Abstract
The complete sequence of two plasmids, pHS-Tet (5.1 kb) and pHS-Rec (9.5 kb), isolated from Haemophilus parasuis strain HS1543 has been obtained. Plasmid pHS-Tet contains four open reading frames including a tet(B) tetracycline resistance gene which unusually did not have an associated tetR repressor gene. From a total of 45 H. parasuis isolates surveyed (15 international reference strains, 15 field isolates selected for their genetic diversity, and 15 recent Australian field isolates), 2 tetracycline-resistant field isolates (HS226 and HS1859) were identified. Analysis of three additional isolates from the same disease outbreak as strain HS1859 revealed a further tetracycline-resistant H. parasuis strain (HS1857, serovar 8) and a tetracycline-resistant Actinobacillus pleuropneumoniae strain (HS1861). An approximately 10.6-kb plasmid was identified in field isolate HS226 and outbreak strains HS1857, HS1859, and HS1861. Southern hybridization analysis of these plasmids showed that the Tet B determinant was present, and restriction digest comparisons suggest that these plasmids are related. This is believed to be the first report of native H. parasuis plasmids and Tet B-mediated tetracycline resistance in this microorganism.
Haemophilus parasuis is the causative agent of Glässer's disease in swine. Common symptoms of this disease include anorexia, pyrexia, and lameness, though some pigs may die suddenly during acute outbreaks (19).
The use of antibiotics in animal production as treatment or prophylaxis against common infections or at subtherapeutic levels in feed to promote growth is under increasing scrutiny (11). Tetracycline has a long history of use in the swine industry (17), and its use generates a strong selective pressure that has resulted in the exchange of tetracycline resistance genes associated with plasmids or transposons within and between bacterial species (13). Tetracycline resistance determinants Tet B, Tet H, and Tet M have previously been found in other members of the Pasteurellaceae (7, 9). Tetracycline-resistant isolates of H. parasuis have previously been reported in Austria (15) and Denmark (1) although the mechanism of this resistance has not been elucidated.
In this study, we have identified and sequenced two native H. parasuis plasmids, one of which encoded the Tet B tetracycline resistance determinant. A survey of tetracycline resistance was undertaken in 45 H. parasuis strains, identifying tet(B) plasmid-mediated tetracycline resistance in two additional H. parasuis strains. One of these tetracycline-resistant field isolates was isolated from a disease outbreak involving both H. parasuis and Actinobacillus pleuropneumoniae. Tetracycline resistance determinant tet(B) was identified in an A. pleuropneumonie and a further H. parasuis isolate from this outbreak.
MATERIALS AND METHODS
Bacteria.
Escherichia coli strains were cultured in Luria-Bertani medium (LB) with appropriate antibiotics. Strain XL1-Blue MR (Stratagene) was used in electroporations using plasmid pHS-Tet, and strain TST-1 (E. coli Genetic Stock Center, Yale University) was used to make the tetR probe. All H. parasuis, Haemophilus paragallinarum (HP31 and 221), and A. pleuropneumoniae (HS1861) strains used in this study were obtained from the culture collection at the Animal Research Institute, Brisbane, Australia. H. parasuis strain HS19, H. paragallinarum strains 221 and HP31, and Haemophilus influenzae strain Rd were used in electroporations using plasmid preparations containing both plasmids pHS-Tet and pHS-Rec. The H. parasuis strains surveyed for tetracycline resistance, including additional outbreak strains, are listed in Table 2. All Pasteurellaceae strains used in this study were grown on TM/SN medium prepared as described previously (20). Briefly, TM/SN is prepared from a basal medium known as TM which contains 1% biosate peptone, 1% NaCl, 0.1% starch, 0.05% glucose, and 1.5% agar. Immediately before pouring, TM/SN medium is supplemented with the following sterile additives: 0.0025% NADH, 0.005% thiamine, 1% heat-inactivated horse serum, and 5% oleic acid bovine serum albumin complex consisting of 4.75% bovine serum albumin (fraction V) in normal saline (with the normal saline containing 0.06% oleic acid and 5% 0.05N NaOH). When required, a liquid medium version of TM/SN, termed TMB, was prepared by omitting the agar from TM/SN.
