activity and the increase of hydrophobicity, the reduction of polar angles (measure of the amphipathic degree in an α-helical using the vector sum of hydrophobicities) is also correlated positively to the antimicrobial activities [72, 96]. AMPs of Leptodactylus species have the propensity to adopt an α-helical conformation in a membrane mimetic system [73], which is typical behavior for them, acquiring an active conformation in the membrane surface contact [60].
The first AMPs isolated in Leptodactylidae were the peptides ocellatins 1, 2, and 3, found in the secretion of Leptodactylus ocellatus (Table 2) [65]. In addition to ocellatins, other groups of AMPs described in Leptodactylidae were evaluated for a range of bacteria and fungi, as listed in Table 3. Generally, the studies with antimicrobial activity of the AMPs from anurans performed their sequencing and production by solid-phase peptide synthesis to expand the biological and pharmacological properties. Considering the potential antimicrobial of peptides, the minimum inhibitory concentration (MIC) lower than 30 µM are noticed for at least 18 peptides of Leptodactylidae, such as leptoglycin, nattererin-1, nattererin-2, ocellatin-5, ocellatin-6, ocellatin-F1, ocellatin-P1, ocellatin-S/Syphaxin (1-22), ocellatin-S (1-16), thaulin-1 and its derivative Gly-thaulin-1, P1-Ll-1577, P2-Ll-1298, P3-Ll-2085, PEP1_N4, PEP2_N5, PEP4_N6 and PEP5_N7. Among them, PEP4_N6 showed potent antimicrobial activity against the gram-negative Escherichia coli ATCC25922 (MIC = 2 µM) and Klebsiella pneumoniae ATCC 13883 (MIC = 2 µM), followed by PEP2_N5 and ocellatin-S/Syphaxin (1-22) with MIC of 4 µM for E. coli ATCC25922, besides PEP1_N4, PEP2_N5, and PEP5_N7 exhibited MIC of 4 µM for K. pneumoniae. These antimicrobial activities evidence the potential of anuran peptides, which demonstrated potent activities for gram-positive and gram-negative bacteria. For instance, ocellatin S (1-22), P3-Ll-208, and ocellatin-6 showed activity for gram-positive Staphylococcus aureus ATCC29213 with MIC values of 14.6, 15 and 28 µM, respectively. These results demonstrate that the studies of new antimicrobial peptides from skin sections of anurans are promising.
Table 3. MIC values for microorganisms tested with peptides and extracts from the skin secretion of the Leptodactylidae family.
| Species | Substance or extract | Pathogen | Gram | MIC (µM) | Reference |
|---|---|---|---|---|---|
| Engystomops pustulosus | Tigerinin-1EP | Escherichia coli ATCC 35218 | Negative | >125 µM | [55] |
| Staphylococcus aureus ATCC 12600 | Positive | >125 µM | [55] | ||
| pustulosin-1 | Escherichia coli ATCC 35218 | Negative | 125 µM | [55] | |
| Staphylococcus aureus ATCC 12600 | Positive | >125 µM | [55] | ||
| pustulosin-3 | Escherichia coli ATCC 35218 | Negative | 125 µM | [55] | |
| Staphylococcus aureus ATCC 12600 | Positive | >125 µM | [55] | ||
| Leptodactylus fallax | LASP | Escherichia coli | Negative | - | [56] |
| Staphylococcus aureus | Positive | - | [56] | ||
| Ocellatin-F1/Fallaxin | Batrachochytrium dendrobatidis | - | 100 | [69] | |
| Candida albicans ATCC 90028 | Positive | >160 | [57] | ||
| Enterobacter cloacae NHTCC 53001 | Negative | 20 | [57] | ||
| Escherichia coli ATCC 25922 | Negative | 40 | [57] | ||
| Klebsiella pneumoniae KK3 9904 | Negative | 80 | [57] | ||
| Proteus mirabilis ATCC 25933 | Negative | >160 | [57] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | 80 | [57] | ||
| Staphylococcus aureus NCTC 8325 | Positive | >160 | [57] | ||
| Leptodactylus insularum | Ocellatin-1I | Enterococcus faecalis ATCC 51299 | Positive | >250 | [58] |
| Enterococcus faecium ATCC 19434 | Positive | - | [58] | ||
| Escherichia coli ATCC 35218 | Negative | 62.5 | [58] | ||
| Klebsiella pneumoniae ATCC 49472 | Negative | 125 | [58] | ||
| Klebsiella pneumoniae ATCC BAA-2814 | Negative | >125 | [58] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | - | [58] | ||
| Salmonella typhimurium ATCC 14028 | Negative | 250 | [58] | ||
| Staphylococcus aureus ATCC 12600 | Positive | 250 | [58] | ||
| Staphylococcus aureus ATCC BAA-2312 | Positive | 250 | [58] | ||
| Ocellatin-2I | Enterococcus faecalis ATCC 51299 | Positive | >250 | [58] | |
| Enterococcus faecium ATCC 19434 | Positive | 250 | [58] | ||
| Escherichia coli ATCC 35218 | Negative | 62.5 | [58] | ||
| Klebsiella pneumoniae ATCC 49472 | Negative | 125 | [58] | ||
| Klebsiella pneumoniae ATCC BAA-2814 | Negative | 125 | [58] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | >125 | [58] | ||
| Salmonella typhimurium ATCC 14028 | Negative | 125 | [58] | ||
| Staphylococcus aureus ATCC 12600 | Positive | >250 | [58] | ||
| Staphylococcus aureusATCC BAA-2312 | Positive | >250 | [58] | ||
| Leptodactylus labyrinthicus | Ocellatin-F1/Fallaxin | Aggregatibacter actinomycetemcomitans ATCC 29522 | Negative | 24.84 | [60] |
| Candida lusitaniae ATCC 56936 | - | 50.25 | [60] | ||
| Escherichia coli ATCC 25922 | Negative | 397.45 | [60] | ||
| Staphylococcus aureusATCC 25923 | Positive | 109.91 | [60] | ||
| Ocellatin-LB1 | Aggregatibacter actinomycetemcomitans ATCC 29522 | Negative | 222.37 | [60] | |
| Candida albicans ATCC 18804 | - | 233.55 | [60] | ||
| Candida lusitaniae ATCC 56936 | - | 233.55 | [60] | ||
| Escherichia coli ATCC 25922 | Negative | 114.04 | [60] | ||
| Ocellatin-LB2 | Aggregatibacter actinomycetemcomitans ATCC 29522 | Negative | 210.04 | [60] | |
| Leptodactylus laticeps | Ocellatin-L1 | Candida albicans ATCC 90028 | Positive | >200 | [62] |
| Enterobacter cloacae HNTCC 53001 | Negative | 50 | [62] | ||
| Enterococcus faecalis ATCC 29212 | Positive | >200 | [62] | ||
| Escherichia coli ATCC 25922 | Negative | 50 | [62] | ||
| Klebsiella pneumoniae KK3 9904 | Negative | 100 | [62] | ||
| Proteus mirabilis ATCC 25933 | Negative | >200 | [62] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | 100 | [62] | ||
| Staphylococcus aureus NCTC 8325 | Positive | >200 | [62] | ||
| Staphylococcus epidermidis RP62A | Positive | >200 | [62] | ||
| Ocellatin-L2 | Escherichia coli ATCC 25726 | Negative | >500 | [62] | |
| Staphylococcus aureusATCC 25923 | Positive | >500 | [62] | ||
| Plasticin-L1 | Escherichia coli ATCC 25726 | Negative | >500 | [62] | |
| Staphylococcus aureus ATCC 25923 | Positive | >500 | [62] | ||
| Leptodactylus latrans | Ocellatin-5 | Escherichia coli ATCC 25922 | Negative | 64 µg/ml | [64] |
| Staphylococcus aureusATCC 29213 | Positive | 128 µg/ml | [64] | ||
| Ocellatin-6 | Escherichia coli ATCC 25922 | Negative | 32 µg/ml | [64] | |
| Staphylococcus aureusATCC 29213 | Positive | 64 µg/ml | [64] | ||
| Leptodactylus luctator | Fraction >1kDa | Bacillus cereus DBFIQB28 | Positive | - | [97] |
| Escherichia coli DBFIQ Ec9 | Negative | - | [97] | ||
| Mycobacterium tuberculosis H37Rv | - | 187.5 µg/mL | [97] | ||
| Pseudomonas sp DBFIQ P 55 | Negative | - | [97] | ||
| Staphylococcus aureusDBFIQ S 21 | Positive | - | [97] | ||
| Fraction >2kDa | Bacillus cereus DBFIQB28 | Positive | - | [97] | |
| Escherichia coli DBFIQ Ec9 | Negative | - | [97] | ||
| Mycobacterium tuberculosis H37Rv | - | NI | [97] | ||
| Pseudomonas sp DBFIQ P 55 | Negative | - | [97] | ||
| Staphylococcus aureusDBFIQ S 21 | Positive | - | [97] | ||
| Methanol extract | Bacillus cereus DBFIQB28 | Positive | - | [97] | |
| Escherichia coli DBFIQ Ec9 | Negative | - | [97] | ||
| Mycobacterium tuberculosis H37Rv | - | 187.5 µg/mL | [97] | ||
| Pseudomonas sp DBFIQ P 55 | Negative | - | [97] | ||
| Staphylococcus aureus DBFIQ S 21 | Positive | - | [97] | ||
| Ocellatin-1 | Escherichia coli ATCC 25922 | Negative | - | [65] | |
| Ocellatin-2 | Escherichia coli ATCC 25922 | Negative | - | [65] | |
| Ocellatin-3 | Escherichia coli ATCC 25922 | Negative | - | [65] | |
| Ocellatin-4 | Escherichia coli ATCC 25922 | Negative | 64 | [67] | |
| Staphylococcus aureus ATCC 29213 | Positive | 64 | [67] | ||
| P1-Ll-1577 | Escherichia coli ATCC 25922 | Negative | 20 | [68] | |
| Staphylococcus aureusATCC 25923 | Positive | 40.5 | [68] | ||
| P2-Ll-1298 | Escherichia coli ATCC 25922 | Negative | 24.6 | [68] | |
| Staphylococcus aureusATCC 25923 | Positive | 49 | [68] | ||
| P3-Ll-2085 | Escherichia coli ATCC 25922 | Negative | 15 | [68] | |
| Staphylococcus aureusATCC 25923 | Positive | 15 | [68] | ||
