Abstract
The mammalian gut secretes a family of multifunctional peptides that affect appetite, intestinal secretions, and motility, while some regulate the microbiota. We have found that peptide YY (PYY1–36), but not endocrine PYY3–36, acts as an antimicrobial peptide (AMP) expressed by gut epithelial Paneth Cells (PC). PC-PYY is packaged into secretory granules and is secreted into and retained by surface mucus, which optimizes PC-PYY activity. While PC-PYY shows some antibacterial activity, it displays selective antifungal activity against virulent Candida albicans hyphae, but not the yeast form. PC-PYY is a cationic molecule that interacts with the anionic surfaces of fungal hyphae to cause membrane disruption and transcriptional reprogramming that selects for the yeast phenotype. Hence, PC-PYY is an antifungal AMP that contributes to the maintenance of gut fungal commensalism.
Keywords: Intestinal epithelial cells, antimicrobial peptide, antifungal, fungal virulence, gut mycobiome, mucus, mucosal barrier, commensalism, innate immunity
One Sentence Summary:
Paneth cell derived peptide YY, in its full-length form, checks the invasive potential of commensal fungi.
The coevolution of metazoans and microbes has been fundamental to the development of mutualistic beneficial relationships, not least in the digestive tract of animals. Gut microbes form region-specific stable and resilient communities essential for processes such as immune and metabolic development and overall intestinal homeostasis (1). Gut bacteria have been investigated in some detail, but less is known about gut fungi and archaea. The microbiota appear to be regulated in a healthy host, although, little is known about host surveillance and control of fungal populations in the gut.
Peptide YY (PYY) is a satiety hormone that is expressed and secreted by enteroendocrine cells (EECs) (2). In this work we found gut-specific Paneth cells (PCs) also express a form of PYY that functions as an antimicrobial peptide (AMP) with selective activity against Candida albicans. This fungus is found commonly as a commensal yeast, but becomes pathogenic upon transformation into its hyphal form (3). The antimicrobial function of PC-PYY plays a role in maintaining fungal commensalism by inhibiting the yeast-to-hypha transformation of opportunistic Candida spp..
Results:
Presence of PYY observed in Paneth cells:
While visualizing mucosal enteroendocrine L-cells in murine distal ileum, we serendipitously observed the satiety regulating peptide PYY, immunostained with lysozyme (LYZ1) expressing PCs (Fig. 1A and 1B). This finding was surprising, as PCs are gut mucosal epithelial cells found in most mammals, which secrete AMPs against pathogens and regulate the local gut microbiota (4). We confirmed PYY immunolocalization in healthy human adult ileal PCs (Fig. S1A), observed antibody epitope specificity through recombinant peptide quenching (Fig. S1B,C), and identified PYY mRNA in PCs via fluorescent in situ hybridization (FISH; Fig. 1B). To achieve higher resolution, stimulated emission depletion (STED) and SP8 confocal microscopy were used to show that PYY and LYZ1 are packaged into discrete granules in the jejunum and ileum (Figs. 1C,D and S2A,B and Movie S1), raising the possibility that these peptides have different roles, sorting pathways, and regulation (5). Laser capture micro-dissection in the murine ileum followed