TABLE 2.
Tetracycline resistance of bacterial strains used in this study
| Groupa | Bacterial strainb | Originc | Associated pathologyd | Serovare | Tetracycline MIC (μg/ml) |
|---|---|---|---|---|---|
| I | No. 4 | Japan | Healthy | 1 | 4 |
| SW 140 | Japan | Healthy | 2 | 4 | |
| SW 114 | Japan | Healthy | 3 | 4 | |
| SW 124 | Japan | Healthy | 4 | 4 | |
| Nagasaki | Japan | Septicemia | 5 | 4 | |
| 131 | Switzerland | Healthy | 6 | 4 | |
| 174 | Switzerland | Healthy | 7 | 4 | |
| C5 | Sweden | NK | 8 | 4 | |
| D74 | Sweden | NK | 9 | 2 | |
| H367 | Germany | Healthy | 10 | 2 | |
| H465 | Germany | Pneumonia | 11 | 4 | |
| H425 | Germany | Polyserositis | 12 | 4 | |
| IA-84-17975 | United States | NK | 13 | 8 | |
| IA-84-22113 | United States | NK | 14 | 4 | |
| SD-84-15995 | United States | Pneumonia | 15 | 4 | |
| II | HS306 | VIC | Septicemia | NT | 2 |
| HS68 | QLD | Septicemia | 5 | 4 | |
| HS202 | VIC | Septicemia | 4 | 4 | |
| HS14 | QLD | NK | 5 | 4 | |
| HS357 | SA | Septicemia | 5 | 2 | |
| HS356 | SA | Septicemia | 13 | 8 | |
| HS333 | VIC | Septicemia | 5 | 4 | |
| HS16 | QLD | Septicemia | 5 | 4 | |
| HS307 | VIC | Septicemia | NT | 4 | |
| HS72 | QLD | Healthy | 9 | 4 | |
| HS145 | TAS | Septicemia | 1 | 4 | |
| HS45 | WA | Respiratory | 4 | 4 | |
| HS58 | WA | Respiratory | NT | 4 | |
| HS29 | VIC | NK | 2 | 4 | |
| HS226 | VIC | Septicemia | NT | 64 | |
| III | HS1919 | NSW | Trachea | 4 | 4 |
| HS1911 | QLD | NK | 4 | 8 | |
| HS1913 | NK | NK | 14 | 2 | |
| HS1906 | VIC | Pneumonia | 13 | 4 | |
| HS1910 | QLD | NK | 1 | 2 | |
| HS1895 | NK | Pneumonia | 2 | 2 | |
| HS1859* | VIC | NK | 8/9 | 64 | |
| HS1854 | WA | NK | 4 | 4 | |
| HS1848 | VIC | Pneumonia | 3 | 4 | |
| HS1939 | NK | NK | 5 | 4 | |
| HS1915 | VIC | NK | 5 | 4 | |
| HS1905 | QLD | NK | NT | 2 | |
| HS1889 | VIC | Septicemia | NT | 4 | |
| HS1921 | WA | Pneumonia | 4 | 4 | |
| HS1916 | NSW | Pneumonia | 4 | 2 | |
| IV | HS1857* | VIC | NK | 8/9 | 64 |
| HS1858* | VIC | NK | 5 | 4 | |
| HS1543 | VIC | NK | 10 | 16 | |
| HS1861* | VIC | NK | NK | 64 | |
| ATCC 27090 | NK | NK | NK | 4 |
Group I, international reference strains; group II, genetically diverse Australian isolates; group III, recent Australian field isolates; group IV, additional strains used in this study.