| TAS | Bacillus cereus DBFIQB28 | Positive | - | [97] | |
| Escherichia coli DBFIQ Ec9 | Negative | - | [97] | ||
| Mycobacterium tuberculosis H37Rv | - | NI | [97] | ||
| Pseudomonas sp DBFIQ P 55 | Negative | - | [97] | ||
| Staphylococcus aureusDBFIQ S 21 | Positive | - | [97] | ||
| Leptodactylus macrosternum | Fatty Extract | Candida albicans ICB 12 | - | >1040 | [98] |
| Candida krusei ATCC 6258 | - | 512 | [98] | ||
| Escherichia coli ATCC 10532 | Negative | >1040 | [98] | ||
| Klebsiella pneumoniae ATCC 4362 | Negative | >1040 | [98] | ||
| Pseudomonas aeruginosa ATCC 15442 | Negative | 256 | [98] | ||
| Staphylococcus aureusATCC 25923 | Positive | >1040 | [98] | ||
| Leptodactylus nesiotus | Ocellatin-1N | Enterococcus faecalis ATCC 51299 | Positive | >250 | [58] |
| Enterococcus faecium ATCC 19434 | Positive | 250 | [58] | ||
| Escherichia coli ATCC 35218 | Negative | 62.5 | [58] | ||
| Klebsiella pneumoniae ATCC 49472 | Negative | 125 | [58] | ||
| Klebsiella pneumoniae ATCC BAA-2814 | Negative | 125 | [58] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | >125 | [58] | ||
| Salmonella typhimurium ATCC 14028 | Negative | 250 | [58] | ||
| Staphylococcus aureus ATCC BAA-2312 | Positive | 250 | [58] | ||
| Staphylococcus aureusATCC 12600 | Positive | 250 | [58] | ||
| Ocellatin-3N | Enterococcus faecalis ATCC 51299 | Positive | 250 | [58] | |
| Enterococcus faecium ATCC 19434 | Positive | 62.5 | [58] | ||
| Escherichia coli ATCC 35218 | Negative | 31.25 | [58] | ||
| Klebsiella pneumoniae ATCC 49472 | Negative | 62.5 | [58] | ||
| Klebsiella pneumoniae ATCC BAA-2814 | Negative | 62.5 | [58] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | 62.5 | [58] | ||
| Salmonella typhimurium ATCC 14028 | Negative | 62.5 | [58] | ||
| Staphylococcus aureus ATCC 12600 | Positive | 31.25 | [58] | ||
| Staphylococcus aureusATCC BAA-2312 | Positive | 31.25 | [58] | ||
| Leptodactylus pentadactylus | Ocellatin-P1/ Pentadactylin | Candida albicans ATCC 90028 | Positive | >200 | [70] |
| Enterobacter cloacae HNTCC 53001 | Negative | 50 | [70] | ||
| Enterococcus faecalis ATCC 29212 | Positive | 200 | [70] | ||
| Escherichia coli ATCC 25922 | Negative | 25 | [70] | ||
| Klebsiella pneumoniae KK3 9904 | Negative | 100 | [70] | ||
| Proteus mirabilis ATCC 25933 | Negative | >200 | [70] | ||
| Pseudomonas aeruginosa ATCC 27853 | Negative | 100 | [70] | ||
| Staphylococcus aureus NCTC 8325 | Positive | 200 | [70] | ||
| Staphylococcus epidermidis RP62A | Positive | 100 | [70] | ||
| Streptococcus Group B HNTCC 80130 | Positive | 50 | [70] | ||
| Leptodactylus pustulatus | Ocellatin-PT1 | Escherichia coli ATCC 25922 | Negative | 300 | [18] |
| Klebsiella pneumoniae ATCC 700603 | Negative | >300 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | >300 | [18] | ||
| Staphylococcus aureusATCC 29313 | Positive | >300 | [18] | ||
| Ocellatin-PT2 | Escherichia coli ATCC 25922 | Negative | >310 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | >310 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | >310 | [18] | ||
| Staphylococcus aureus ATCC 29313 | Positive | >310 | [18] | ||
| Ocellatin-PT3 | Escherichia coli ATCC 25922 | Negative | 320 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | >320 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | >320 | [18] | ||
| Staphylococcus aureus ATCC 29313 | Positive | >320 | [18] | ||
| Ocellatin-PT4 | Escherichia coli ATCC 25922 | Negative | 80 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | 310 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | 310 | [18] | ||
| Staphylococcus aureusATCC 29313 | Positive | >310 | [18] | ||
| Ocellatin-PT5 | Escherichia coli ATCC 25922 | Negative | 300 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | >300 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | >300 | [18] | ||
| Staphylococcus aureus ATCC 29313 | Positive | >300 | [18] | ||
| Ocellatin-PT6 | Escherichia coli ATCC 25922 | Negative | 120 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | >240 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | >240 | [18] | ||
| Staphylococcus aureusATCC 29313 | Positive | >240 | [18] | ||
| Ocellatin-PT7 | Escherichia coli ATCC 25922 | Negative | 60 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | >240 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | 240 | [18] | ||
| Staphylococcus aureusATCC 29313 | Positive | 240 | [18] | ||
| Ocellatin-PT8 | Escherichia coli ATCC 25922 | Negative | 60 | [18] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | 240 | [18] | ||
| Salmonella choleraesuis ATCC 14028 | Negative | 240 | [18] | ||
| Staphylococcus aureusATCC 29313 | Positive | 240 | [18] | ||
| Leptodactylus syphax | Syphaxin (1-16) | Escherichia coli ATCC 25922 | Negative | 10.6 | [71] |
| Staphylococcus aureus ATCC 29213 | Positive | 40.5 | [71] | ||
| Syphaxin (1-22) | Escherichia coli ATCC 25922 | Negative | 40.5 | [71] | |
| Staphylococcus aureus ATCC 29213 | Positive | 14.6 | [71] | ||
| Leptodactylus validus | Ocellatin-V1 | Escherichia coli ATCC 25923 | Negative | >200 | [72] |
| Staphylococcus aureus ATCC 25726 | Positive | >200 | [72] | ||
| Ocellatin-V2 | Escherichia coli ATCC 25923 | Negative | >200 | [72] | |
| Staphylococcus aureus ATCC 25726 | Positive | >200 | [72] | ||
| Ocellatin-V3 | Escherichia coli ATCC 25923 | Negative | >200 | [72] | |
| Staphylococcus aureus ATCC 25726 | Positive | >200 | [72] | ||
| Fat-Extract | Candida albicans ICB 12 | - | >1040 | [98] | |
| Candida krusei ATCC 6258 | - | 256 | [98] | ||
| Escherichia coli ATCC 10532 | Negative | >1040 | [98] | ||
| Klebsiella pneumoniae ATCC 4362 | Negative | >1040 | [98] | ||
| Pseudomonas aeruginosa ATCC 15442 | Negative | 512 | [98] | ||
| Staphylococcus aureusATCC 25923 | Positive | >1040 | [98] | ||
| Leptoglycin | Candida albicans CEMM 01-3-075 | - | >200 | [73] | |
| Candida tropicalis CEMM 01-2-078 | - | >200 | [73] | ||
| Citrobacter freundii ATCC 8090 | Negative | 75 | [73] | ||
| Enterococcus faecalis ATCC 29912 | Positive | >200 | [73] | ||
| Escherichia coli ATCC 28922 | Negative | 50 | [73] | ||
| Micrococcus luteus ATCC 29912 | Positive | >200 | [73] | ||
| Microporum canis CEMM 01-2-133 | - | >200 | [73] | ||
| Pseudomonas aeruginosa ATCC 9027 | Negative | 8 | [73] | ||
| Staphylococcus aureusATCC 25.923 | Positive | >200 | [73] | ||
| Trichophyton rubrum CEMM0 1-1-100 | - | >200 | [73] | ||
| Ocellatin-K1 (1-21) | Escherichia coli ATCC 25922 | Negative | 125 μg/ml | [74] | |
| Staphylococcus aureus ATCC 25923 | Positive | NI | [74] | ||
| Ocellatin-K1(1-16) | Escherichia coli ATCC 25922 | Negative | 125 μg/ml | [74] | |
| Staphylococcus aureus ATCC 25923 | Positive | 31.25μg/ml | [74] | ||
| Physalaemus nattereri | Antioxidin-I | Enterococcus faecalis ATCC 29212 | Positive | 256 µg/ml | [2] |
| Escherichia coli ATCC 25922 | Negative | >1024 µg/ml | [2] | ||
| Pseudomonas aeruginosa ATCC 27853 ATCC 27853 | Negative | >1024 µg/ml | [2] | ||
| Staphylococcus aureusATCC 25923 | Positive | >1024 µg/ml | [2] | ||
| Escherichia coli ATCC 25922 | Negative | 10 | [79] | ||
| Nattererin-2 | Escherichia coli ATCC 25922 | Negative | 10 | [79] | |
| PEP1_N4 | Candida albicans ATCC 14053 | - | >128 | [77] | |
| Escherichia coli ATCC 25922 | Negative | 8 | [77] | ||
| Klebsiella pneumoniae ATCC 13883 | Negative | 4 | [77] | ||
| Staphylococcus aureus ATCC 25923 | Positive | 32 | [77] | ||
| Staphylococcus epidermidis ATCC 12228 | Positive | 64 | [77] | ||
| PEP2_N5 | Escherichia coli ATCC 25922 | Negative | 4 | [77] | |
| Klebsiella pneumoniae ATCC 13883 | Negative | 4 | [77] | ||
| Staphylococcus aureus ATCC 25923 | Positive | 64 | [77] | ||
| Staphylococcus epidermidis ATCC 12228 | Positive | 64 | [77] | ||
| PEP4_N6 | Candida albicans ATCC 14053 | - | >128 | [77] | |
| Escherichia coli ATCC 25922 | Negative | 2 | [77] | ||
| Klebsiella pneumoniae ATCC 13883 | Negative | 2 | [77] | ||
| Staphylococcus aureus ATCC 25923 | Positive | ND | [77] | ||
| Staphylococcus epidermidis ATCC 12228 | Positive | ND | [77] | ||
| PEP5_N7 | Candida albicans ATCC 14053 | - | >128 | [77] | |
| Escherichia coli ATCC 25922 | Negative | 32 | [77] | ||
| Klebsiella pneumoniae ATCC 13883 | Negative | 4 | [77] | ||
| Staphylococcus aureus ATCC 25923 | Positive | ND | [77] | ||
| Staphylococcus epidermidis ATCC 12228 | Positive | 128 | [77] | ||