by quantitative RT-PCR revealed robust PYY transcript levels in basal crypts in line with other PC-specific AMP transcripts (Figs. 1E and 1F). Reanalysis of a publicly deposited murine small intestinal epithelial single-cell RNAseq dataset identified PYY expression in mature EEC populations that co-expressed with Scg1, Cck, and Glp1, but also in PCs, where it was co-expressed with Dfa17, Atg16l1, Defa5, and Lyz1 (Figs. S3A,B) (6). The Human Protein Atlas single cell RNA database was searched for PYY, showing detection in PCs (Fig. S3C).
Fig. 1. Peptide YY (PYY) localizes to ileal Paneth cells.
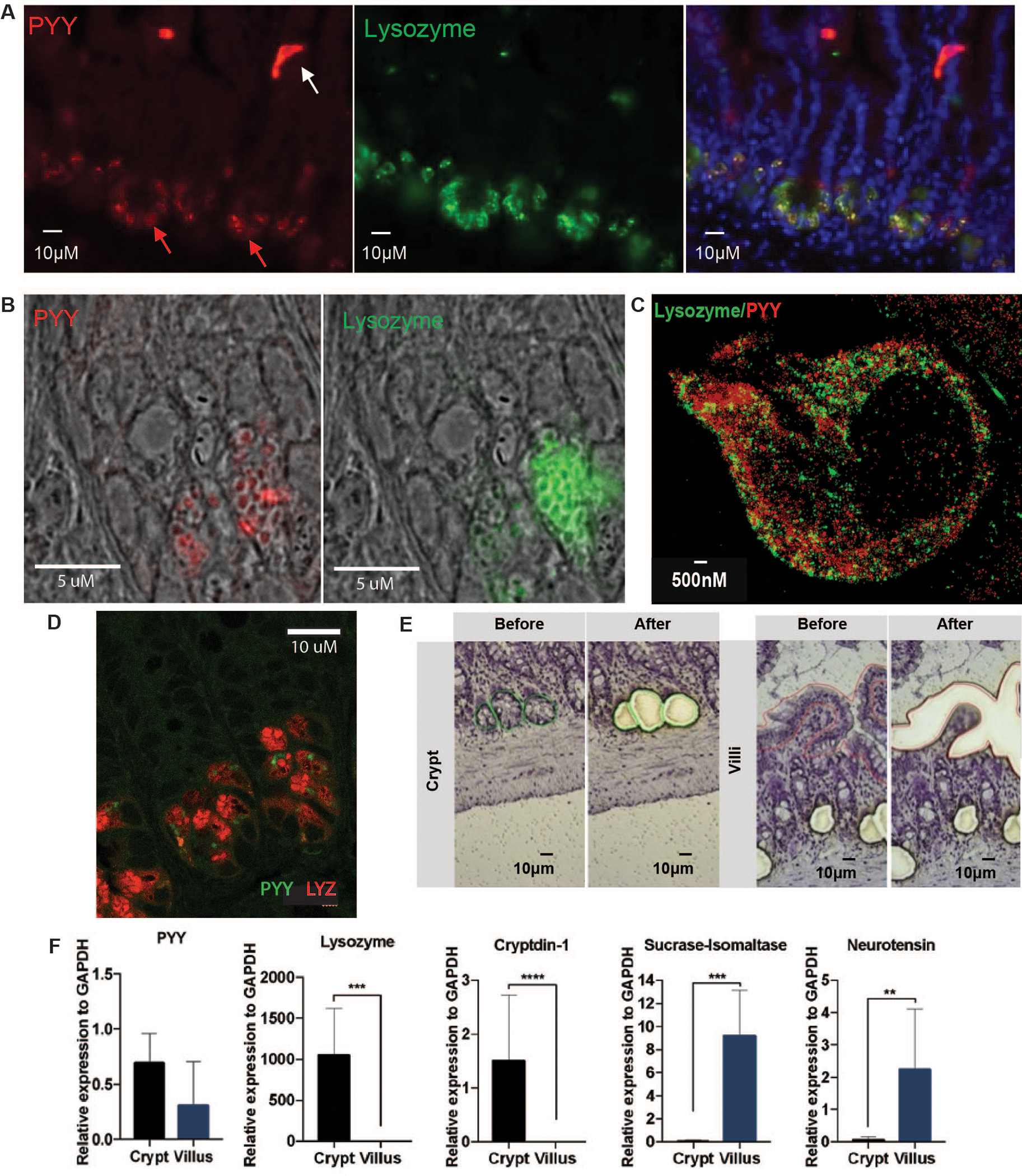
(A) PYY (red) detection in Paneth cells (PC, red arrow) and L-cells (white arrow) via anti-PYY antibody immunofluorescence (IF). PC PYY staining co-localized (right panel) with PC Lysozyme (LYZ, green; middle panel). (B) Confirmation of PC PYY localization via mRNA Fluorescent In Situ Hybridization (red, left panel) and counterstaining with anti-LYZ antibody (green, right panel). (C) High-resolution stimulated emission depletion IF microscopy of PC PYY and LYZ packaging in discrete secretory granules within the cytosol. (D) SP8 confocal microscopy of PYY (green) and LYZ (red) in ileum PCs. (E) Representative extractions from ileal crypts and villus epithelial cells via Laser-capture microdissection (LCM). (F) Relative expression of PYY and marker mRNA for crypt secretion compared with villus epithelium secretion estimated from LCM. Lysozyme and Cryptdin-1 are AMPs that are also secreted from PCs; Sucrase-isomaltase is released from the villus surface brush border; Neurotensin is a marker for enteroendocrine cells. Significance was determined using t-test (n=6, repeated twice, *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Anti-microbial functions of Paneth cell PYY:
PC expression of PYY indicated it might have an antimicrobial function. The predicted structure of PYY resembles the alpha-helical, amphipathic AMP, magainin-2 (Figs. 2A,B), produced in the skin of Xenopus laevis. PC-PYY is strictly the unmodified, full length PYY1–36, whereas the circulating endocrine form, PYY3–36, is formed by removal of two N-terminal amino acids by dipeptidyl peptidase IV (DPP-IV) (7). We initially assessed the antimicrobial activity of full length PYY peptide against representative Gram-positive and -negative bacteria, which showed less activity compared with more effective magainin-2 (Fig. 2L–N). We then tested full length PC-PYY on C. albicans growth and viability (8, 9). We induced transition of yeast into hyphal forms in the presence of 2.5% w/v porcine mucin. Propidium iodide (PI) was used to measure membrane permeabilization of PYY1–36-treated C. albicans hyphae. Permeabilization of hyphae was observed at 30 μM PYY1–36, but yeast were unaffected (Fig. 2C and Fig. S4A). PYY1–36 - fluorescein isothiocyanate (FITC) tagged peptide co-localized with the PI signal within hyphae (Fig. 2C). We synthesized PYY using D-amino acids to generate a peptide stereoisomer, which disrupted hyphae in the same way as the native L-isoform (Fig. S4B). Less pronounced permeabilization was observed with the shorter endocrine PYY3–36 (Fig. S4B,C). Similar staining with PI was found with hyphal forms of C. tropicalis and C. dubliniensis treated with PYY1–36 (Fig. S5).
Fig. 2. PYY displays antimicrobial activity towards Candida albicans hyphae and some bacteria.