Asterisk, strains isolated from the same outbreak. Underlined strains are tetracycline resistant. Strains HS1861 and ATCC 27090 are A. pleuropneumoniae.
NSW, New South Wales; QLD, Queensland; SA, South Australia; TAS, Tasmania; VIC, Victoria; WA, Western Australia.
NK, not known.
NT, nontypeable.
DNA techniques.
Restriction and DNA modification enzymes were purchased from New England Biolabs. Taq polymerase for amplifying probes by PCR was purchased from Fisher Biotech. Molecular biology methods used were as previously described (3). QIAGEN plasmid midi kits were used for large-scale plasmid purifications.
DNA sequencing, analysis, and annotation of plasmids.
DNA fragments from a midi-prep (QIAGEN) isolation of plasmids derived from strain HS1543 digested with either Sau3A (cloned fragment sizes, 0.3 and 0.6 kb) or MspI (cloned fragment sizes, 1.0 and 1.6 kb) were cloned into pUC19 digested with BamHI or AccI, respectively, and sequenced using universal M13 forward and reverse primers. Oligonucleotides were designed at the ends of the cloned fragments. Purified plasmid DNA from strain HS1543 was used as a template in subsequent sequencing reactions. After each round of sequencing, new primers were designed until a complete double-stranded sequence of the plasmid was obtained. ABI Prism BigDye Terminator version 3.1 (PE Applied Biosystems) was used for DNA sequencing. Following EDTA/ethanol precipitation, samples were sent to the Australian Genome Research Facility for automated sequencing using an ABI 3730xl 96-capillary automatic sequencer (PE Applied Biosystems).
Sequence data were aligned and annotated using MacVector version 7.2 (Accelrys). Open reading frames (ORFs) were identified using MacVector, and gene identities were assigned using searches against the nucleotide and protein databases at the National Center for Biotechnology Information using the tBLAST-n algorithm (2). Similarity between ORFs was calculated using BLAST analysis of two sequences with filters off.
Electroporation of pHS-Tet into H. parasuis, H. paragallinarum, H. influenzae strain Rd, and E. coli.
pHS-Tet was isolated from strain HS1543 using a QIAGEN midi-prep kit. H. paragallinarum strains 221 and HP31 and H. parasuis strains HS19 and HS29 were grown to an optical density at 600 nm of 0.5 in TMB medium, washed three times with sucrose/glycerol (SG) buffer (15% [vol/vol] glycerol, 272 mM sucrose), and resuspended in a final volume of 0.5 ml of SG buffer. H. influenzae strain Rd was heavily inoculated onto 10 brain heart infusion (BHI) plates, harvested into 50 ml of SG buffer and washed as described above. Cells (100 μl) were mixed with 10 μg of pHS-Tet DNA. After electroporation (Bio-Rad micropulser electroporator, 2.5 kV, 0.2-cm cuvettes), cells were suspended in 1 ml of TMB (H. parasuis and H. paragallinarum) medium or BHI broth supplemented with 10 μg/ml Hemin and 2 μg/ml NAD (H. influenzae) and grown for 1 h at 37°C before plating on TM/SN (H. parasuis and H. paragallinarum) or BHI (H. influenzae) plates supplemented with 5 μg/ml tetracycline and incubated overnight at 37°C with 5% CO2. Tetracycline-resistant colonies were inoculated into appropriate liquid media and grown overnight, and plasmid DNA was isolated using a mini-prep kit. Restriction digests using AccI and ClaI confirmed the presence of pHS-Tet.
One microgram of plasmid pHS-Tet isolated from an H. parasuis HS19/pHS-Tet transformant was electroporated in E. coli strain XL1-Blue MR as described above. Cells were resuspended in 1 ml of LB broth and incubated for 1 h at 37°C with shaking and then plated on LB agar with tetracycline (10 μg/ml). The presence of pHS-Tet in transformants was confirmed using restriction digests as described above.
Southern hybridization analysis.