| PEP2_N5 | Candida albicans ATCC 14053 | - | >128 | [77] | |
| Pleurodema somuncurense | somuncurin-1 | Escherichia coli ATCC 25922 | Negative | 250µg/ml | [80] |
| Staphylococcus aureusATCC 29213 | Positive | 500µg/ml | [80] | ||
| somuncurin-2 | Escherichia coli ATCC 25922 | Negative | 600µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| somuncurin-4.2 | Escherichia coli ATCC 25922 | Negative | >700 µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| somuncurin-4.2a | Escherichia coli ATCC 25922 | Negative | >700 µg/ml | [80] | |
| Staphylococcus aureus ATCC 29213 | Positive | >700 µg/ml | [80] | ||
| somuncurin-4.3 | Escherichia coli ATCC 25922 | Negative | >700 µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| somuncurin-4.3a | Escherichia coli ATCC 25922 | Negative | >700 µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| thaulin-3 | Escherichia coli ATCC 25922 | Negative | 600µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| thaulin-Sl | Escherichia coli ATCC 25922 | Negative | >700 µg/ml | [80] | |
| Staphylococcus aureusATCC 29213 | Positive | >700 µg/ml | [80] | ||
| Pleurodema thaul | Gly-Thaulin-1 | Escherichia coli ATCC 25922 | Negative | 62.5 µg/ml | [81] |
| Klebsiella pneumoniae ATCC 700603 | Negative | 125 µg/ml | [81] | ||
| Staphylococcus aureusATCC 29213 | Positive | 500 µg/ml | [81] | ||
| Thaulin-1 | Escherichia coli ATCC 25922 | Negative | 62.5 µg/ml | [81] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | 125 µg/ml | [81] | ||
| Staphylococcus aureusATCC 29213 | Positive | 500 µg/ml | [81] | ||
| Thaulin-2 | Escherichia coli ATCC 25922 | Negative | NI | [81] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | NI | [81] | ||
| Staphylococcus aureusATCC 29213 | Positive | NI | [81] | ||
| Thaulin-3 | Escherichia coli ATCC 25922 | Negative | NI | [81] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | NI | [81] | ||
| Staphylococcus aureusATCC 29213 | Positive | NI | [81] | ||
| Thaulin-4 | Escherichia coli ATCC 25922 | Negative | NI | [81] | |
| Klebsiella pneumoniae ATCC 700603 | Negative | NI | [81] | ||
| Staphylococcus aureusATCC 29213 | Positive | NI | [81] |
MIC values are presented in µM or µg/ml. NI: Non Inhibition.
Leptoglycin (MW: 1761.0) exhibited a MIC of 8 µM for the gram-negative Pseudomonas aeruginosa, while ocellatin-F1 (fallaxin) was only active in gram-negative Enterobacter cloacae (MIC = 20 µM) and Aggregatibacter actinomycetemcomitans (MIC = 25 µM). Although ocellatin-F1 shows potential activity only for two bacteria strains, it is relevant to notice that this peptide reveals a broad spectrum of action at concentrations lower than 110 µM against diverse gram-negative and positive bacteria and was active against pathogenic fungi (Table 3). Despite the high diversity of ocellatins, only six of them presented antimicrobial activity at concentrations lower than 30 µM which are the following: ocellatin S/Syphaxin (1-22) (MW = 2189.40 Da), ocellatin S (1-16) (MW = 1577.8 Da), ocellatin-5 (MW = 23113.0 Da), ocellatin-6 (MW = 22732.8 Da), ocellatin-P1 (MW = 26374.7 Da), and ocellatin-F1 (MW = 2547.5 Da) (Table 3). Peptides from the skin of Leptodactylidae species have similar inhibition of E. coli than ampicillin, azithromycin, cefotaxime, and nalidixic acid; all exhibited a MIC around 4 µM [86]. AMPs from Leptodactylidae with potential activity against E. coli were a fraction contained both nattererin-1, nattererin-2, the peptides ocellatin-5, ocellatin-6, ocellatin-P1, ocellatin S (1-16), Gly-thaulin-1, thaulin-1, P1-Ll-1577, P2-Ll-1298, and P3-Ll-208, which showed MIC varying between 10 to 28 µM (Table 3).
In addition to the antimicrobial potentials represented by MIC values of peptides, they are also investigated concerning their hemolytic properties. Since the main mechanism of action of these peptides is the interaction with bacterial membranes, some of them can also affect the cellular membrane of mammals [99]. As a result, if a peptide shows a potent antimicrobial activity, but hemolysis of human erythrocytes and/or cytotoxicity in murine fibroblasts occurs at the concentration of MIC value, this peptide is considered poorly selective, and it can be rejected as a potential candidate for therapeutic application [99]. In this way, we can emphasize that most peptides from skin secretions of Leptodactylidae have reported no hemolytic effect, highlighting their selectivity [57, 60, 68, 70, 71, 73, 79, 81]. However, P3-Ll-2085, a mix of two other peptides, caused 100% hemolysis at 40 µM, which can limit the use of this molecule [68]. There is no information about the hemolytic properties of ocellatin-5 and ocellatin-6 [64].
Although several peptides reported from frog secretions have no antimicrobial activities for the human pathogenic microorganism strains evaluated [71, 86], it is important to highlight that wild microorganisms, in general, are more susceptible to the action of antimicrobial substances [60]. Also, it is common to find more than one type of peptide in the skin secretion of frogs that could present activity by synergistic effects, and they can be efficient in protecting the amphibian [89].
Therefore, beyond the active antimicrobial peptides from skin frogs, some peptides demonstrate low or absent antimicrobial properties but have shown selectivity for microorganisms. Additionally, these peptides can act by synergism or represent a change of permeability membrane when, in combination with antibiotics, assisting the access of the antibiotics into pathogenic microorganisms [18]. These appointments highlight the potential of peptides from skin frogs even for the peptides with low or absent antimicrobial properties, but future investigations are still required to understand them, including in vivo experiments. Additionally, the inactive peptides of frogs can be involved in other essential functions, such as amphibian survival or modulating the immune system response [53, 99].
Origin and evolution of peptides in anurans
In anurans, the origins of peptides go back 150 million years [100] from a series of genes involved in other skin functions in front of a scenario of conquering new land environments and fulfilling all new necessities [82]. Evidence from Phyllomedusidae, Pelodryadidae, and Ranidae families show that encoding genes come from a large and unique family of genes with several duplication events resulting in an evolutionary divergence and producing more than 100.000 different peptides [100, 101]. Gene family is well conserved with origin from a common ancestor before the fragmentation of Gondwana during the late Jurassic and early Cretaceous, and they do not follow speciation [10, 100].
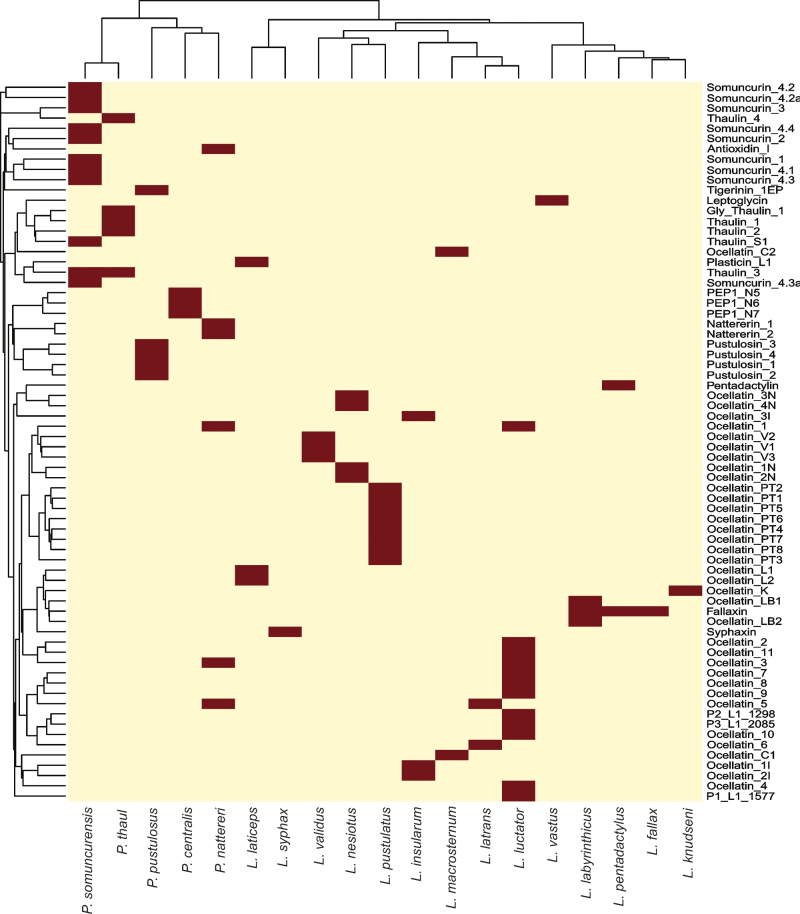
Peptide-encoding genes display different mutation rates, even so, genes remain similar when compared to species phylogenetically distant [100]. Although conservative, peptides are rapid response systems for a faster pathogenic answer and depend on direct contact with pathogens, thus peptide encoding genes evolution does not follow speciation [82, 83]. We observed the same pattern when comparing a phylogenetic species tree with a ClustalW2 phylogeny of the antimicrobial peptides, where we can realize how little the peptides similarities reflected the phylogenetic relationship of the species (Figure 3). Caerulein, for example, is a peptide shared by several species from two species groups of Leptodactylus (L. pentadactylus and L. fuscus), which may indicate the origin of the peptide in a common ancestor of the group separation. Besides caerulein, ocellatin-F1 and ocellatin-K1 are the only peptides shared by the species of the L. pentadactylus group. The peptides of the L. melanonotus group are all exclusive, and no species share peptides. A similar situation occurs for Physalaemin, a peptide present in the skin of seven species from two different genera, and its origin must be a common ancestor of Physalaemus and Engystomops. Only two peptides are shared by Physalaemus and Leptodactylus (genus from different subfamilies), ocellatin-1 and ocellatin-3, both shared by P. nattereri and L. luctator. Sheared peptides have two possible explications; they can indicate an ancient origin previous to speciation or convergent evolution.