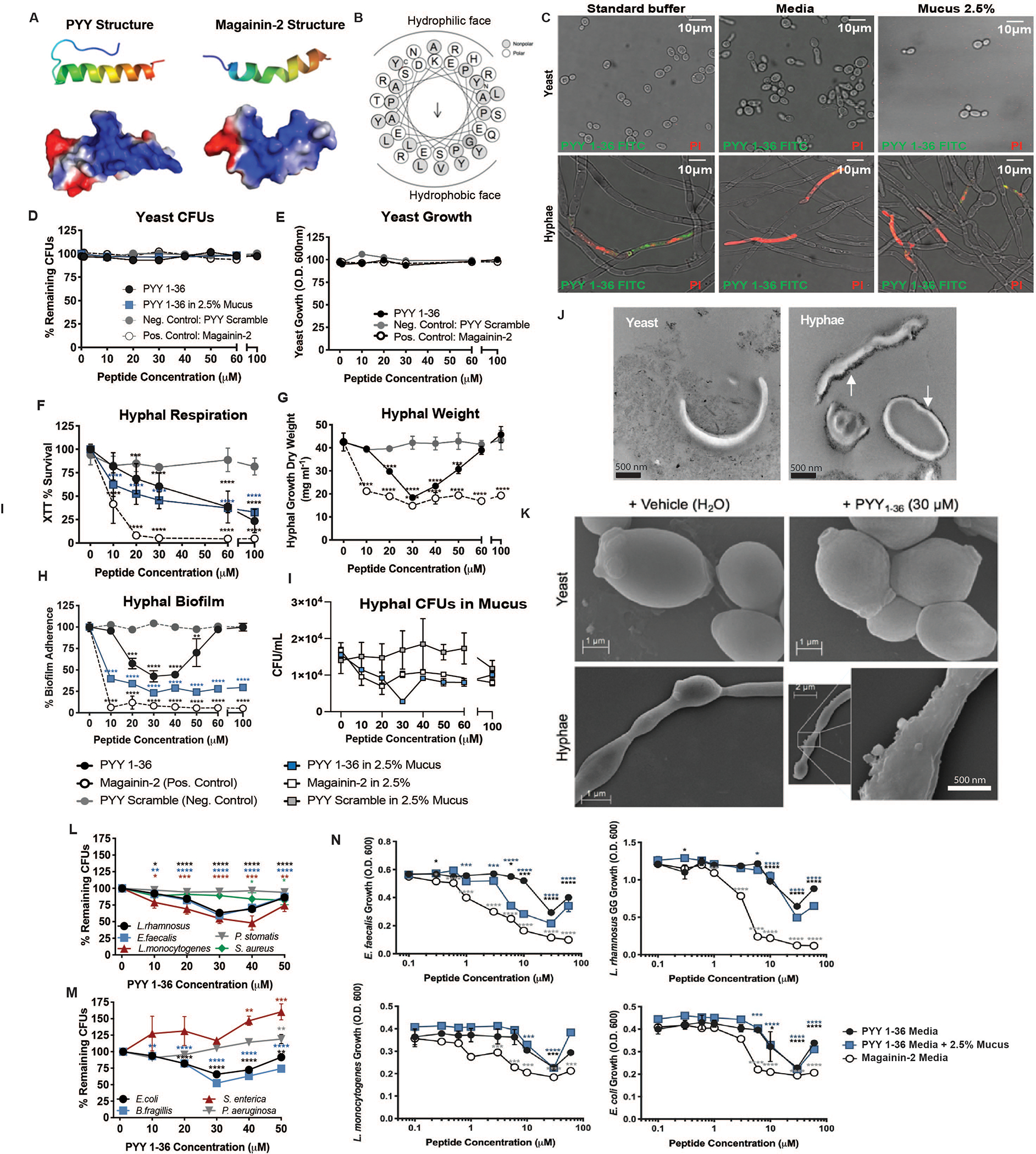
(A) Ribbon diagram (top) and space-filling model indicating electrostatic surface charge (bottom; red negative, blue positive) for amphipathic alpha-helix structures of PYY1–36 and Magainin-2. (B) Amphipathic PYY1–36 helical wheel projection displaying surface localization of residues and calculated hydrophobic dipole moment (μH 0.208, arrow). (C) Propidium iodide (PI) staining of C. albicans yeast and hyphae following exposure to PYY1–36-FITC (green) in standard antimicrobial peptide assay buffer (AMP), preferred growth media for each form (YPD-yeast or RPMI-hyphae), or standard buffer + 2.5% w/v porcine mucus. C. albicans yeast (D) survival (Colony Forming Units, CFUs) and (E) growth (Optical density, O.D.) following exposure to PYY1–36 +/− mucus, PYY scramble, and Magainin-2 peptides. Pathogenic C. albicans hyphal (F) respiration by tetrazolium salt (XTT) assay (G) dry weight (H) biofilm adherence and (I) CFUs following exposure to PYY1–36 +/− mucus, PYY scramble, and Magainin-2 peptides. (J) Transmission electron microscopy for visualization of cationic surfaces (black probe accumulation) in C. albicans yeast and hyphae membranes. (K) Scanning electron microscopy of C. albicans yeast and hyphae following 2-hour exposure to vehicle (H2O) or PYY1–36. Dose-dependent killing of Gram-positive (L) and Gram-negative (M) bacteria induced by PYY1–36. CFUs remaining were normalized to 0 μM PYY. Significance was determined via ANOVA. (N) Impact of varying concentrations of PYY1–36 +/− mucus versus Magainin-2 on bacteria growth. O.D. values were normalized to wells containing 0 μM PYY. All assays were performed in duplicate three times. Significance was determined via ANOVA (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Given the observed hyphal specificity, we subjected C. albicans yeast and hyphae to increasing PYY1–36 concentrations in media with or without 2.5% w/v mucin. PYY1–36 did not affect yeast numbers or density (Fig. 2D and 2E); however, it elicited a decrease in hyphal respiration (Fig. 2F). Similar results were obtained for C. tropicalis (Fig. S6). The enteroendocrine form, PYY3–36, showed less activity (Fig. S7A). Hyphal growth and survival were examined under increasing concentrations of PYY1–36, where magainin-2 (positive) and scrambled (negative) peptides were used as controls. In aqueous conditions, hyphal growth and biofilm formation were significantly reduced at 20–50 μM PYY1–36 (Figs. 2G and 2H). We observed a biphasic dose-response of PYY1–36 action in aqueous (RPMI) buffer, similar to observations of other AMPs, with peak effect at ~30 μM and loss of activity by 100 μM (10, 11). We attribute this observation to concentration-dependent self-aggregation, or multimerization, of amphipathic alpha-helical AMP precipitates in aqueous medium, reducing bioactivity. Consistent with this hypothesis, a sigmoidal dose-response curve was observed in biofilm and colony forming units (CFUs) when 2.5% w/v mucin is present (Fig. 2H and 2I). No differences were observed for biofilm with PYY3–36 (Fig. S7B). We surmise that mucin prevents self-aggregation of PYY1–36, which allows its activity to be maintained.