Bacterial genomic DNA was isolated as previously described (3). Restriction endonuclease (EcoRV or AccI for strain HS1858 only)-digested plasmid DNA [for tet(B) blot] or MspI-digested H. parasuis strain 1543 genomic DNA and EcoRV-digested pHS-Tet ad pHS-Tet/pHS-Rec plasmid preparations (for tetR blot) were separated on 0.7% agarose gels and transferred to GeneScreen Hybridization Transfer membrane (NEN Life Science Products) by capillary action (3). A DNA fragment containing the tet(B) gene was amplified from pHS-Tet using primers BF and BR as previously described (22). A DNA fragment containing the Tn10 tetR gene was amplified from E. coli strain TST-1 genomic DNA using primers TetRfwd (5′-ATGATGTCTAGATTAGATAAAAGTAAAG) and TetRrev (5′-TTAAGACCCACTTTCACATTTAAGTTG). Blots were hybridized with digoxigenin (DIG)-labeled PCR products for 16 h at 68°C. Washes and detection were carried out (DIG DNA Labeling and Detection Kit; Roche) as recommended by the manufacturer.
Antibiotic susceptibility assays.
The tetracycline MIC test described by Blackall (5) was used. Isolates were regarded as being resistant to tetracycline if the MIC was ≥16 μg/ml and sensitive if the MIC was ≤4 μg/ml (5). A. pleuropneumoniae strain ATCC 27090 was used as the control strain. Serial dilutions of bacterial dilutions were also grown on TM/SN medium with and without 5 μg/ml tetracycline to confirm the MIC results.
Nucleotide sequence accession numbers.
The sequences of plasmids pHS-Tet and pHS-Rec have been deposited in the GenBank database under accession numbers AY862435 and AY862436, respectively.
RESULTS AND DISCUSSION
Sequence analysis and genetic organization of plasmids pHS-Tet and pHS-Rec.
Examination of whole genomic DNA preparations of H. parasuis strain HS1543 digested with MspI revealed distinct DNA bands that were confirmed to be plasmids by restriction endonuclease digests of small-scale plasmid isolation preparations.
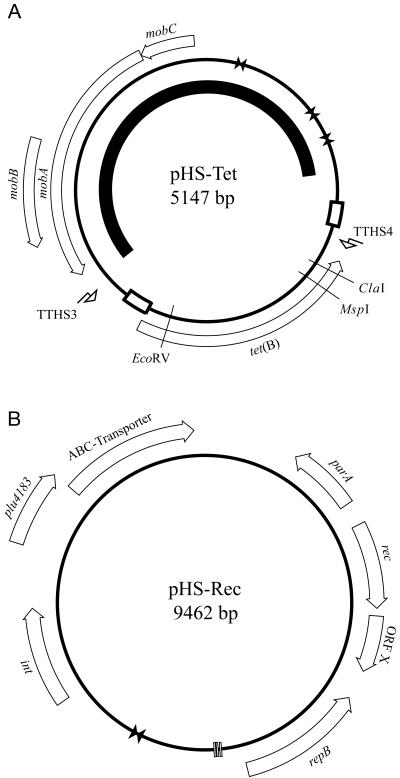
The genetic map of plasmid pHS-Tet is shown in Fig. 1. The plasmid is 5,147 bp in size, and detailed analysis of the sequence revealed a tet(B) tetracycline resistance gene and mobABC plasmid mobilization genes (see Table 1). The plasmid is similar in structure to the Mannheimia haemolytica plasmid pAB2 (23), which differs from pHS-Tet by containing a β-lactamase (bla) resistance gene in place of tet(B). The tet(B) gene has 99% amino acid similarity to the Salmonella enterica serovar Typhi Tn10 tet(B) efflux pump and is flanked by a duplicated 53-bp sequence. Unusually, there is no tetR gene encoding the Tn10-associated repressor protein located downstream of the pHS-Tet tet(B) gene. This is in contrast to plasmid pPAT2 (14), previously isolated from Pasteurella aerogenes, that contains the tet(B) gene and its repressor tetR. Tn10 and plasmid pHS-Tet diverge 15 bp upstream and 78 bp downstream of the tet(B) gene. Plasmid pHS-Tet has a single partial repressor binding site, in contrast to the two binding sites necessary for full gene repression located between the tet(B) and tetR genes in Tn10. Southern hybridization analysis of the HS1543 genome (including plasmids pHS-Tet and pHS-Rec) using a tetR probe confirmed the absence of tetR either on pHS-Tet, pHS-Rec, or in the chromosome (data not shown). Constitutive expression of Tn10-like tetracycline