Abstract
The genotypic inhibitory quotient (GIQ) has been proposed as a way to integrate drug exposure and genotypic resistance to protease inhibitors and can be useful to enhance the predictivity of virologic response for boosted protease inhibitors. The aim of this study was to evaluate the predictivity of the GIQ in 116 protease inhibitor-experienced patients treated with lopinavir-ritonavir. The overall decrease in human immunodeficiency virus type 1 (HIV-1) RNA from baseline to month 6 was a median of −1.50 log10 copies/ml and 40% of patients had plasma HIV-1 RNA below 400 copies/ml at month 6. The overall median lopinavir study-state Cmin concentration was 5,856 ng/ml. Using univariate linear regression analyses, both lopinavir GIQ and the number of baseline lopinavir mutations were highly associated with virologic response through 6 months. In the multivariate analysis, only lopinavir GIQ, baseline HIV RNA, and the number of prior protease inhibitors were significantly associated with response. When the analysis was limited to patients with more highly mutant viruses (three or more lopinavir mutations), only lopinavir GIQ remained significantly associated with virologic response. This study suggests that GIQ could be a better predictor of the virologic response than virological (genotype) or pharmacological (minimal plasma concentration) approaches used separately, especially among patients with at least three protease inhibitor resistance mutations. Therapeutic drug monitoring for patients treated by lopinavir-ritonavir would likely be most useful in patients with substantially resistant viruses.
The development of human immunodeficiency virus (HIV) drug resistance during antiretroviral therapy can compromise the efficacy of subsequent regimens following virologic failure. Several studies have shown that changes in baseline viral genotype, compared to that of wild-type virus, adversely affect the virologic response of antiretroviral-experienced subjects with a subsequent regimen (3, 4, 19). However, the efficacy of antiretroviral treatment can be impaired by several factors, including poor adherence to treatment regimens, suboptimal antiviral potency and/or drug concentrations, and of course selection of antiretrovirus agent-resistant HIV quasispecies.
More information on the effect of these parameters should be beneficial for optimizing the use of protease inhibitors in salvage therapy. The inhibitory quotient, mainly used for the protease inhibitors, has been proposed as a way to integrate drug exposure and viral susceptibility. Defined as the ratio between the trough concentration of a drug in a patient and the susceptibility of the virus in that patient to that drug, the inhibitory quotient has been associated with virologic response to protease inhibitor-based antiretroviral therapy in several studies (6, 17; J. Hellinger, A. B. Morris, S. Piscitelli, D. Gordon, K. Foy, L. Jackson-Pope, D. Cordeiro, M. Peeters, R. Hoetelmans, P. J. de Caprariis, and C. J. Cohen, Abstr. 9th Conf. Retrovir. Opport. Infect., abstr. 451W, 2002).
The susceptibility of the virus has been initially expressed as the plasma protein-corrected in vitro 50% inhibitory concentration, as determined by a phenotypic assay or alternatively by the virtual phenotype. However, genotypic scores (based on the number of baseline mutations out of a cumulative number of mutations that were found to be associated with lowered rates of virologic response) are other ways to evaluate the magnitude of protease inhibitor resistance (8). Recently, in a cohort of protease inhibitor-experienced patients treated with an amprenavir-ritonavir-based regimen, the genotypic inhibitory quotient (GIQ, ratio of trough amprenavir concentration to number of baseline protease inhibitor mutations) was shown to be a significant predictor of the virological response to a protease inhibitor-containing regimen (11).
Lopinavir is a protease inhibitor that is rapidly metabolized in vitro. However, coadministration with ritonavir (an inhibitor of the CYP P450 3A4 isoenzyme) inhibits lopinavir metabolism, significantly increasing plasma concentrations of the drug and affording high and consistent levels of lopinavir. Thus, a coformulation of lopinavir (lopinavir-ritonavir) has been developed for clinical use in treatment-naive and in protease inhibitor-experienced patients (1, 8, 14). Previously, 11 mutations in HIV protease were found to be associated with significant changes in in vitro susceptibility to lopinavir (9). These 11 mutations constitute the original lopinavir mutation score. In study 957, virologic response to lopinavir-ritonavir, efavirenz, and nucleoside reverse transcriptase inhibitors in multiple protease inhibitor-experienced, nonnucleoside reverse transcriptase inhibitor-naive patients was highest in those patients with a baseline lopinavir mutation score of 5 or less. Intermediate response was observed in patients with a baseline mutation score of 6 to 7, and the lowest response was observed in patients with 8 or more of the above 11 mutations at baseline (8). Similar data were obtained in another study, suggesting that the lopinavir mutation score is relevant for predicting the virologic response to lopinavir-ritonavir (13); however, the de novo analysis of response with respect to genotype has not been adequately characterized. Furthermore, the original lopinavir mutation score was based on an analysis of 112 viral isolates from protease inhibitor-experienced patients. Mutations that were underrepresented in that panel of isolates are not currently included in this score, despite the possibility that some may contribute to significantly reduced susceptibility to lopinavir (16).