To test whether the cationic properties of PYY1–36 (as shown in Figs. 2A and 2B) allow binding to the anionic surface charge of fungi (5, 12) we used a cationic ferritin probe and transmission electron microscopy to identify sites of anionic surface charge (Fig. 2J, white arrows). The cationic probe decorated the surface of hyphae, while the surfaces of yeast cells were not labelled. To further investigate charge interactions, we repeated PI membrane permeability assays with PYY1–36 in the presence of sodium sulfate, to provide anionic quenching of cationic PYY1–36 charge, which reduced PYY1–36 induced permeability (Fig. S4D). Finally, scanning electron microscopy of yeast and hyphal forms of C. albicans showed that hyphal membranes exposed to PYY1–36 develop surface blebbing and irregularities, while yeast membranes remain unaffected (Fig. 2K).
RNA-seq was used to measure the transcriptional response to PYY1–36. The hyphal phenotype showed downregulation of cell wall synthesis, biofilm formation, and ribosome biogenesis pathways (Figs. S8A,B). By contrast, yeast cells showed fewer changes in gene expression, and those mostly affected were genes involved in yeast-to-hyphal transition and adhesion. In all, our evidence indicates PYY1–36 has selective antimicrobial activity against the virulent, invasive hyphae of C. albicans, with limited effect on the blastoconidia.
Paneth cell PYY release and mucus localization:
PCs secrete AMPs into the gut lumen in response to the presence of microbial products (5). Although PYY1–36 is produced by L-cells where it is released systemically, the ubiquitous serine protease DPP-IV rapidly cleaves it into endocrine PYY3–36. We used ex vivo murine distal ileal loops to examine luminal PYY release (Fig. S9A–C). Following exposure to C. albicans, PYY1–36 was measured by LC-MS (Fig. S10) in the lumen contents, the surface mucus, and mucosal tissue (Fig. 3A). At baseline, micromolar amounts of PYY1–36 were detected in mucus following control incubations in this model (Fig. S11). However, introduction of C. albicans hyphae into ileal loops significantly increased PYY1–36 levels. Less PYY1–36 was observed when yeast were present or when cell free spent media were applied from cultures of either fungal morphology. PYY3–36 was not detected in any of the lumen contents, mucus, or tissue compartments by MS, similar to previous findings (13, 14). These data indicate PC-PYY is released into overlying mucus where it is retained specifically in response to the presence of C. albicans hyphae. We confirmed this response using in vivo ileal ligated loops (Fig. 3B). Immunofluorescent studies confirmed in vivo depletion of PC PYY in the presence of hyphae (Fig. 3C; Fig. S12).