resistance has previously been reported in Haemophilus parainfluenzae, mediated by an inactivating point deletion mutation in the tetR gene (10). Tetracycline sensitivity profiling revealed that strain HS1543 was tetracycline resistant, indicating that the tet(B) gene product is functional. Tetracycline resistance determinants Tet A and Tet B have previously been reported in transposons on a conjugative multidrug-resistant plasmid in E. coli of the normal enteric flora of pigs (21). The tetracycline resistance determinant tet(B), the most common determinant found in Enterobacteriaceae (12), has also been found in Pasteurella multocida isolates from swine (13). In addition, it has been demonstrated that in chickens given feed with tetracycline, the transfer of tetracycline resistance genes occurred between E. coli strains (16). Hence, it is possible that the use of tetracycline as a therapeutic agent and in-feed additive in the swine industry may select for H. parasuis strains resistant to tetracycline.
FIG. 1.
Schematic map of plasmid pHS-Tet (A) and pHS-Rec (B). White arrows indicate putative ORFs, vertical bars indicate direct repeats, bow ties (opposing triangles) indicate inverted repeats, and bent arrows indicate primer binding sites. The black arc inside the map of pHS-Tet indicates the region of pHS-Tet sharing similarity with plasmid pAB2. The white boxes on the map of pHS-Tet indicate regions of duplicated sequence.
TABLE 1.
Summary of genes found on the pHS-Tet and pHS-Rec plasmids of H. parasuis
| Plasmid and genes | Nearest homology | Nucleotide accession no. | Putative function | % DNA identity | % Protein identity/similarity |
|---|---|---|---|---|---|
| Plasmid pHS-Tet genesa | |||||
| tet(B) | S. enterica serovar Typhi Tn10 | AY150213 | Tetracycline efflux pump | 99 | 99/99 |
| MobA | MobA pAB2 | Z21724 | Plasmid mobilization | 99 | 99/100 |
| MobB | MobB pAB2 | Z21724 | Plasmid mobilization | 99 | 98/98 |
| MobC | MobC pAB2 | Z21724 | Plasmid mobilization | 100 | 100/100 |
| Plasmid pHS-Rec genesb | |||||
| rec | Haemophilus somnus | NC_002947 | Recombinase | 44 | 45/54 |
| int | H. paragallinarum p250 | AY300023 | Integrase | 42 | 68/78 |
| repB | H. paragallinarum p250 | AY300023 | Replication | 67 | 23/38 |
| ABC-transporter | Pseudomonas putida KT2440 | NC_002947 | Transporter | 46 | 29/55 |
| parA | Actinobacillus actinomycetemcomitans | AF302424 | DNA partitioning | 42 | 28/48 |
| plu4183 | Photorhabdus luminescens | NC_005126 | Unknown | 57 | 11/16 |
| ORFX | None | Unknown |
The plasmid pHS-Tet also contains genes encoding proteins homologous to the MobABC family of proteins that facilitate plasmid mobilization. These genes share 99% nucleotide identity (1,666/1,668; one gap) with the mobilization genes in plasmid pAB2, which was shown to use host proteins to conjugate E. coli and Pasteurella haemolytica although it was unable to mediate transfer out of E. coli (8). The same region shares 72% identity (1,197/1,668) with plasmid pIG1, a streptomycin- and sulfonamide-resistant plasmid isolated from P. multocida (24). The presence of mobility proteins has implications for disease outbreaks on farms where treatment with antibiotics gives rise to a selective pressure that favors resistant strains that are capable of transferring resistance within and between bacterial species.