In order to further characterize genotypic resistance to lopinavir-ritonavir and to quantitatively assess the effect of individual mutations within the mutation score on virologic response, data from the lopinavir-ritonavir authorization for use (expanded-access) program were analyzed. In these highly antiretroviral agent-experienced patients, mutations at 10 positions in the protease (10, 20, 24, 33, 36, 47, 48, 54, 82, and 84) were found to be associated with lowered rates of virologic response (J. D. Isaacson, D. J. Kempf, V. Calvez, I. Cohen-Codar, D. Descamps, E. Guillevic, B. Bernstein, E. Sun, J. P. Chauvin, and R. A. Rode, Abstr. 9th Conf. Retrovir. Opport. Infect., abstr. 559, 2002). The total number of these mutations appeared to predict virologic response better than any individual mutation and better than the original lopinavir mutation score for this data set.
The aim of the present study was to integrate the descriptors of genotypic resistance to lopinavir with pharmacological parameters to construct a GIQ that is predictive of virologic response to lopinavir-ritonavir in protease inhibitor-experienced patients. In addition, we analyzed whether the GIQ was more predictive of response than genotype alone.
(This study was presented at the second International AIDS Society meeting in July 2003, Paris, France, abstract 827.)
MATERIALS AND METHODS
Study population.
From March 2000 to April 2001, the French Drug Agency authorized the prescription of lopinavir-ritonavir in the setting of a temporary authorization for use. Highly antiretroviral agent-experienced patients (n = 792) were enrolled in an observational cohort. At the initiation of lopinavir-ritonavir therapy, demographic data, prior and current antiretroviral regimen, HIV-1 RNA, CD4 cell count, as well as HIV-1 protease and reverse transcriptase gene sequences were collected. Patients were followed up at months 1, 3, and 6 with HIV-1 RNA and CD4 measurements. Within this cohort, 116 patients with complete genotype, including mutation polymorphisms in the protease gene and during-study viral load and pharmacokinetic data available were extensively analyzed. For inclusion in the analysis, all patients were required to have at least one plasma lopinavir Cmin value and had an HIV-1 RNA value at month 3 to 6. Baseline patient characteristics are presented in Table 1.
TABLE 1.
Baseline patient characteristicsa
| Parameter | Value |
|---|---|
| Male, no. (%) | 96 (83) |
| Median age, y (range) | 40 (27-68) |
| CDC stage C, no. (%) | 67 (58) |
| Median plasma HIV-1 RNA, log copies/ml (range) | 4.9 (2.3-6.9) |
| Median CD4 cell count/mm3 (range) | 120 (2-626) |
| Median no. of previous: | |
| Antiretroviral agents (range) | 9 (3-14) |
| Nucleoside analogues (range) | 5 (1-6) |
| Nonnucleoside analogues (range) | 1 (0-3) |
| Protease inhibitors (range) | 3 (1-5) |
| Protease inhibitor exposure, no. of patients (%) | |
| Indinavir | 95 (82) |
| Nelfinavir | 76 (66) |
| Ritonavir | 81 (70) |
| Saquinavir | 79 (68) |
| Amprenavir | 36 (31) |
Total number of study patients = 116.
Virological analyses.
Quantification of plasma HIV-1 RNA was performed using the Cobas Amplicor Monitor assay v1.5 (Roche Diagnostics, Bazel, Switzerland) with a detection limit of 400 copies/ml.