Fig 3. Exposure to C. albicans hyphae but not yeast enhances PYY localization into ileal mucus.
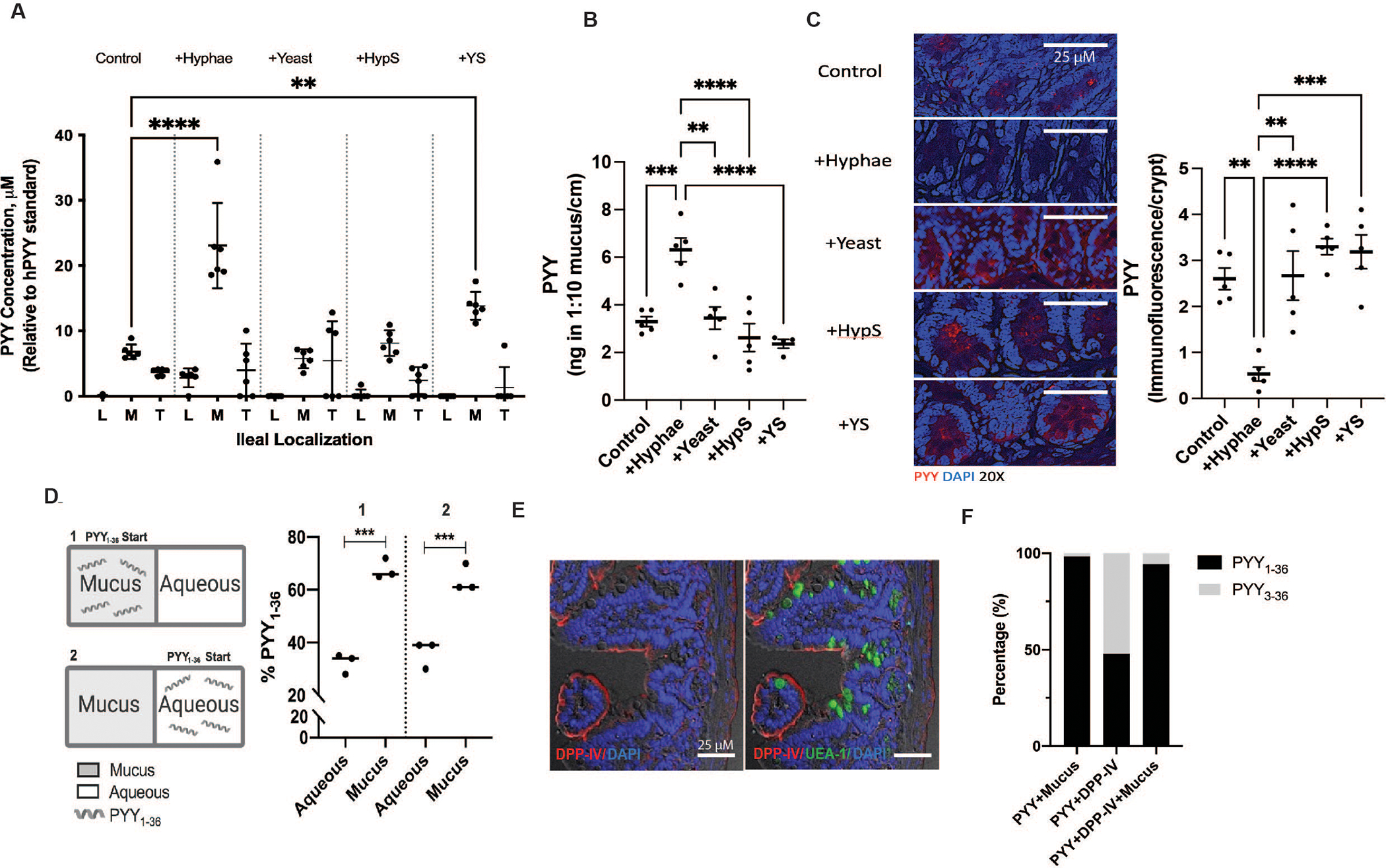
(A) PYY1–36 quantification using liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) in lumen (L), mucus (M), and tissue (T) of ex vivo ileal loops (n=6 /treatment) and in (B) mucus of in vivo ileal loops (n=5 /treatment) following stimulation with vehicle (Control), C. albicans hyphae (+Hyphae), C. albicans yeast (+Yeast), hyphae conditioned media (CM) supernatant (+HypS), or yeast CM supernatant (+YS). (C) Representative staining and quantification of PC PYY protein (red) following in vivo ileal loop stimulation with vehicle control, +Hyphae, +Yeast, +HypS, or +YS. Significance was measured using ANOVA and Dunnett’s Multiple Comparisons Test; **p<0.01, *** p<0.005, **** p<0.001. (D) PYY mucus vs. aqueous partitioning in vitro. PYY1–36 was added to either mucus (1) or aqueous (2) wells separated by a 10 kDa MWCO filter. Significance was measured using t-test; ***p<0.001. (E) Dipeptidyl peptidase IV (DDP-IV) immunofluorescence (IF) in murine ileum brush border. Lectin UEA-I-FITC (Ulex europaeus) counterstain = ileal mucus; DAPI = nuclei. (F) PYY1–36 and PYY3–36 exposure to DPP-IV +/− mucus to assess peptide degradation via LC-ESI-MS (n=3/treatment), expressed as a percent of total peptide. All experiments were done in triplicate with 3 replicate experiments.
We used a 10 kDa MWCO filter to separate mucus and aqueous compartments to test whether the cationic amphipathic properties of PYY1–36 has affinity to mucus (Fig. 3D). We found PYY1–36 preferentially partitions into mucus, likely through charge interactions, hence explaining its retention on the mucosal surface. Since DPP-IV is present in the mucosal brush border (15) (Fig. 3E), we questioned why PYY3–36 was not detected in overlying mucus. We incubated DPP-IV and PYY1–36 with and without mucin, which showed PYY3–36 generation in the absence of mucin, but none in the presence of mucin, suggesting proteolytic protection of PYY1–36 within mucus layers. (Fig. 3F)
The effect of PYY on Candida at the intestinal mucosa:
Green fluorescent protein-tagged C. albicans hyphae (Fig. S13) were co-cultured with and without PYY1–36 in human epithelial Caco-2 cells. In the presence of PYY1–36, the numbers of hyphae attached to Caco-2 cells were significantly reduced (Figs. 4A, B).
Fig 4. PYY reduces gastrointestinal colonization of C. albicans in vitro and in vivo.