Figure 3. Heatmap representing the presence (brown) and absence of antimicrobial peptides in Leptodactylidae species. Phylogenetic tree of Leptodactylidae species (up) and ClustalW2 Phylogeny of the antimicrobial peptides (left).
Peptides exclusive for one species do not bring evolutionary information since they could either have an ancient origin that has been conserved until today by only one species or a recent origin that emerged after speciation. However, the first option seems less probable for species from the same groups. That is the case for most peptides, including ocellatins from L. validus and L. pustulatus, as well as L. latrans, L. luctator, and L. macrosternum. Another species with several exclusive peptides is Pleurodema thaul, but since there are no other studies with Pleurodema species, we cannot assure the exclusivity of these peptides. Despite all of the current knowledge, no phylogenetic comparative analyses are available, and genes involved in peptide productions remain unknown, as well as the mechanisms of expression.
Ecological functions of skin secretions
Defensive secretion against predators can be classified as Odoriferous, Adhesive Noxious, and Slippery [102]. Additionally, these substances can have synergic actions with defensive behaviors, such as body-raising or thanatosis, to name a few [102]. For instance, L. labyrinthicus and L. vastus stretch the legs and lift the pelvis, while leaving the snout close to the ground, inguinal, and dorsal lateral skin presents bright colorations in red and yellow tones to a potential aggressor [78,103]. Besides the chemical defenses, the skin substances can act as cues and signals for many interactions including aggregation, territory defending, predator-prey interactions, mate attraction, and parental care [104, 105].
Leptodactylus fallax is a large frog from the Caribbean with restricted distribution [20]. Males are territorial and fight to defend the best call locations [106]. A peptide named Leptodactylus aggression-stimulating peptide (LASP) is used for males to stimulate other male aggressive behavior. This peptide has no action over females suggesting an exclusive agonist function [56].
Lithodytes lineatus is an Amazonian frog that can use the leaf-cutting ants' nest during reproduction without any consequences by mimicking ant chemical cues [107]. The leaf-cutting ants nest provides better environmental conditions to avoid egg drying and offers protection against terrestrial predators [107].
Multiple species of L. latrans and L. melanonotus groups display parental care behaviors, such as schooling guidance to sheltered places by pumping behavior (e.g. L. insularum, L. podicipinus, and L. macrosternum) [108-111]. Attending females call their tadpole schools by hitting the water with their pelvis to produce waves from a maximum distance of 18 cm. Consequently, schooling follows attending females through the ponds [110, 112]. Waves presumably transfer chemical signals that the tadpoles identify to follow attending females and to encourage tadpole schooling behavior [112, 113]. Inside the parental care context, the chemical signals and the biological mechanism remained unknown.
Additional medicinal applications for the peptides of Leptodactylidae
In addition to the antibiotic activity, other applications are known for the secretions and peptides from the skin of amphibians, as well as for the secretions of Leptodactylidae species [53]. Biological and pharmacological applications of skin secretion from Leptodactylidae include immunomodulation, treatment of degenerative and zoonotic diseases, anticancer, antioxidant, and antifungal activities, control of arboviruses vectors, mosquito larvae control, and rabies control (Table 4) [2, 89, 91, 114, 115].
Table 4. Species of Leptodactylidae with pharmacological or biological properties.
| Species name | Substance/Extract | Property | Reference |
|---|---|---|---|
| Leptodactylus laticeps | Plasticin-L1 | Immunomodulatory | [91] |
| Leptodactylus fallax | Ocellatin-S1/ Syphaxin | Antiviral | [69] |
| Leptodactylus knudseni | crude secretion | Insecticidal | [114] |
| Leptodactylus labyrinthicus | Ocellatin-F1 and bufotenine | Anti-rabies | [115] |
| Leptodactylus luctator | Skin extract | Multi-target agents for Alzheimer Disease (AChE, MAOB) and DPPH | [116] |
| Leptodactylus macrosternum | Skin extract | Multi-target agents for Alzheimer Disease (BChE, MAOB) and DPPH | [116] |
| Leptodactylus mystacinus | Skin extract | Multi-target agents for Alzheimer Disease (MAOB) | [116] |
| Leptodactylus pentadactylus | Pentadactylin | Anti-proliferative | [117] |
| Physalaemus nattereni | Secretion | Anticancer | [118] |
| Physalaemus nattereni | Antioxidin-I | Antioxidant | [2] |
| Physalaemus santafecinus | Skin extract | Multi-target agents for Alzheimer Disease (AChE, BChE, MAOB) and DPPH | [116] |
| Pseudopaludicula falcipes | Skin extract | Multi-target agents for Alzheimer Disease (AChE, BChE, MAOB) | [116] |
AChE: acetylcholinesterase; BChE: butyrylcholinesterase; MAOB: monoamine oxidase B.
One of the most relevant applications is cancer treatment. Pentadactylin from Leptodactylus pentadactylus and a crude secretion from Physalaemus nattereri (Figure 1) skin demonstrated a significant reduction of growth and proliferation of melanoma cells [118, 119]. Another application is on Alzheimer’s disease treatment, a neurodegenerative disorder of the brain and a major public health problem with 50 million cases worldwide [116, 120]. Extracts of P. santafecinus, and P. falcipes skin have shown inhibition of acetylcholinesterase, an enzyme that hydrolysis acetylcholine, which is a common factor associated with Alzheimer’s disease, and no haemolytic activity was observed for these extracts [116]. In addition, Leptodactylus macrosternum secretion shows antioxidant activity, which is associated with several diseases, including Alzheimer’s disease [116]
Plasticin-L1, a helical peptide rich in glycine and leucine from L. laticeps, has shown immunomodulatory properties since it stimulates cytokine production in macrophages from frog skin [91]. Immunomodulation was also reported for several amines listed in Table 1.
The compounds obtained from Leptodactylidae have also been evaluated to control virus vectors. Arboviruses, which are viruses transmitted through arthropods such as mosquitoes, are a major public health concern in tropical and subtropical countries, disseminating Dengue fever and resulting in over 100 million cases yearly [121, 122]. Therefore, the control of the Dengue vectors is crucial for the prevalence of tropical diseases [121]. Aedes aegypti is the main vector of Yellow Fever, Dengue, Chikungunya, and Zika [123], and Anopheles darling is the vector of malaria [124], two very important diseases in tropical countries. The crude skin secretion of L. knudseni exhibits insecticidal activity for A. aegypti and A. darling. The frog secretion affects adults and larvae of both species, and the ingestion of the secretion increases the dipterans mortality [114].
At least 16 species known of Rabies viruses are the cause of zoonotic neurotropic disease in mammals [125, 126]. Viruses attack and kill defensive T cells (lymphocytes) and stay in the nervous system, avoiding cell host apoptosis that results in encephalitic illness and posterior death [127]. Agency WHO estimates 59,000 rabies cases annually by dog-mediation, with higher prevalence in Asia and Africa [128]. In this manner, ocellatin-F1, a peptide found in L. fallax, L. pentadactylus, and L. labyrinthicus [57, 60, 70], revealed antiviral activity against rabies virus [115]. Ocellatin-F1, in combination with bufotenine, an alkaloid from Rhinella jimi, showed synergistic activity in inhibiting viral penetration into BHK-21 cells, thereby restraining the infection [115]. These substances were also evaluated separately, and inhibitions lower than 25% were observed [115].
Future considerations
Despite their high diversity and potential, only 9% of the species from the Leptodactylidae family were studied concerning chemical, biological, and pharmacological properties, which are relative to four genera (Engystomops, Leptodactylus, Physalaemus, and Pleurodema). This percentage is likely to decrease as the number of species in the family continues to grow, with nine species added to the family only in 2020, for example [20]. All the evaluated species belong to Leptodactyline and Leiuperine, and species of Paratelmatobiinae have not been studied yet. Therefore, there is a huge potential to be discovered from Leptodactylidae, as well as many ecological and evolutionary relationships to understand.
The OMICS techniques (e.g. proteomics, transcriptomics, and metabolomics) have provided opportunities for investigations more holistic from frog skin secretions [129]. These techniques combined with bioassays will allow better comprehension of the ecological issues and functionalities of the chemical signals and cues. Intra and interspecific frog communication are not limited to acoustic calls or visual signaling [129], instead chemical signaling plays several roles in social interaction like courtship, territoriality, and parental care, but this area has been underexplored in Leptodactylidae.
RNA-seq analysis is another applicable technique with multiple advantages, allowing the identification of the entire transcriptomes and the quantification of the gene expression, making it possible for comparisons in particular scenarios such as stages of development, ecological situations, and/or environmental conditions [130]. Additionally, the rapid and harmless identification of alkaloids in poison frogs has been proved by the MasSpec Pen technique that applies mass spectrometry and represents an opportunity to discover new bioactive substances with an easy and fast method without sample preparation, since the data is obtained directly from tissue [131]
Leptodactylidae species reveal many antimicrobial peptides (AMPs) with potent activity against pathogenic bacteria. On the other hand, there is a significant number of species without any study, and highlights the potential source for new antimicrobial molecules from them. AMPs from Leptodactylidae species are majority cationic α-helical (positive charge +1 to +6 at pH 7) with hydrophobic amino acids (40 to 70%), being able to act by different mechanisms of action, presenting a broad spectrum of activities [87, 99]. Thus, these AMPs can interact with bacterial and fungal cell membranes and change, for example, the permeability, inducing the death of microorganisms [89, 99]. Since the AMPs act in cell membranes, which are highly conservated organs, it is difficult for pathogens to develop resistance against these substances [99]. Currently, antibiotic resistance is a worldwide public health issue [121]. This resistance is a natural process in which the microorganisms develop mechanisms to resist harmful substances from the environment as an adaptation to environmental pressure or threat [132]. Thus, the reach for new potent antibiotics to combat infections by clinical antibiotic resistance led traditional research to alternative sources such as animal species with natural exposure to pathogens like amphibians [1]. Natural exposure to pathogens, combined with diversity and live history, gives amphibians great potential to treat human diseases with skin secretion, an ecosystem service not well known [1, 16, 19].