Plasma HIV-1 RNA was used for sequence analysis of the reverse transcriptase (reverse transcriptase) gene (codons 1 to 240) and protease gene (codons 1 to 99). Plasma HIV-1 RNA was amplified by a one-step reverse transcription-PCR using the TITAN one tube reverse transcription PCR kit (Boehringer Mannheim) followed by a nested PCR with AmpliTaq Gold (Applied Biosystems, Foster City, California). All primers used were described previously (7, 10, 15). Direct sequencing of the PCR product was performed using the d-rhodamine terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA). Sequencing reaction products were analyzed on an ABI 377 Genetic Analyzer (PE Applied Biosystems). The sequences were analyzed using the Sequence Navigator software (PE Applied Biosystems) by comparing the sense and antisense strands of each fragment with the wild-type virus HXB2 sequence. For each patient at baseline, the number of lopinavir mutations was defined as those occurring at the following 10 amino acid positions: 10, 20, 24, 33, 36, 47, 48, 54, 82, and 84 (J. D. Isaacson, D. J. Kempf, V. Calvez, I. Cohen-Codar, D. Descamps, E. Guillevic, B. Bernstein, E. Sun, J. P. Chauvin, and R. A. Rode, Abstr. 9th Conf. Retrovir. Opport. Infect., abstr. 559, 2002).
Pharmacological analyses.
Blood samples were collected to determine lopinavir and ritonavir plasma concentrations at steady state. Intervals between the last drug intake and sampling were recorded. The time frame after dose was 12 ± 2 h. Lopinavir and ritonavir minimal plasma concentrations (Cmin) were determined by reversed-phase high-pressure liquid chromatography using a slightly modified method previously described for ritonavir (12). The lopinavir and ritonavir methods were validated over plasma concentration ranges of 30 to 15,000 ng/ml and 30 to 7,500 ng/ml, respectively, with lower limits of quantification of 30 and 30 ng/ml, respectively. The between-assay bias for lopinavir and ritonavir was below 9% and 10% for all assays, respectively.
Statistical analyses.
The efficacy variable was HIV-1 RNA change from baseline to month 6. For each patient, the HIV-1 RNA value during months 3 to 6 that was closest to month 6 was used. The effects of the number of baseline lopinavir mutations, lopinavir Cmin, and lopinavir genotypic inhibitory quotient (lopinavir GIQ) were assessed by linear regression. For each patient, lopinavir Cmin was defined as the median of all available lopinavir concentrations from months 1 to 6. The lopinavir GIQ was defined as the ratio of lopinavir Cmin to the number of baseline lopinavir mutations, as described for amprenavir (11), with the exception that patients with 0 baseline protease inhibitor mutations were assigned a lopinavir GIQ equal to their Cmin value. In addition to lopinavir GIQ, covariates considered included baseline HIV-1 RNA level, baseline CD4 cell count, number of prior protease inhibitors, number of active nucleoside reverse transcriptase inhibitors (NRTIs), non-NRTIs (NNRTIs) used in an NNRTI-naive patient, gender, Centers for Disease Control class C status, and age.
Quantitative independent variables were evaluated to determine whether the relationship with HIV-1 RNA change from baseline was linear. For most variables, the assumption of a linear relationship was reasonable; however, for lopinavir GIQ a nonlinear relationship was observed that suggested this variable should be log-transformed for analysis. The number of active NRTIs was determined using genotypic resistance testing criteria derived from the French ANRS AC11 algorithm (http://www.hivfrenchresistance.org). Each NRTI used by a patient was assigned a value of 1 (susceptible), 0.5 (possible resistance) or 0 (resistant).
To confirm the effects observed in the univariate analysis, a multiple linear regression analysis was conducted using a forward stepwise selection procedure with a significance level of 0.10 to enter and remain in the model. Variables considered statistically significant or marginally significant in the univariate model (P < 0.10) were included in the multiple linear regression model.
Based on observations that viruses with 0 to 2 mutations often display wild type susceptibility (9), and because differences in Cmin would not be expected to correlate with initial response in patients with lopinavir-sensitive HIV variants, we repeated the analyses described above in the subset of 67 patients with 3 or more baseline mutations (i.e., those whose baseline variants are more likely to have substantially reduced susceptibility to lopinavir).
If undetectable HIV-1 RNA is achieved by a substantial proportion of patients, analysis of the mean change from baseline has the potential to produce misleading results due to censoring of HIV-1 RNA values. Therefore, using logistic regression, we conducted a sensitivity analysis to assess the association of lopinavir GIQ and number of baseline lopinavir mutations with HIV-1 RNA < 400 copies/ml at month 6.