There was approximately 1.2 kb of DNA sequence obtained from plasmid pHS-Tet which had 99% similarity (1,192/1,200; 6 gaps) to the replication region of plasmid pAB2 (23) and 88% similarity (1,060/1,200; 12 gaps) to the replication region of plasmid pIG1 (24). In this region, pHS-Tet contained 20-, 16-, and 38-bp inverted repeats, similar to the 20-, 16-, and 38-bp inverted repeats found in the pIG1 replication region. The absence of large ORFs, with the presence of inverted repeats, and homology to known replication origins suggest that the origin of replication for plasmid pHS-Tet may be located in this region. Like plasmids pAB2 and pIG1, plasmid pHS-Tet is able to replicate in E. coli after electroporation into strain XL1-Blue MR.
The genetic map of plasmid pHS-Rec is shown in Fig. 1. The plasmid is 9,462 bp in size, and detailed analysis of the sequence revealed eight ORFs (Table 1). Although plasmid pHS-Rec does not share any DNA or protein similarity to any known plasmid families, the association between repB and parA has been previously reported in the backbone of self-replicating plasmids (4). There appears to be no antibiotic resistance marker or tetR repressor gene on pHS-Rec.
Survey of tetracycline resistance in H. parasuis.
A survey of the prevalence of tetracycline resistance was undertaken using a panel of H. parasuis strains including the 15 international reference strains, a genetically diverse collection of Australian field isolates (15 strains) selected by electropherotype (6), and a group of 15 recent Australian field isolates of different serovars (Table 2). No formally approved methodology exists for a MIC test with H. parasuis. The method we have used has been previously used on another difficult bacterium (H. paragallinarum) and has been shown to give the expected MIC results with formal control strains (5). The modifications to this procedure were validated by the A. pleuropneumoniae control strain ATCC 27090 (18).
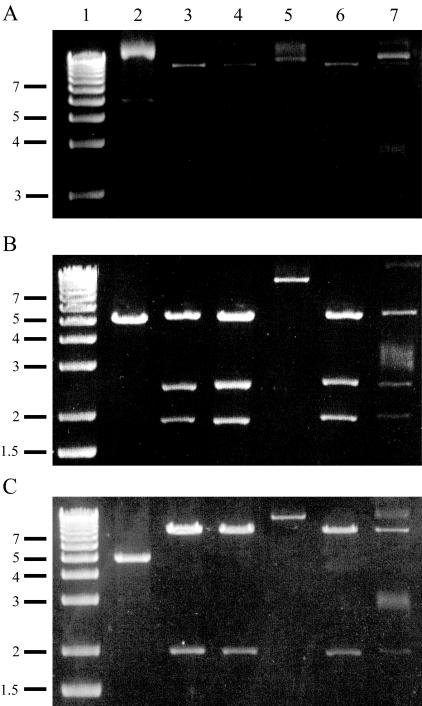
H. parasuis strains HS226 and HS1859 were tetracycline resistant (MIC of 64 μg/ml) as was strain HS1543 (MIC of 16 μg/ml). Additional clinical isolates from the same disease outbreak as strain HS1859 were examined, and a further tetracycline-resistant H. parasuis strain (HS1857) and a resistant A. pleuropneumoniae strain (HS1861) were identified along with a tetracycline-sensitive H. parasuis strain (HS1858). An approximately 10.6-kb plasmid was identified in these strains, and characterization by restriction digests (Fig. 2) suggests that the plasmids from strains HS226 and HS1861 are related. Similarly, plasmids from strains HS1857 and HS1859 appear related, though these two strains are both serovar 8/9, suggesting possible clonality.
FIG. 2.