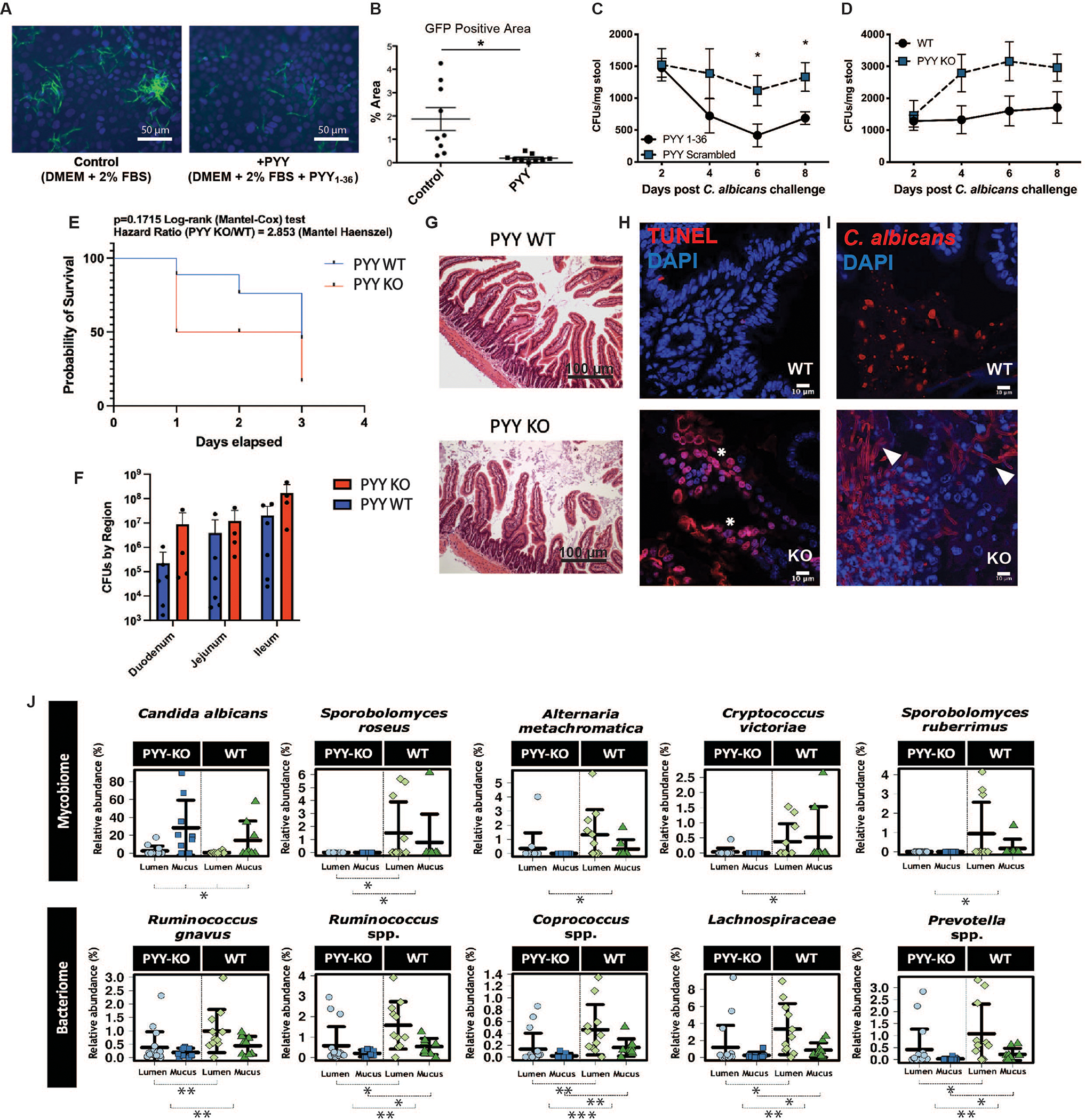
(A, B) C. albicans hyphae abundance (Hyphae-GFP) on confluent Caco2 cells exposed to vehicle (Control) or PYY1–36 (20μm) after 6 hours. (C) Fungal colony forming units (CFUs) in wild-type C57BL/6 mice (WT) +/− exogenously administered PYY1–36 or scrambled peptide via oral gavage 8 days post C. albicans (4×106 CFU) challenge (n=8/group, *p<0.05 vs control). (D) C. albicans colonization (4×106 CFUs) in PYY-KO relative to WT mice (n=8–9/timepoint/group). (E) 72-hour PYY-KO and WT mouse survival after gavage with C. albicans (2×107 CFU; n=8/group, repeated twice with both males and females) under cefoxitin and clindamycin (2 WT were humanely euthanized). (F) CFUs recovered from in small intestinal regions of PYY-KO and WT animals gavaged with C. albicans (2×107 CFU) (G) Intestinal mucosal gross morphology remained intact, while (H) epithelial cell apoptosis (red = TUNEL; white asterisk) and (I) virulent Candida morphology (hyphae; white arrows) was increased vs yeast in the mucosa of PYY-KO vs WT animals (36.3±12.1 vs. 11.77±3.8 %, p<0.04) following oral gavage challenge under clindamycin (2×107 CFU/mouse; n=8/group; repeated twice with both males and females, red = Candida). (J) Differentially altered fungal (top panel, n=39 samples) and bacterial (bottom panel, n=46 samples) populations from small intestinal mucus and lumen of PYY-KO vs. WT animals without Candida challenge as determined via Wilcoxon rank test (between groups) and Kruskal-Wallis test (across groups) *: p<0.05, **: p<0.01, ***: p<0.001. To account for uneven sequencing depth, data were transformed into relative abundances based on total sum scaling.
Antibiotic induced chronic C. albicans intestinal colonization was established via oral gavage (4×106 CFU) in specific pathogen-free wild-type (WT) or PYY gene-deficient (PYY-KO) mice and used to investigate the effect of PYY on fungal populations in vivo. Oral gavage of colonized WT mice with PYY1–36 decreased fungal titers in stool and intestinal contents as compared to mice given a scrambled peptide control (Fig. 4C and Fig. S14). Conversely, fungal colonization of PYY-KO mice was 2–3-fold higher than in WT counterparts (Fig. 4D). Total fecal output remained similar between groups, indicating comparable gut motility was maintained. A second intestinal colonization challenge experiment was performed in PYY-KO and WT animals with a higher C. albicans inoculation (2×107 CFU) under systemic and oral antibiotics to induce greater virulence. Here, PYY-KO animals exhibited elevated small intestinal C. albicans counts, more pronounced mucosal apoptosis, and significantly greater hyphal vs. yeast (36.3±12.1%) forms at the mucosal surface relative to WT counterparts (11.77±3.8%, P<0.04) (Fig 4E–I).