RESULTS
Baseline characteristics.
The 116 patients enrolled in this study had at least one plasma lopinavir Cmin value and had an HIV-1 RNA value at month 6. Baseline characteristics indicated a relatively advanced population, with a median of 9 prior antiretroviral agents including a median of 3 prior protease inhibitors, and a median baseline CD4 cell count of 120 cells/mm3 (Table 1). Patients had a median (range) of 3 (0 to 7) baseline lopinavir mutations. A majority of patients received concurrent treatment with two NRTIs and 29% of patients used an NNRTI. The median number of active NRTIs was 1.
Virologic response to lopinavir-ritonavir containing regimen and pharmacological results.
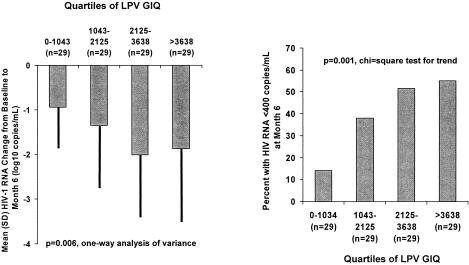
The overall decrease in HIV-1 RNA from baseline to month 6 was a median (range) of −1.50 (+0.73, −4.64) log10 copies/ml, and 46 patients (40%) had plasma HIV-1 RNA below 400 copies/ml at month 6. The overall median (range) lopinavir steady-state Cmin concentration was 5856 (84-16091) ng/ml. Pharmacokinetic measurements and lopinavir GIQ are summarized in Table 2. GIQ values ranged from 36 to 16,091. Virologic response with respect to GIQ, expressed by quartiles, is shown in Fig. 1. The mean decline in HIV RNA increased to a maximum of ca. 2 log10 copies/ml in the patients with GIQ values above the median (GIQ = 2,125). Similarly, >50% of patients with GIQ > 2,125 and achieved HIV RNA < 400 copies/ml, whereas only 15% of patients with GIQ < 1,043 (lowest quartiles) experienced complete virologic response.
TABLE 2.
Summary of pharmacokinetic measurements and lopinavir GIQa
| Parameter | Value |
|---|---|
| No. of Cmin measurements | |
| 1 | 13 (11) |
| 2 | 75 (65) |
| 3 | 24 (21) |
| 4 | 4 (3) |
| LPV Cmin (ng/ml) | |
| Median | 5,856 |
| Interquartile range | 3,920-7,931 |
| Intrasubject variability in LPV Cmin | |
| No. of patients with value >1 | 103 |
| Median %CV | 35 |
| Lopinavir GIQ | |
| Median | 2,125 |
| Interquartile range | 1,043-3,638 |
Cmin, minimum concentration; CV, coefficient of variation; GIQ, genotypic inhibitory quotient; LPV, lopinavir.
FIG. 1.
HIV-1 RNA mean change from baseline to month 6 and proportion of patients with HIV-1 RNA < 400 copies/ml at month 6 by quartiles of lopinavir (LPV) genotypic inhibitory quotient.
Predictors of virologic response.
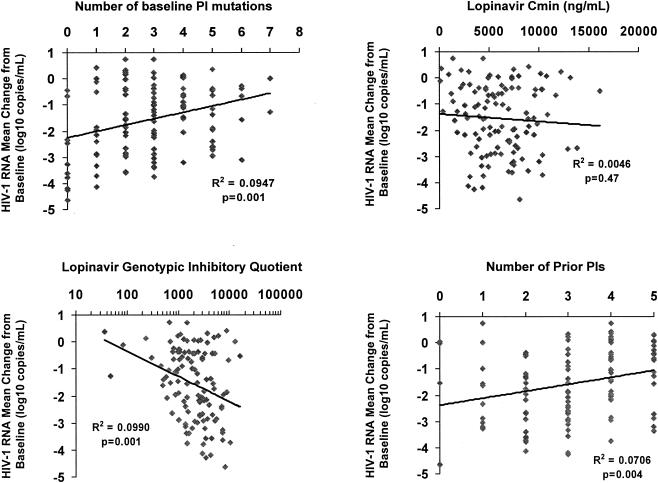
The quantitative relationships between various pharmacological and baseline variables and virologic response were probed using simple linear regression (Fig. 2). As observed previously, no correlation between lopinavir Cmin and response was observed (6). In contrast, having fewer baseline lopinavir mutations and a higher (log transformed) lopinavir GIQ were both significant predictors of a greater decline in HIV-1 RNA. Virologic response was also better among the 9% of patients who were NNRTI-naive and received a NNRTI concomitant with lopinavir-ritonavir. In contrast, the number of active NRTIs was not significantly associated with response in this highly treatment-experienced cohort. Other factors either statistically significantly or marginally associated with response were having fewer prior protease inhibitors (Fig. 2), lower age, and higher baseline HIV-1 RNA (Table 3). The final model from the multiple linear regression analysis using forward stepwise selection included lopinavir GIQ, baseline HIV-1 RNA, and number of prior protease inhibitors.