Conclusion
In summary, the current knowledge regarding the skin secretion of Leptodactylidae is limited compared to the family's diversity. The use of new technologies and reduced sample sizes for substance isolation and description is an advancement in the chemical studies of anuran skin. However, there are unstudied genera yet, as research focused on only the most common species.
The main compounds reported from Leptodactylidae are amines and peptides, mainly classified as neuropeptides and antimicrobial peptides. Ocellatins are the peptides most commonly reported. In addition, glycine (G) and glycine-valine (GV) are frequently observed as C-terminal amino acids, while N-terminal amino acids are observed as glutamic acid (E), lysine (K), and valine (V). The more active peptides against pathogenic bacterial strains (gram-positive and gram-negative) exhibit MIC of 1-15 µM, demonstrating the potential of Leptodactylidae species to search for new active compounds and stimulating the expansion of the investigation from them since they are scarcely explored.
Although several peptides are potent antimicrobials, some inactive peptides could act in synergism, and they can also be combined with traditional antibiotics since they change the permeability of microbial membranes. These studies of the combinations (peptides and antibiotics) are relevant targets to investigate and develop new therapeutic strategies because they are unknown yet. Furthermore, these inactive antimicrobial peptides have been attributed to other ecological functions, including desiccation prevention, reproductive strategies, and the stimulation of aggressive behavior in male frogs.
There are still gaps to fill in terms of ecological context, functions, and evolution. The origin of the encoded genes seems to be before Leptodactylidae divergence, as proved for other families, and there is no reason to believe that it could be different. However, these theories need to be proven for Leptodactylidae. Peptide gene evolution in the family remains unknown, and transcriptomic techniques represent an opportunity to understand this phenomenon.
Acknowledgments
JFCC thanks Priscila Lopes, Jimena Grosso, and Sean Keuroghlian-Eaton for their help with the graphic layout. DBS thanks Instituto Nacional de Ciência e Tecnologia em Áreas Úmidas (INAU).
Footnotes
Availability of data and materials: Not applicable.
Funding: JFCC is the recipient of a Ph.D. fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). DJS is the recipient of the Conselho Nacional de Desenvolvimento Científico e Tecnológico research fellowship (CNPq, process numbers 309420/2020-2) . DBS is the recipient of CNPq research fellowship (CNPq, process numbers 313047/2020-0 and 312194/2023-4) and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul fellowship (FUNDECT, process number 71/000.491/2021).
Ethics approval: Not applicable.
Consent for publication: The authors declare no need for consent for publication.
Reference
- Clarke BT. The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol Ver Camb Philos Soc. 1997;72(3):365–379. doi: 10.1017/s0006323197005045. [DOI] [PubMed] [Google Scholar]
- Barbosa EA, Oliveira A, Plácido A, Socodato R, Portugal CC, Mafud AC, Ombredane AS, Moreira DC, Vale N, Bessa LJ, Joanitti GA, Alves C, Gomes P, Delerue-Matos C, Mascarenhas YP, Marani MM, Relvas JB, Pintado M, Leite JRSA. Structure and function of a novel antioxidant peptide from the skin of tropical frogs. Free Radic Biol Med. 2018;115:68–79. doi: 10.1016/j.freeradbiomed.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Garraffo HM, Andriamaharavo NR, Vaira M, Quiroga MF, Heit C, Spande TF. Alkaloids from single skins of the Argentinian toad Melanophryniscus rubriventris (Anura, Bufonidae): An unexpected variability in alkaloid profiles and a profusion of new structures. Springerplus. 2012;1(1):51. doi: 10.1186/2193-1801-1-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prates I, Antoniazzi MM, Sciani JM, Pimenta DC, Toledo LF, Haddad CFB, Jaret C. Skin glands, poison and mimicry in dendrobatid and leptodactylid amphibians. J Morphol. 2012;273(3):279–390. doi: 10.1002/jmor.11021. [DOI] [PubMed] [Google Scholar]
- Mailho-Fontana PL, Antoniazzi MM, Rodrigues I, Sciani JM, Pimenta DC, Brodie ED, Rodrigues MT, Jared C. Parotoid, radial, and tibial macroglands of the frog Odontophrynus cultripes: Differences and similarities with toads. Toxicon. 2017;129:123–133. doi: 10.1016/j.toxicon.2017.02.022. [DOI] [PubMed] [Google Scholar]
- Garbino GST, da Silva LH, Amaral RG, Rezende GC, Pereira VJA, Culot L. Predation of treefrogs (Anura: Hylidae) with toxic skin secretions by the black lion tamarin (Leontopithecus chrysopygus, Callitrichinae) Primates. 2020;61(4):567–572. doi: 10.1007/s10329-020-00818-1. [DOI] [PubMed] [Google Scholar]
- Duellman E, Trueb L. Biology of amphibians. Baltimore (MD): The Johns Hopkins University Press; 1994. [Google Scholar]
- Sengezer Inceli M, Kaptan E, Sancar S, Murathanoglu O, Suren Castillo S. Localization of prolactin receptor in the dorsal and ventral skin of the frog (Rana ridibunda) Biologia. 2010;65(1):157–163. [Google Scholar]
- Varga JFA, Bui-Marinos MP, Katzenback BA. Frog skin innate immune defences: Sensing and surviving pathogens. Front Immunol. 2019;9:3128. doi: 10.3389/fimmu.2018.03128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmaco M, Mignogna G, Barra D. Antimicrobial peptides from amphibian skin: What do they tell us? Biopolymers. 1998;47(6):435–450. doi: 10.1002/(SICI)1097-0282(1998)47:6<435::AID-BIP3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Conlon JM. Structural diversity and species distribution of host-defense peptides in frog skin secretions. Cell Mol Life Sci. 2011;68(13):2303–2315. doi: 10.1007/s00018-011-0720-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lai R. The chemistry and biological activities of peptides from amphibian skin secretions. Chem Rev. 2015;115(4):1760–1846. doi: 10.1021/cr4006704. [DOI] [PubMed] [Google Scholar]
- Brunetti AE, Hermida GN, Luna MC, Barsotti AMG, Jared C, Antoniazzi MM, RiveraCorrea M, Berneck BVM, Faivovich J. Diversity and evolution of sexually dimorphic mental and lateral glands in Cophomantini treefrogs (Anura: Hylidae: Hylinae) Biol J Linn Soc. 2015;114(1):12–34. [Google Scholar]
- Regueira E, Dávila C, Sassone AG, O’Donohoe MEA, Hermida GN. Post-metamorphic development of skin glands in a true toad: Parotoids versus dorsal skin. J Morphol. 2017;278(5):652–664. doi: 10.1002/jmor.20661. [DOI] [PubMed] [Google Scholar]
- Olea GB, Cheij EO, Curi LM, Cuzziol Boccioni AP, Céspedez JA, Lombardo DM. Histological and immunohistochemical characterization of the integument and parotoids glands Rhinella bergi (Anura: Bufsonidae): Development and differentiation. Acta Histochem. 2019;121(3):277–283. doi: 10.1016/j.acthis.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Toledo R, Jared C. Cutaneous granular glands and amphibian venoms. Comp Biochem Physiol Part A Physiol. 1995;111(1):1–29. [Google Scholar]
- Rollins-Smith LA, Reinert LK, O'Leary CJ, Houston LE, Woodhams DC. Antimicrobial Peptide Defenses in Amphibian Skin. Integr Comp Biol. 2005;45(1):137–142. doi: 10.1093/icb/45.1.137. [DOI] [PubMed] [Google Scholar]
- Marani MM, Dourado FS, Quelemes PV, De Araujo AR, Perfeito MLG, Barbosa EA, Véras LMC, Coelho ALR, Andrade EB, Eaton P, Longo JPF, Azevedo RB, Delerue-Matos C, Leite JRSA. Characterization and Biological Activities of Ocellatin Peptides from the Skin Secretion of the Frog Leptodactylus pustulatus. J Nat Prod American Chemical Society. 2015;78(7):1495–1504. doi: 10.1021/np500907t. [DOI] [PubMed] [Google Scholar]
- Hocking DJ, Babbitt KJ, Hocking DJ. Amphibian contributions to ecosystem services. Herpetol Conserv Biol. 2014;9:1–17. [Google Scholar]
- Frost D. Amphibian Species of the World: an Online Reference. Version 6.1. 2023. [2023 Jan 20]. Available from: https://amphibiansoftheworld.amnh.org/index.php .