Restriction endonuclease characterization of plasmids uncut (A), digested with AccI (B), or digested with ClaI (C). Lanes 1, 1-kb ladder (Invitrogen); lanes, 2, pHS-Tet; lanes 3, HS226 plasmid; lanes 4, HS1857 plasmid; lanes 5, HS1858 plasmid; lanes 6, HS1859 plasmid; lanes 7, HS1861 plasmid.
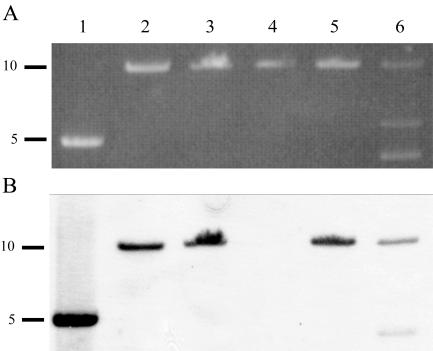
Southern hybridization analysis (Fig. 3) using a tet(B) probe showed that plasmid pHS-Tet and plasmids isolated from strains HS226, HS1857, HS1859, and HS1961 contained the Tet B determinant. However, the Tet B determinant was not found in strain HS1858, which is consistent with the tetracycline-sensitive phenotype of this strain. Strain HS226 was isolated in 1991 from central nervous system tissue of a 24-week-old pig that died from Glässer's disease on farm A. Breeding stock was supplied from farm A to farm B, where strains HS1857, HS1858, HS1859, and HS1861 were isolated from a Glässer's disease outbreak in 2003. Thus, it appears that the same plasmid has been identified from two clinical isolates (H. parasuis HS226 and A. pleuropneumonie HS1861) from epidemiologically connected disease outbreaks occurring 12 years apart on two different farms.
FIG. 3.
Agarose gel of restriction endonuclease linearization of plasmids using EcoRV (except that AccI was used to linearize plasmid from HS1858) (A) and Southern hybridization analysis of gel shown in panel A transferred to nitrocellulose membrane and probed with a DIG-labeled PCR product containing the tet(B) gene (B). Lanes 1, pHS-Tet; lanes 2, HS226 plasmid; lanes 3, HS1857 plasmid; lanes 4, HS1858 plasmid; lanes 5, HS1859 plasmid; lanes 6, HS1861 plasmid. DNA fragment sizes are indicated in kb.
The occurrence of tetracycline resistance among H. parasuis isolates in this study (4 resistant strains from 48 strains examined) is comparable to earlier work (15), where 6.4% of isolates from Austria were tetracycline resistant. However, the occurrence of tetracycline resistance in H. parasuis is considerably lower than that reported in a study of A. pleuropneumoniae, a related pathogenic porcine Pasteurellaceae, where 12.1% of clinical isolates from Austria were tetracycline resistant (15).
Acknowledgments
Work in the Jennings laboratory, Tamsin Terry's position, and John Lancashire's Ph.D. scholarship were supported by an ARC Linkage grant LP0218929 and by Bioproperties (Australia) Pty Ltd.