FIG. 2.
Univariate effects of selected baseline and pharmacologic characteristics on HIV-1 RNA change from baseline to month 6 among all patients. PIs, protease inhibitors.
TABLE 3.
Effects of baseline and pharmacologic characteristics on HIV-1 RNA change from baseline among all patients and patients with three or more baseline protease inhibitor mutationsa
| Variable | All patients
|
Patients with three or more PI mutations
|
||
|---|---|---|---|---|
| P (simple linear regression) | P (multiple linear regression) | P (simple linear regression) | P (multiple linear regression) | |
| No. of LPV mutations | 0.001 | ns | 0.25 | |
| LPV Cmin | 0.47 | 0.06 | ns | |
| Log10 LPV GIQ | 0.001 | 0.008 | 0.03 | 0.04 |
| Baseline HIV-1 RNA | 0.10 | 0.04 | 0.32 | |
| Baseline CD4 count | 0.59 | 0.29 | ||
| No. of prior PIs | 0.004 | 0.04 | 0.17 | |
| No. of active NRTIs | 0.17 | 0.46 | ||
| NNRTI use in an NNRTI-naive patient | 0.05 | ns | 0.06 | 0.09 |
| CDC class C status | 0.27 | 0.18 | ||
| Gender | 0.58 | 0.36 | ||
| Age | 0.05 | ns | 0.56 | |
PI, protease inhibitor; LPV, lopinavir; GIQ, genotypic inhibitory quotient; NRTI; nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; ns, not significant.
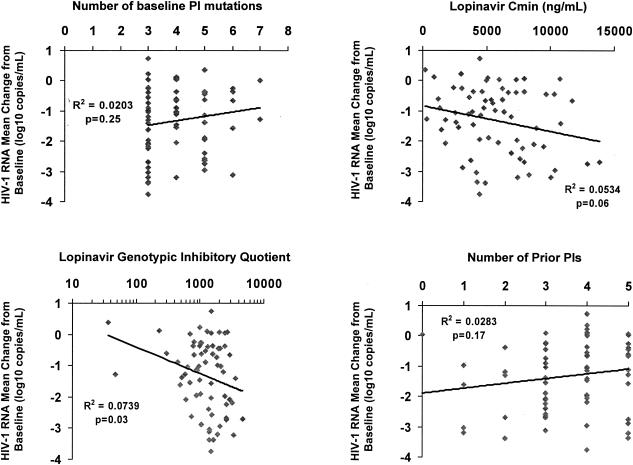
In the subset of 67 patients with three or more baseline lopinavir mutations (i.e., those whose baseline variants are more likely to have substantially reduced susceptibility to lopinavir), using univariate analysis, the number of baseline lopinavir mutations was no longer significantly correlated with HIV-1 RNA change from baseline. The correlation of lopinavir GIQ with response remained statistically significant in this subset. Interestingly, the lopinavir Cmin became marginally associated with response (Table 3, Fig. 3). In the final multiple linear regression model using forward stepwise selection, the lopinavir GIQ and the use of a new NNRTI were significant predictors of HIV-1 RNA decline.
FIG. 3.
Univariate effects of selected baseline and pharmacologic characteristics on HIV-1 RNA change from baseline to month 6 among patients with three or more baseline protease inhibitor (PI) mutations.