- WHO . Global action plan on antimicrobial resistance. World Health Organization; 2015. [DOI] [PubMed] [Google Scholar]
- Kim YG, Lee JH, Raorane CJ, Oh ST, Park JG, Lee J. Herring Oil and Omega Fatty Acids Inhibit Staphylococcus aureus Biofilm Formation and Virulence. Front Microbiol. 2018;9:1241. doi: 10.3389/fmicb.2018.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa T, de Oliveira C, Chambers H, Chatterjee S. PBP4: A New Perspective on Staphylococcus aureus β-Lactam Resistance. Microorganism. 2018;6(3):57. doi: 10.3390/microorganisms6030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livorsi DJ, Chorazy ML, Schweizer ML, Balkenende EC, Blevins AE, Nair R, Samore MH, Nelson RE, Khader K, Perencevich EN. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob Resist Infect Control. 2018;7:55. doi: 10.1186/s13756-018-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I, Rahman S, Jan AT, Siddiqui MT, Mondal AH, Haq QMR. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front Microbiol. 2018;9:2066. doi: 10.3389/fmicb.2018.02066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson TM, Ming D, Ahmad R, Moore LSP, Holmes AH. Antimicrobial use, drug resistant infections and COVID-19. Nat Rev Microbiol. 2020;18:409–410. doi: 10.1038/s41579-020-0395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla-Cuenca L, Gil C, Cuesta S, Rapún-Araiz B, Žiemytė M, Mira A, Lasa I, Valle J. Antibiofilm activity of flavonoids on staphylococcal biofilms through targeting BAP amyloids. Sci Rep. 2020;10(1):18968. doi: 10.1038/s41598-020-75929-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sá ROD, Grant T, Camargo A, Heyer WR, Ponssa ML, Stanley E. Systematics of the neotropical genus Leptodactylus fitzinger, 1826 (Anura: Leptodactylidae): Phylogeny, the relevance of non-molecular evidence, and species accounts. South Am J Herpetol. 2014;9:S1–28. [Google Scholar]
- Da Silva LA, Magalhães FM, Thomassen H, Leite FSF, Garda AA, Brandão RA, Haddad CFB, Giaretta AA, De Carvalho TR. Unraveling the species diversity and relationships in the Leptodactylus mystaceus complex (Anura: Leptodactylidae), with the description of three new Brazilian species. Zootaxa. 2020;4779(2):151–189. doi: 10.11646/zootaxa.4779.2.1. [DOI] [PubMed] [Google Scholar]
- Magalhães F de M, Lyra ML, de Carvalho TR, Baldo D, Brusquetti F, Burella P, Colli GR, Gehara MC, Giaretta AA, Haddad CFB, Langone JA, López JA, Napoli MF, Santana DJ, de Sá RO, Garda AA. Taxonomic Review of South American Butter Frogs: Phylogeny, Geographic Patterns, and Species Delimitation in the Leptodactylus latrans Species Group (Anura: Leptodactylidae) Herpetol Monogr. 2020;34(1):131–177. [Google Scholar]
- Gazoni T, Lyra ML, Ron SR, Strüssmann C, Baldo D, Narimatsu H, Pansonato A, Schneider RG, Giaretta AA, Haddad CFB, Parise-Maltempi PP, Carvalho TR. Revisiting the systematics of the Leptodactylus melanonotus group (Anura: Leptodactylidae): Redescription of L. petersii and revalidation of its junior synonyms. Zool Anz. 2021;290:117–134. [Google Scholar]
- de Amaral M, Ienes-Lima J. Anurans against SARS-CoV-2: A review of the potential antiviral action of anurans cutaneous peptides. Virus Res. 2022;315:198769. doi: 10.1016/j.virusres.2022.198769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron A, Wiens J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol Phylogenet Evol. 2011;61(2):543–583. doi: 10.1016/j.ympev.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Heyer WR. The Adaptive Ecology of the Species Groups of the Genus Leptodactylus (Amphibia, Leptodactylidae) Evolution. 1969;23(3):421–428. doi: 10.1111/j.1558-5646.1969.tb03525.x. [DOI] [PubMed] [Google Scholar]
- Bernal MH, Lynch JD. Review and analysis of altitudinal distribution of the Andean anurans in Colombia. Zootaxa. 2008;1826(1):1–25. [Google Scholar]
- Oliveira EF de, Tolledo J, Feio RN. Amphibia, Anura, Physalaemus rupestris Caramaschi, Carcerelli and Feio, 1991: distribution extension and geographic distribution map. Check List. 2009;5(4):815–818. [Google Scholar]
- Carvalho TR, Leite FSF, Peszzuti TL. A new species of Leptodactylus Fitzinger (Anura, Leptodactylidae, Leptodactylinae) from montane rock fields of the Chapada Diamantina, northeastern Brazil. Zootaxa. 2013;3701(3):349–364. doi: 10.11646/zootaxa.3701.3.5. [DOI] [PubMed] [Google Scholar]
- Erspamer V. Isolation of leptodactyline (m-hydroxyphenylethyltrimethyl-ammonium) from extracts of Leptodactylus skin. Arch Biochem Biophys. 1959;82:431–438. doi: 10.1016/0003-9861(59)90139-0. [DOI] [PubMed] [Google Scholar]
- Biji KB, Ravishankar CN, Venkateswarlu R, Mohan CO, Gopal TKS. Biogenic amines in seafood: a review. J Food Sci Technol. 2016;53(5):2210–2218. doi: 10.1007/s13197-016-2224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasi A, Bertaccini G, Cei JM, De Caro G, Erspamer V, Impicciatore M, Roseghini M. Presence of caerulein in extracts of the skin of Leptodactylus pentadactylus labyrinthicus and of Xenopus laevis. Br J Pharmacol. 1970;38(1):221–228. doi: 10.1111/j.1476-5381.1970.tb10351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erspamer V, Roseghini M, Cei JM. Indole-, imidazole-, and phenyl-alkylamines in the skin of thirteen Leptodactylus species. Biochem Pharmacol. 1964;13:1083–1093. doi: 10.1016/0006-2952(64)90104-2. [DOI] [PubMed] [Google Scholar]
- Cei JM, Erspamer V. Biochemical Taxonomy of South American Amphibians by Means of Skin Amines and Polypeptides. Copeia. 1966;1966(1):74–78. [Google Scholar]
- Erspamer GF, Cei JM. Biogenic amines and active polypeptides in the skin of Leptodactylus vilarsi melin. Biochem Pharmacol. 1970;19(2):321–325. doi: 10.1016/0006-2952(70)90189-9. [DOI] [PubMed] [Google Scholar]
- Erspamer V. Biogenic Amines and Active Polypeptides of the Amphibian Skin. Annu Rev Pharmacol. 1971;11:327–350. doi: 10.1146/annurev.pa.11.040171.001551. [DOI] [PubMed] [Google Scholar]
- Erspamer V, Glasser A. The pharmacological action of (m‐hidroxyphenethyl) trimethylammonium (Leptodactyline) Br J Pharmacol Chemother. 1960;15(1):14–22. doi: 10.1111/j.1476-5381.1960.tb01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseghini M, Erspamer V, Erspamer GF, Cei JM. Indole-, imidazole- and phenylalkylamines in the skin of one hundred and forty American amphibian species other than bufonids. Comp Biochem Physiol Part C Comp Pharmacol. 1986;85(1):139–147. doi: 10.1016/0742-8413(86)90064-2. [DOI] [PubMed] [Google Scholar]
- Deulofeu V. Progress in the Chemistry of Organic Natural Products. Viena: Springer; 1948. The Chemistry of the Constituents of Toad Venoms; pp. 241–266. [Google Scholar]
- Erspamer V, Vitali T, Roseghini M, Cei JM. Occurrence of new imidazolealkylamines (Spinaceamine and 6-Methylspinaceamine) in skin extracts of Leptodactylus pentadactylus labyrinthicus. Experientia. 1963;19:346–347. doi: 10.1007/BF02152309. [DOI] [PubMed] [Google Scholar]
- Roseghini M, Erspamer GF, Severini C. Biogenic amines and active peptides in the skin of fifty-two African amphibian species other than bufonids. Comp Biochem Physiol Part C Comp Pharmacol. 1988;91(2):281–286. doi: 10.1016/0742-8413(88)90030-8. [DOI] [PubMed] [Google Scholar]
- Roseghini M, Endean R, Temperilli A. New and Uncommon Indole- and ImidazoleAlkylamines in Skins of Amphibians from Australia and Papua New Guinea. Zeitschrift für Naturforsch C. 1976;31(3-4):118–120. doi: 10.1515/znc-1976-3-403. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Hoener MC, Berry MD. Trace Amines and Their Receptors. Pharmacol Rev. 2018;70(3):549–620. doi: 10.1124/pr.117.015305. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W. H. Freeman and Company; 2017. [Google Scholar]
- Conlon JM, Mechkarska M, Lukic ML, Flatt PR. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides. 2014;57:67–77. doi: 10.1016/j.peptides.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Erspamer V, Erspamer GF, Cei JM. Active peptides in the skins of two hundred and thirty American amphibian species. Comp Biochem Physiol C Comp Pharmacol Toxicol. 1986;85(1):125–137. doi: 10.1016/0742-8413(86)90063-0. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Guilhaudis L, Attoub S, Coquet L, Leprince J, Jouenne T, Mechkarska M. Purification, conformational analysis and cytotoxic activities of host-defense peptides from the Tungara frog Engystomops pustulosus (Leptodactylidae; Leiuperinae) Amino Acids. 2023;55(10):1349–1359. doi: 10.1007/s00726-023-03312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JD, Rollins-Smith LA, Nielsen PF, John A, Conlon JM. Characterization of a peptide from skin secretions of male specimens of the frog, Leptodactylus fallax that stimulates aggression in male frogs. Peptides. 2005;26(4):597–601. doi: 10.1016/j.peptides.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Rollins-Smith LA, King JD, Nielsen PF, Sonnevend A, Conlon JM. An antimicrobial peptide from the skin secretions of the mountain chicken frog Leptodactylus fallax (Anura:Leptodactylidae) Regul Pept. 2005;124(1-3):173–178. doi: 10.1016/j.regpep.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Barran G, Kolodziejek J, Coquet L, Leprince J, Jouenne T, Nowotny N, Conlon JM, Mechkarska M. Peptidomic analysis of skin secretions of the Caribbean frogs Leptodactylus insularum and Leptodactylus nesiotus (Leptodactylidae) identifies an ocellatin with broad spectrum antimicrobial activity. Antibiotics. 2020;9(10):718. doi: 10.3390/antibiotics9100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo-Filho JL. Caracterização estrutural de peptídeos antimicrobianos da secreção cutânea de Leptodactylus knudseni e Leptodactylus chaquensis (Anura: Leptodactylidae) Universidade Federal de Rondônia; 2011. [Google Scholar]