Given the current evidence that full-length PC-PYY localizes to the mucus layer, we isolated lumen- and mucus-associated samples from PYY-KO and WT littermates for mycobiota characterization under standard colony conditions. Young animals were used since PYY KO animals develop insulinemia and weight gain by 14 weeks of age, which could itself confound mycobiome results (2). Compared with the variable lumen fungal communities from either WT or PYY-KO, the mucus-associated layer harbored a distinct fungal population enriched for Candida (Figs. 4J and S15A–C). Within the mucus, PYY-KO animals, compared with WT counterparts, displayed elevated relative abundances of Candida, as well as of other members of Saccharomycetaceae (Fig. 4J, top panel and Fig. S15C). In contrast, bacteriome analysis showed minor relative abundance changes in Ruminococcus, Coprococcus, Lachnospiraceae, and Prevotella, as well as significantly altered beta diversity (Fig. 4, bottom panel and Fig. S15B). No differences were observed in EEC or PC density between genotypes (Fig. S16).
Discussion:
Fungi are normally present in the “healthy” gut microbiome. The yeast Candida albicans is found in 70% of humans (16), but it can transition into an opportunistic pathogen (17). PYY is highly conserved in vertebrate species (18) and its only known function to date is endocrine (19, 20). Here, we report that PC-PYY provides functionally-specific AMP activity against the invasive hyphal phenotype of C. albicans but not the yeast phenotype of this fungus, and limited antibacterial activity. Moreover, PC-PYY is packaged into secretory granules, distinct from those carrying Lyz1, and are released if mucosal surfaces are exposed to fungal hyphae, but not yeast. The specificity of cationic PC-PYY appears to lie in its electrostatic interaction with the anionic surface charge of C. albicans hyphae, determined by cationic probe imaging of the hyphal and yeast surface, and charge quenching experiments were performed with sulfate, which inhibited PC-PYY activity. Once secreted, PC-PYY is retained in the overlying mucus of the gut lumen. Finally, we show PC-PYY drives transcriptional programming in C. albicans hyphae consistent with cell death and downregulation of virulence, whereas commensal yeast respond by downregulating pathways that promote the yeast-to-hypha transition. Thus, microbial selectivity, activation, and mucus compartmentalization of PC-PYY distinguishes it from other AMPs, which have broader ranges of activity against microorganisms (4, 21).
Our studies show that PYY is active against virulent forms of C. albicans and less so on other commensal microbes. We observed similar membrane disrupting effects of PC-PYY in other species of Candida, including C. dubliniensis and C. tropicalis, but not the respiratory tract pathogen, Aspergillus fumigatus, leading us to conclude these observations may be specific to intestinal fungal pathobionts. This property arguably plays a role in maintaining the gut mycobiome in a state of commensalism. This finding complements recent work showing anti-virulence properties of intestinal mucin glycans (22) and specific mucosal IgA responses to C. albicans hyphae, which similarly inhibit virulence and maintain commensalism (23).
This study has several important limitations. First, our in vivo model utilized a global PYY KO, which has disrupted endocrine PYY functions that could confound results compared with a PC specific PYY mutant. To limit this, young animals were used prior to metabolic disruption. Second, while PYY is found in several single cell RNAseq databases, not all report its presence, suggesting expression is at low abundance or regionally variable, consistent with our data. Finally, we employed in vitro PYY concentrations of 20–30 μM, whereas PYY was found to be 8–10 μM in intestinal mucus at baseline, but becomes elevated upon C. albicans challenge.
In summary, PC-PYY is a dual function intestinally secreted peptide, with an AMP structure that differs from its endocrine counterpart in biological action and function. The regulation and selectivity of PYY against gut invasive fungal phenotypes distinguishes it from other AMPs, implicating its function in gut mycobiome regulation.
Supplementary Material
Acknowledgments:
We acknowledge the MRSEC Shared User Facilities at the University of Chicago (NSF DMR-1420709) and the Mass Spectrometry Facility (NSF CHE-1048528), as well as the University of Wisconsin-Madison Dairy Innovation Hub Histology Resource. The authors thank Daina L. Ringus, Hyoann Choi, Mirae Lee, Tahliyah S. Mims and Monika Krezalek for helpful discussions and guidance, and acknowledge Vytas Bindokas at the Integrated Light Microscopy Core at the University of Chicago (RRID: SCR_019197).
Funding:
This project was funded by NIH R01 DK113788 (EBC), NIH RC2DK122394 (EBC), NIH DDRCC P30DK44086 (EBC) and NIH T32DK007074 (EBC), NIH F32DK105728 (JFP), NIH K01DK111785 (VAL), NIH R01 AI134796 (BMP), NIH R01 GM062344-21 (JA) and the University of Chicago Gastro-Intestinal Research Foundation.
Footnotes
Competing interests: EBC, JFP, and KGH are co-inventors of Patent PCT/US18/32997, Publication No 20200062823, “Compositions and methods for treating and/or preventing pathogenic fungal infection and for maintenance of microbiome commensalism”.
Data and materials availability:
All data is available in the main text or the supplementary materials, including lead contact, materials, data, and code availability. Raw data for Candida RNAseq transcriptomics is available at NCBI GEO #GSE229566. Raw amplicon sequencing data for 16S and ITS is available at NCBI Short read achieve under BioProject IS: PRJNA631955.