A second analysis of the predictors of response was conducted using HIV-1 RNA < 400 copies/ml at month 6 as the response variable. Results were similar to the analysis of HIV-1 RNA change from baseline. Among all patients, lopinavir GIQ and number of baseline lopinavir mutations were significantly associated with HIV-1 RNA < 400 copies/ml at month 6 (P ≤ 0.001 for each by univariate logistic regression). Among patients with 3 or more baseline lopinavir mutations, lopinavir GIQ (P = 0.039) was significantly associated with HIV-1 RNA < 400 copies/ml, but number of baseline lopinavir mutations (P = 0.159) was not associated with response.
DISCUSSION
In this study, we analyzed the response to lopinavir-ritonavir in a cohort of highly antiretroviral agent-experienced patients with respect to various pharmacological and baseline parameters. Using univariate linear regression analyses, both lopinavir GIQ and the number of baseline lopinavir mutations were highly associated with virologic response through 6 months. In the multivariate analysis, only lopinavir GIQ, baseline HIV RNA and the number of prior protease inhibitors were significantly associated with response. When the analysis was limited to patients with more highly mutant virus (3 or more lopinavir mutations), the number of mutations no longer predicted response, but the lopinavir Cmin was marginally associated. Taken together, these results suggest that variability in lopinavir pharmacokinetics are likely to be most important when significant resistance is present at baseline. Consequently, higher doses of lopinavir-ritonavir might be useful in some patients with protease inhibitor-resistant HIV (R. Bertz, J. Li, M. King, D. J. Kempf, D. Podzamczer, C. Flexner, C. Katlama, D. V. Havlir, S. Letendre, J. Eron, L. Weiss, J. Gatell, A. Simon, K. Robinson, and S. Brun, Abstr. 11th Conf. Retrovir. Opport. Infect., abstr. 134, 2004). The loss of the association of baseline genotype with response in the above subset also suggests that as more mutations accumulate (and the number of possible combinations of mutations greatly expands), the variability in the phenotypic susceptibility of variants with a given number of mutations increases compared to viruses with just one or two mutations.
Notably, according to the present method, the change in GIQ values for a given Cmin is incrementally smaller as more baseline mutations are present (for example, between one and two patients, GIQ changes by 50% at a constant Cmin whereas the incremental changes only 20% between four and five mutations). The observation that the log-transformed GIQ but not the GIQ itself (data not shown) was predictive of response suggests that the average change in lopinavir susceptibility is relatively constant with increasing numbers of mutations. This general relationship has been observed in studies characterizing the relationship of in vitro lopinavir phenotype and HIV genotype (8, 9).
The overall median decrease from baseline in HIV-1 RNA in our study was −1.50 log10 copies/ml in these highly pretreated, NNRTI- and multiple protease inhibitor-experienced patients, with a high number of protease inhibitor resistance mutations at baseline. This efficacy of lopinavir-ritonavir in protease inhibitor experienced patients was observed previously in clinical trials or in other expanded access datasets and in the whole cohort of 792 patients (8, 13) (V. Calvez, I. Cohen-Codar, A. G. Marcelin, E. Guillevic, J. Isaacson, R. Rode, B. Bernstein, E. Sun, D. Kempf, J. P. Chauvin. Abstr. 5th Int. Work. HIV Drug Resist. Treat. Strat., abstr. 90, 2001). Previous studies evidenced the effect of protease inhibitor Cmin on the virologic response among protease inhibitor experienced patients (5, 11). This correlation has been also suggested for lopinavir-ritonavir (2);D. Gonzales de Requena, O. Gallego, F. Blanco, L. Valer, T. Garcia-Benayas, C. De Mendoza, I. Jimenez-Nacher, and V. Soriano. Abstr. 10th Conf. Retrovir. Opport. Infect., abstr. 525, 2003).
In the current retrospective analysis, we demonstrated that inhibitory quotient evaluated using genotypic test results was a better predictor of the virologic response than virological (genotype) or pharmacological (Cmin) approaches used separately, especially among patients with at least 3 protease inhibitor resistance mutations. The observation of predictivity of the GIQ, even among patients with substantially reduced genotypic susceptibility to lopinavir-ritonavir, is in accordance with its pharmacokinetic profile, in which mean trough levels (5,500 ng/ml) are sustained far above (>75-fold when dosed at 400/100 mg twice-daily) the human serum-adjusted 50% inhibitory concentration for wild-type virus (inhibitory quotient > 75). This finding also suggests that therapeutic drug monitoring, which is essential for calculation of GIQ, is likely to be most useful in lopinavir-ritonavir-treated patients with substantially resistant viruses.