- Gusmão KAG, dos Santos DM, Santos VM, Cortés ME, Reis PVM, Santos VL, PilóVeloso D, Verly RM, de Lima ME, Resende JM. Ocellatin peptides from the skin secretion of the South American frog Leptodactylus labyrinthicus (Leptodactylidae): Characterization, antimicrobial activities and membrane interactions. J Venom Anim Toxins incl Trop Dis. 2017;23:4. doi: 10.1186/s40409-017-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cei JM. Taxonomic and evolutionary significance of peptides in amphibian skin. Peptides. 1985;6(3):13–16. doi: 10.1016/0196-9781(85)90344-4. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Al-Ghaferi N, Abraham B, Sonnevend A, King JD, Nielsen PF. Purification and properties of Laticeptin, an Antimicrobial Peptide from Skin Secretions of the South American Frog Leptodactylus laticeps. Protein Pept Lett. 2006;13(4):411–415. doi: 10.2174/092986606775974410. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Iwamuro S, King JD. Dermal cytolytic peptides and the system of innate immunity in anurans. Ann N Y Acad Sci. 2009;1163:75–82. doi: 10.1111/j.17496632.2008.03618.x. [DOI] [PubMed] [Google Scholar]
- Leite JMA, Silva LP, Silva-Leite RR, Ferrari AS, Noronha SE, Silva HR, Bloch JC, Leite JRSA. Leptodactylus ocellatus (Amphibia): Mechanism of defense in the skin and molecular phylogenetic relationships. J Exp Zool Part A Ecol Genet Physiol. 2010;313(1):1–8. doi: 10.1002/jez.551. [DOI] [PubMed] [Google Scholar]
- Nascimento ACC, Zanotta LC, Kyaw CM, Schwartz ENF, Schwartz CA, Sebben A, Sousa MV, Fontes W, Castro MS. Ocellatins: New Antimicrobial Peptides from the Skin Secretion of the South American Frog Leptodactylus ocellatus (Anura: Leptodactylidae) Protein J. 2004;23(8):501–508. doi: 10.1007/s10930-004-7877-z. [DOI] [PubMed] [Google Scholar]
- Marani MM, Aguilar S, Cuzziol Boccioni AP, Cancelarich NL, Basso NG, Albericio F. Identification of new ocellatin antimicrobial peptides by cDNA precursor cloning in the frame of this family of intriguing peptides. Antibiotics. 2020;9(11):751. doi: 10.3390/antibiotics9110751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento A, Chapeaurouge A, Perales J, Sebben A, Sousa MV., Fontes W, Castro MS. Purification, characterization and homology analysis of ocellatin 4, a cytolytic peptide from the skin secretion of the frog Leptodactylus ocellatus. Toxicon. 2007;50(8):1095–1104. doi: 10.1016/j.toxicon.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Rollins-Smith LA, Conlon JM. Antimicrobial peptide defenses against chytridiomycosis, an emerging infectious disease of amphibian populations. Dev Comp Immunol. 2005;29(7):589–598. doi: 10.1016/j.dci.2004.11.004. [DOI] [PubMed] [Google Scholar]
- King JD, Al-Ghaferi N, Abraham B, Sonnevend A, Leprince J, Nielsen PF, Conlon JM. Pentadactylin: An antimicrobial peptide from the skin secretions of the South American bullfrog Leptodactylus pentadactylus. Comp Biochem Physiol - C Toxicol Pharmacol. 2005;141(4):393–397. doi: 10.1016/j.cbpc.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Dourado FS, Leite JRSA, Silva LP, Melo JAT, Bloch C, Schwartz EF. Antimicrobial peptide from the skin secretion of the frog Leptodactylus syphax. Toxicon. 2007;50(4):572–580. doi: 10.1016/j.toxicon.2007.04.027. [DOI] [PubMed] [Google Scholar]
- King JD, Leprince J, Vaudry H, Coquet L, Jouenne T, Conlon JM. Purification and characterization of antimicrobial peptides from the Caribbean frog, Leptodactylus validus (Anura: Leptodactylidae) Peptides. 2008;29(8):1287–1292. doi: 10.1016/j.peptides.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Sousa JC, Berto RF, Gois EA, Fontenele-Cardi NC, Honório-Júnior JER, Konno K, Richardson M, Rocha MFG, Camargo AACM, Pimenta DC, Cardi BA, Carvalho KM. Leptoglycin: A new Glycine/Leucine-rich antimicrobial peptide isolated from the skin secretion of the South American frog Leptodactylus pentadactylus (Leptodactylidae) Toxicon. 2009;54(1):23–32. doi: 10.1016/j.toxicon.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Sousa NA, Oliveira GAL, de Oliveira AP, Lopes ALF, Iles B, Nogueira KM, Araújo TSL, Souza LKM, Araújo AR, Ramos-Jesus J, Plácido A, Amaral C, Campelo YDM, Barbosa EA, Portugal CC, Socodato R, Lobo A, Relvas J, Bemquerer M, Eaton P, Leite JRSA, Medeiros JVR. Novel Ocellatin Peptides Mitigate LPS-induced ROS Formation and NF-kB Activation in Microglia and Hippocampal Neurons. Sci Rep. 2020;10(1):2696. doi: 10.1038/s41598-020-59665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasi A, Erspamer V, Cei JM. Isolation Active and Amino Acid Sequence of Physalaemin, the Main Polypeptide of the Skin of Physalaemus fuscumaculatus. Arch Biochem Biophys. 1964;108(2):341–348. doi: 10.1016/0003-9861(64)90395-9. [DOI] [PubMed] [Google Scholar]
- Bertaccini G, Cei JM, Erspamer V. Occurrence of Physalaemin in extracts of the skin of Physalaemus fuscumaculatus and its pharmacological actions on extravascular smooth muscle. Br J Pharmacol Chemother. 1965;25(2):363–379. doi: 10.1111/j.1476-5381.1965.tb02056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa JM. Bioprospecção de peptídeos antimicrobianos presentes na secreção cutânea do anfíbio Physalaemus centralis (anura: leptodactylidae) Brazilia (BR): Universidade de Brasília; 2020. [Google Scholar]
- Castro MS, Pires OR, Júnior, Fontes W, Roepstorff P. Analysis of skin derived peptides from the Cuyaba Dwarf Frog Physalaemus nattereri by off-line LC MALDI MS/MS. Int J Mass Spectrom. 2017;416:90–95. [Google Scholar]
- Barbosa EA, Iembo T, Martins GR, Silva LP, Prates MV, Andrade AC, Blosh JC. Skin secretion peptides: The molecular facet of the deimatic behavior of the four-eyed frog, Physalaemus nattereri (Anura, Leptodactylidae) Rapid Commun Mass Spectrom. 2015;29(21):206–218. doi: 10.1002/rcm.7313. [DOI] [PubMed] [Google Scholar]
- Cancelarich NL, Wilke N, Fanani ML, Moreira DC, Pérez LO, Alves Barbosa E, Plácido A, Socodato R, Portugal CC, Relvas JB, de la Torre BG, Albericio F, Basso NG, Leite JR, Marani MM. Somuncurins: Bioactive Peptides from the Skin of the Endangered Endemic Patagonian Frog Pleurodema somuncurense. J Nat Prod. 2020;83(4):972984. doi: 10.1021/acs.jnatprod.9b00906. [DOI] [PubMed] [Google Scholar]
- Marani MM, Perez LO, de Araujo AR, Plácido A, Sousa CF, Quelemes PV, Oliveira M, Gomes-Alves AG, Pueta M, Gameiro P, Tomás AM, Delerue-Matos C, Eaton P, Camperi SA, Basso NG, Leite JRSA. Thaulin-1: The first antimicrobial peptide isolated from the skin of a Patagonian frog Pleurodema thaul (Anura: Leptodactylidae: Leiuperinae) with activity against Escherichia coli. Gene. 2017;605:70–80. doi: 10.1016/j.gene.2016.12.020. [DOI] [PubMed] [Google Scholar]
- König E, Bininda-Emonds ORP, Shaw C. The diversity and evolution of anuran skin peptides. Peptides. 2015;63:96–117. doi: 10.1016/j.peptides.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Pukala TL, Bowie JH, Maselli VM, Musgrave IF, Tyler MJ. Host-defence peptides from the glandular secretions of amphibians: Structure and activity. Nat Prod Rep. 2006;23(3):368–393. doi: 10.1039/b512118n. [DOI] [PubMed] [Google Scholar]
- Basir YJ, Knoop FC, Dulka J, Conlon JM. Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim Biophys Acta - Protein Struct Mol Enzymol. 2000;1543(1):95–105. doi: 10.1016/s0167-4838(00)00191-6. [DOI] [PubMed] [Google Scholar]
- Erspamer V. In: Amphib Biol Integument. Heatwole H, Bartholameus G, editors. Surrey Beatty and Sons; 1994. Bioactive secretions of the amphibian integument; pp. 178–350. [Google Scholar]
- Ebbensgaard A, Mordhorst H, Overgaard MT, Nielsen CG, Aarestrup FM, Hansen EB. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS One. 2015;10:e0144611. doi: 10.1371/journal.pone.0144611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omardien S, Brul S, Zaat SAJ. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: current progress made in understanding the mode of action and the response of bacteria. Front Cell Dev Biol. 2016;4:111. doi: 10.3389/fcell.2016.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M-D, Won H-S, Kim J-H, Mishig-Ochir T, Lee B-J. Antimicrobial Peptides for Therapeutic Applications: A Review. Molecules. 2012;17(10):12276–12286. doi: 10.3390/molecules171012276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon JM, Mechkarska M, Lukic ML, Flatt PR. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides. 2014;57:67–77. doi: 10.1016/j.peptides.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Gomes KAGG, dos Santos DM, Santos VM, Piló-Veloso D, Mundim HM, Rodrigues LV, Lião LM, Verly EM, de Lima ME, Resende JM. NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus. Peptides. 2018;103:72–83. doi: 10.1016/j.peptides.2018.03.016. [DOI] [PubMed] [Google Scholar]