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, and O. Angen. 2004. Antimicrobial sensitivity of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Vet. Microbiol. 101:143-146. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. 1994. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotech. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 5.Blackall, P. J. 1988. Antimicrobial drug resistance and the occurrence of plasmids in Haemophilus paragallinarum. Avian Dis. 32:742-747. [PubMed] [Google Scholar]
- 6.Blackall, P. J., D. J. Trott, V. Rapp-Gabrielson, and D. J. Hampson. 1997. Analysis of Haemophilus parasuis by multilocus enzyme electrophoresis. Vet. Microbiol. 56:125-134. [DOI] [PubMed] [Google Scholar]
- 7.Chaslus-Dancla, E., M.-C. Lesage-Descauses, S. Leroy-Sétrin, J.-L. Martel, and J.-P. Lafont. 1995. Tetracycline resistance determinants, Tet B and Tet M, detected in Pasteurella haemolytica and Pasteurella multocida from bovine herds. J. Antimicrob. Chemother. 36:815-819. [DOI] [PubMed] [Google Scholar]
- 8.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, L. M., P. C. Blanchard, and D. C. Hirsh. 1996. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob. Agents Chemother. 40:1558-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heuer, C., R. K. Hickman, M. S. Curiale, W. Hillen, and S. B. Levy. 1987. Constitutive expression of tetracycline resistance mediated by a Tn10-like element in Haemophilus parainfluenzae results from a mutation in the repressor gene. J. Bacteriol. 169:990-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joint Expert Advisory Committee on Antibiotic Resistance (JETACAR). 1999. The use of antibiotics in food-producing animals: antibiotic-resistant bacteria in animals and humans. Commonwealth Department of Health and Aged Care and Commonwealth Department of Agriculture, Fisheries and Forestry, Canberra, Australia.
- 12.Kehrenberg, C., S. A. Salmon, J. L. Watts, and S. Schwarz. 2001. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMHT1. J. Antimicrob. Agents Chemother. 48:631-640. [DOI] [PubMed] [Google Scholar]
- 13.Kehrenberg, C., G. Schulze-Tanzil, J. L. Martel, E. Chaslus-Dancla, and S. Schwarz. 2001. Antimicrobial resistance in Pasteurella and Mannheimia: epidemiology and genetic basis. Vet. Res. 32:323-339. [DOI] [PubMed] [Google Scholar]
- 14.Kehrenberg, C., and S. Schwarz. 2001. Molecular analysis of tetracycline resistance in Pasteurella aerogenes. Antimicrob. Agents Chemother. 45:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kofer, J., F. Hinterdorfer, and M. Awad-Masalmeh. 1992. Vorkommen und Resistenz gegen Chemo-therapeutika von lungenpathogenen Bakterien aus Sektionsmaterial beim Schwein. Tierarztl. Prax. 20:600-604. [PubMed] [Google Scholar]
- 16.Levy, S. B., G. B. Fitzgerald, and A. B. Macone. 1976. Spread of antibiotic resistance plasmids from chicken to chicken and from chicken to man. Nature 260:400-421. [DOI] [PubMed] [Google Scholar]
- 17.Mathew, A. G., W. G. Upchurch, and S. E. Chattin. 1998. Incidence of antibiotic resistance in fecal Escherichia coli isolated from commercial swine farms. J. Anim. Sci. 76:429-434. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A2, 2nd ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Nicolet, J. 1992. Haemophilus parasuis, p. 526-528. In A. Leman, B. Straw, W. Mengeling, S. D'Allaire, and D. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames, Iowa.
- 20.Reid, G., and P. J. Blackall. 1987. Comparison of adjuvants for an inactivated infectious coryza vaccine. Avian Dis. 31:59-63. [PubMed] [Google Scholar]
- 21.Sunde, M., and H. Sorum. 2001. Self-transmissible multidrug resistance plasmids in Escherichia coli of the normal intestinal flora of healthy swine. Microb. Drug Resist. 7:191-196. [DOI] [PubMed] [Google Scholar]
- 22.Wilkerson, C., M. Samadpour, N. van Kirk, and M. C. Roberts. 2004. Antibiotic resistance and distribution of tetracycline genes in Escherichia coli 0157:H7 isolates from humans and bovines. Antimicrob. Agents Chemother. 48:1066-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood, A. R., F. A. Lainson, F. Wright, G. D. Baird, and W. Donachie. 1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res. Vet. Sci. 58:163-168. [DOI] [PubMed] [Google Scholar]
- 24.Wright, C. L., R. A. Strugnell, and A. L. Hodgson. 1997. Characterisation of a Pasteurella multocida plasmid and its use to express recombinant proteins in P. multocida. Plasmid 37:65-79. [DOI] [PubMed] [Google Scholar]