References and Notes:
- 1.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R, Bacterial community variation in human body habitats across space and time. Science. 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterham RL, Cowley MA, Small CJ, Herzog H, Cohen MA, Dakin CL, Wren AM, Brynes AE, Low MJ, Ghatei MA, Cone RD, Bloom SR, Gut hormone PYY(3–36) physiologically inhibits food intake. Nature. 418, 650–4 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Witchley JN, Penumetcha P, Abonv N, Woolford C, Michell A, Noble SM, Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe. 25, 432–443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevins CL, Salzman NH, Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 9, 356–68 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Clevers HC, Bevins CL, Paneth cells: Maestros of the small intestinal crypts. Annu Rev Physiol. 75 (2013), pp. 289–311. [DOI] [PubMed] [Google Scholar]
- 6.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A, A single-cell survey of the small intestinal epithelium. Nature. 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandt D, Dahms P, Schimiczek M, Eysselein VE, Reeve JR, Mentlein R, Proteolytic processing by dipeptidyl aminopeptidase IV generates receptor selectivity for peptide YY (PYY). Med Klin (Munich). 88, 143–145 (1993). [PubMed] [Google Scholar]
- 8.Colina AR, Aumont F, Deslauriers N, Belhumeur P, De Repentigny L, Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 64, 4514–4519 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Repentigny L, Aumont F, Bernard K, Belhumeur P, Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect Immun. 68, 3172–3179 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan BL, Ganz T, Lehrer RI, Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology. 87, 131–149 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Phadke SM, Lazarevic V, Bahr CC, Islam K, Stolz DB, Watkins S, Tencza SB, Vogel HJ, Montelaro RC, Mietzner TA, Lentivirus lytic peptide 1 perturbs both outer and inner membranes of Serratia marcescens. Antimicrob Agents Chemother. 46, 2041–2045 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Weerden NL, Bleackley MR, Anderson MA, Properties and mechanisms of action of naturally occurring antifungal peptides. Cellular and Molecular Life Sciences. 70, 3545–3570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greeley GH, Hill FLC, Spannagel A, Thompson JC, Distribution of peptide YY in the gastrointestinal tract of the rat, dog, and monkey. Regul Pept. 19, 365–372 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Keire DA, Whitelegge JP, Souda P, Faull KF, Bassilian S, Reidelberger RD, Haver AC, Reeve JR, PYY(1–36) is the major form of PYY in rat distal small intestine: Quantification using high-resolution mass spectrometry. Regul Pept. 165, 151–157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darmoul D, Voisin T, Couvineau A, Rouyer-Fessard C, Salomon R, Wang Y, Swallow DM, Laburthe M, Regional expression of epithelial dipeptidyl peptidase IV in the human intestines. Biochem Biophys Res Commun. 203, 1224–1229 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Wang C, Tang C, He Q, Li N, Li J, Dysbiosis of Gut Fungal Microbiota is Associated With Mucosal Inflammation in Crohn’s Disease. J Clin Gastroenterol. 48, 513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noble SM, Gianetti BA, Witchley JN, Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 15 (2017), pp. 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conlon JM, The origin and evolution of peptide YY (PYY) and pancreatic polypeptide (PP). Peptides (N.Y.). 23, 269–278 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Batterham RL, Ffytche DH, Rosenthal JM, Zelaya FO, Barker GJ, Withers DJ, Williams SCR, PYY modulation of cortical and hypothalamic brain areas predicts feeding behaviour in humans. Nature. 450, 106–109 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Meguid MM, Glade MJ, Middleton FA, Weight regain after Roux-en-Y: a significant 20% complication related to PYY. Nutrition. 24, 832–42 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Bevins CL, The Paneth cell and the innate immune response. Curr Opin Gastroenterol. 20, 572–80 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Takagi J, Aoki K, Turner BS, Lamont S, Lehoux S, Kavanaugh N, Gulati M, Valle Arevalo A, Lawrence TJ, Kim CY, Bakshi B, Ishihara M, Nobile CJ, Cummings RD, Wozniak DJ, Tiemeyer M, Hevey R, Ribbeck K, Mucin O-glycans are natural inhibitors of Candida albicans pathogenicity. Nat Chem Biol. 18, 762–773 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, Bell R, Catanzaro JR, Song D, Singh S, Call DH, Hwang-Wong E, Hanson KE, Valentine JF, Christensen KA, O’Connell RM, Cormack B, Ibrahim AS, Palm NW, Noble SM, Round JL, Adaptive immunity induces mutualism between commensal eukaryotes. Nature. 596, 114–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batterham RL, Heffron H, Kapoor S, Chivers JE, Chandarana K, Herzog H, Le Roux CW, Thomas EL, Bell JD, Withers DJ, Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 4, 223–233 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Luong PM, Shogan BD, Zaborin A, Belogortseva N, Shrout JD, Zaborina O, Alverdy JC, Emergence of the P2 phenotype in Pseudomonas aeruginosa PAO1 strains involves various mutations in mexT or mexF. J. Bacteriol. 196, 504–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ovesny M, K i ek P, Borkovec J, vindrych Z, Hagen GM, ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley LA, Sternberg MJE, Protein structure prediction on the web: A case study using the phyre server. Nat. Protoc. 4, 363–373 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Gille C, Fähling M, Weyand B, Wieland T, Gille A, Alignment-Annotator web server: rendering and annotating sequence alignments. Nucleic Acids Res. 42, W3–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thévenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tufféry P, PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 40, W288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnenfeld EM, Beveridge TJ, Koch AL, Doyle RJ, Asymmetric distribution of charge on the cell wall of Bacillus subtilis. J. Bacteriol. 163, 1167 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gautier R, Douguet D, Antonny B, Drin G, HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24, 2101–2 (2008). [DOI] [PubMed] [Google Scholar]
- 32.HeliQuest ComputParam form version2, (available at http://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParamsV2.py). [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9 (2012), pp. 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parada AE, Needham DM, Fuhrman JA, Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K, The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White TJ, Bruns T, Lee S, Taylor J, AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS. PCR Protoc., 315–322 (1990). [Google Scholar]
- 38.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, V Melnik A, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP, DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials, including lead contact, materials, data, and code availability. Raw data for Candida RNAseq transcriptomics is available at NCBI GEO #GSE229566. Raw amplicon sequencing data for 16S and ITS is available at NCBI Short read achieve under BioProject IS: PRJNA631955.