- Scorciapino MA, Manzo G, Rinaldi AC, Sanna R, Casu M, Pantic JM, Lukic ML, Conlon M. Conformational analysis of the frog skin peptide, plasticin-L1 and its effects on the production of proinflammatory cytokines by macrophages. Biochemistry. 2013;52(41):7231–7241. doi: 10.1021/bi4008287. [DOI] [PubMed] [Google Scholar]
- Conlon JM, Abdel-Wahab YHA, Flatt PR, Leprince J, Vaudry H, Jouenne T, Vaudry H, Jouenne T, Condamine E. A glycine-leucine-rich peptide structurally related to the plasticins from skin secretions of the frog Leptodactylus laticeps (Leptodactylidae) Peptides. 2009;30:888–892. doi: 10.1016/j.peptides.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol Rev. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Nielsen SL, Frimodt‐Møller N, Kragelund BB, Hansen PR. Structure-activity study of the antibacterial peptide fallaxin. Protein Sci. 2007;16:1969–1976. doi: 10.1110/ps.072966007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangaspero A, Sandri L, Tossi A. Amphipathic α helical antimicrobial peptides. Eur J Biochem. 2001;268(21):5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Siano A, Gatti PI, Imaz MS, Zerbini E, Simonetta AC, Lajmanovich R, Tonarelli GG. A Comparative Study of the Biological Activity of Skin and Granular Gland Secretions of Leptodactylus latrans and Hypsiboas pulchellus from Argentina. Nat. Prod. 2014;8(2):128–135. [Google Scholar]
- Cabral MES, Dias DDQ, Sales DL, Oliveira OP, Teles DA, A JA, Filho, Sousa JGG, Coutinho HDM, da Costa JGM, Kerntopf MR, Alves RRN, Almeida WO. Evaluations of the antimicrobial activities and chemical compositions of body fat from the amphibians Leptodactylus macrosternum Miranda-Ribeiro (1926) and Leptodactylus vastus Adolf Lutz (1930) in Northeastern Brazil. Evidence-based Complement Altern Med. 2013;2013:913671. doi: 10.1155/2013/913671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Sun L, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He Q. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11(7):3919–3931. [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Vanhoye D, Amiche M. Molecular strategies in biological evolution of antimicrobial peptides. Peptides. 2003;24(11):1669–1680. doi: 10.1016/j.peptides.2003.08.017. [DOI] [PubMed] [Google Scholar]
- Duda TF, Vanhoye D, Nicolas P. Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol Biol Evol. 2002;19(6):858–864. doi: 10.1093/oxfordjournals.molbev.a004143. [DOI] [PubMed] [Google Scholar]
- Toledo LF, Sazima I, Haddad CFB. Behavioural defences of anurans: an overview. Ethol Ecol Evol. 2011;23(1):1–25. [Google Scholar]
- Toledo L, Tozetti A, Zina J. Leptodactylus labyrinthicus (Pepper frog): repertoire of defensive behaviour. Herpetol Bull. 2005;90:29–31. [Google Scholar]
- Wyatt T. In: Pheromones and Animal Behavaviour. Wyatt T, editor. Cambridge: Cambridge University Press; 2003. Animals in a chemical world; pp. 1–22. [Google Scholar]
- Schulte LM, Yeager J, Schulte R, Veith M, Werner P, Beck LA, Lötters S. The smell of success: choice of larval rearing sites by means of chemical cues in a Peruvian poison frog. Anim Behav. 2011;81(6):1147–1154. [Google Scholar]
- Gibson RC, Buley KR. Maternal Care and Obligatory Oophagy in Leptodactylus fallax: A New Reproductive Mode in Frogs. Copeia. 2004;2004(1):128–135. [Google Scholar]
- Schlüter A, Löttker P, Mebert K. Use of an active nest of the leaf cutter ant Atta cephalotes (Hymenoptera: Formicidae) as a breeding site of Lithodytes lineatus (Anura: Leptodactylidae) Herpetol Notes. 2009;2:101–105. [Google Scholar]
- Ponssa ML. Cuidado parental y comportamiento de cardumen de larvas en Leptodactylus insularum (Anura, Leptodctylidae) Alytes. 2001;19:183–195. [Google Scholar]
- Rodrigues AP, Giaretta AA, da Silva DR, Facure KG. Reproductive features of three maternal-caring species of Leptodactylus (Anura: Leptodactylidae) with a report on alloparental care in frogs. J Nat Hist. 2011;45(33-34):2037–2047. [Google Scholar]
- Martins IA. Parental care behaviour in Leptodactylus podicipinus (Cope, 1862) (Anura, Leptodactylidae) Herpetol J. 2001;11(1):29–32. [Google Scholar]
- Carrillo JFC, Santana DJ, Prado CPA. An overview of parental care in the foam-nesting frogs of the genus Leptodactylus (Anura: Leptodactylidae): current knowledge and future directions. Amphibia-Reptilia. 2023;44(33):301–311. [Google Scholar]
- Wells KD, Bard KM. Parental Behavior of an Aquatic-Breeding Tropical Frog, Leptodactylus bolivianus. J Herpetol. 1988;22(3):361–364. [Google Scholar]
- Hoffmann H. Observations on behaviour and parental care of Leptodactylus melanonotus (Hallowell) in Costa Rica. Salamandra. 2006;42(2/3):109–116. [Google Scholar]
- Trindade FTT, Soares ÂA, de Moura AA, Rego TB, Soares AM, Stábeli RG, Calderon LA, Silva AA. Insecticidal activity of Leptodactylus knudseni and Phyllomedusa vaillantii crude skin secretions against the mosquitoes Anopheles darlingi and Aedes aegypti. J Venom Anim Toxins incl Trop Dis. 2014;20(1):28. doi: 10.1186/1678-9199-20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha R dos S, Neto, Vigerelli H, Jared C, Antoniazzi MM, Chaves LB, da Silva ACR, Melo LR, Sciani JM, Pimenta DC. Synergic effects between ocellatin-F1 and bufotenine on the inhibition of BHK-21 cellular infection by the rabies virus. J Venom Anim Toxins incl Trop Dis. 2015;21:50. doi: 10.1186/s40409-015-0048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli R, Aimaretti FM, López JA, Siano AS. Amphibian skin extracts as source of bioactive multi-target agents against different pathways of Alzheimer’s disease. Nat Prod Res. 2021;35(4):686–689. doi: 10.1080/14786419.2019.1591396. [DOI] [PubMed] [Google Scholar]
- Libério MS, Joanitti GA, Azevedo RB, Cilli EM, Zanotta LC, Nascimento AC, Sousa MV, P OR, Júnior, Fontes W, Castro MS. Anti-proliferative and cytotoxic activity of pentadactylin isolated from Leptodactylus labyrinthicus on melanoma cells. Amino Acids. 2011;40(1):51–59. doi: 10.1007/s00726-009-0384-y. [DOI] [PubMed] [Google Scholar]
- Carvalho AC, Márquez CAP, Azevedo RB, Joanitti GA, Pires OR, Júnior, Fontes W, Castro MS. Cytotoxic activity and antiproliferative effects of crude skin secretion from Physalaemus nattereri (Anura: Leptodactylidae) on in vitro melanoma cells. Toxins (Basel) 2015;7(10):3989–4005. doi: 10.3390/toxins7103989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libério MDS, Bastos IMD, Pires OR, Fontes W, Santana JM, Castro MS. The crude skin secretion of the pepper frog Leptodactylus labyrinthicus is rich in metallo and serine peptidases. PLoS One. 2014;9(6):e96893. doi: 10.1371/journal.pone.0096893. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:1–10. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Global strategy for dengue prevention and control 2012-2020. WHO; Geneva: 2012. [Google Scholar]
- Gutiérrez EHJ, Walker KR, Ernst KC, Riehle MA, Davidowitz G. Size as a proxy for survival in Aedes aegypti (Diptera: Culicidae) mosquitoes. J Med Entomol. 2020;57(4):1228–1238. doi: 10.1093/jme/tjaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascini TV, Ramalho-Ortigão M, Ribeiro JM, Jacobs-Lorena M, Martins GF. Transcriptional profiling and physiological roles of Aedes aegypti spermathecal-related genes. BMC Genomics. 2020;21:143. doi: 10.1186/s12864-020-6543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos AN, Ferreira FA da S, Cunha HB da, Tadei WP. Characterization of artificial larval habitats of Anopheles darlingi (Diptera: Culicidae) in the Brazilian Central Amazon. Rev Bras Entomol Rev Bras Entomo. 2018;62(2):267–274. [Google Scholar]
- Walker PJ, Blasdell KR, Calisher CH, Dietzgen RG, Kondo H, Kurath G, Longdon B, Stone DM, Tesh RBT, Tordo N, Vasilakis N, Whitfield AE. ICTV Virus Taxonomy Profile: Rhabdoviridae. J Gen Virol. 2018;99(4):447–448. doi: 10.1099/jgv.0.001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieracci EG, Brown JA, Bergman DL, Gilbert A, Wallace RM, Blanton JD, VelascoVilla A, Morgan CN, Lindquist S, Chipman RB. Evaluation of species identification and rabies virus characterization among bat rabies cases in the United States. J Am Vet Med Assoc. 2020;256(1):77–84. doi: 10.2460/javma.256.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon M. In: Advances in Virus Research. Jackson Alan C., editor. San Diego(CA): Academic Press; 2011. Evasive Strategies in Rabies Virus Infection; pp. 33–53. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Expert Consultation on Rabies, third report. World Health Organization; Genova: 2018. [Google Scholar]
- Brunetti AE, Carnevale F, Neto, Vera MC, Taboada C, Pavarini DP, Bauermeister A, Lopes NP. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem Soc Rev. 2018;47(5):1574–1591. doi: 10.1039/c7cs00368d. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger A, Povilaitis S, Gowda P, O’Connell L, Eberlin L. Noninvasive Detection of Chemical Defenses in Poison Frogs Using the MasSpec Pen. ACS Meas Sci Au. 2022;2:475–484. doi: 10.1021/acsmeasuresciau.2c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DCD . Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention; Atlanta (GA); 2019. [Google Scholar]
- Valencia-Aguilar A, Cortés-Gómez AM, Ruiz-Agudelo CA. Ecosystem services provided by amphibians and reptiles in Neotropical ecosystems. Int J Biodivers Sci Ecosyst Serv Manag. 2013;9(3):257–272. [Google Scholar]