The correlation of GIQ with response is consistent with findings of other observational cohorts using amprenavir/ritonavir, saquinavir/ritonavir and atazanavir (11) (V. Soriano, L. Valer, D. Gonzales de Requena, C. De Mendoza, and J. Gonzalez-Lahoz. Abstr. 43rd ICAAC, abstr. H-1999, 2003; A. Barrios, A. Rendon, P. Rios, L. Martin-Carbonero, D. Conzalez de Requena, O. Gallego, L. Valer, I. Maida, I. Jimenez-Nacher, J. Gonzalez-Laboz, and V. Soriano. Abstr. 11th Conf. Retrovir. Opport. Infect., abstr. 606, 2004). Use of GIQ, which is based on a relatively inexpensive and accessible genotypic assay, may have practical advantages over inhibitory quotient approaches based on phenotypic assays. Notably, the GIQ will be subject to the inherent limitations of the rules-based genotypic algorithm upon which it is based (18). For example, the 10 lopinavir mutations considered in this study do not include some low-prevalence mutations known to be associated with reduced lopinavir susceptibility (16). However, the present findings suggest that the GIQ may enhance the predictivity of genotype interpretation. Thus, this approach might be useful in therapeutic drug monitoring to define the target plasma concentration required to control replication of viruses at different stages of protease inhibitor resistance measured by the number of protease inhibitor resistance mutations. Consequently, the predictability of response by GIQ should be validated in other datasets and in prospective clinical trials.
Acknowledgments
This study was supported by Abbott Laboratories, France.
REFERENCES
- 1.Benson, C. A., S. G. Deeks, S. C. Brun, R. M. Gulick, J. J. Eron, H. A. Kessler, R. L. Murphy, C. Hicks, M. King, D. Wheeler, J. Feinberg, R. Stryker, P. E. Sax, S. Riddler, M. Thompson, K. Real, A. Hsu, D. Kempf, A. J. Japour, and E. Sun. 2002. Safety and antiviral activity at 48 weeks of lopinavir/ritonavir plus nevirapine and 2 nucleoside reverse-transcriptase inhibitors in human immunodeficiency virus type 1-infected protease inhibitor-experienced patients. J. Infect. Dis. 185:599-607. [DOI] [PubMed] [Google Scholar]
- 2.Boffito, M., I. Arnaudo, R. Raiteri, S. Bonora, A. Sinicco, A. Di Garbo, H. E. Reynolds, P. G. Hoggard, D. J. Back, and G. Di Perri. 2002. Clinical use of lopinavir/ritonavir in a salvage therapy setting: pharmacokinetics and pharmacodynamics. AIDS 16:2081-2083. [DOI] [PubMed] [Google Scholar]
- 3.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 5.Durant, J., P. Clevenbergh, R. Garraffo, P. Halfon, S. Icard, P. Del Giudice, N. Montagne, J. M. Schapiro, and P. Dellamonica. 2000. Importance of protease inhibitor plasma levels in HIV-infected patients treated with genotypic-guided therapy: pharmacological data from the Viradapt Study. AIDS 14:1333-1339. [DOI] [PubMed] [Google Scholar]
- 6.Hsu, A., J. Isaacson, S. Brun, B. Bernstein, W. Lam, R. Bertz, C. Foit, K. Rynkiewicz, B. Richards, M. King, R. Rode, D. J. Kempf, G. R. Granneman, and E. Sun. 2003. Pharmacokinetic-pharmacodynamic analysis of lopinavir-ritonavir in combination with efavirenz and two nucleoside reverse transcriptase inhibitors in extensively pretreated human immunodeficiency virus-infected patients. Antimicrob Agents Chemother. 47:350-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung, M., H. Agut, D. Candotti, D. Ingrand, C. Katlama, and J. Huraux. 1992. Suceptibility of HIV-1 isolates to zidovudine: correlation between widely applicable culture test and PCR analysis. J. Acquir. Immune Defic. Syndr. 5:359-364. [PubMed] [Google Scholar]
- 8.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, J. Sylte, B. Richards, B. Bernstein, R. Rode, and E. Sun. 2002. Analysis of the virologic response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir. Ther. 7:165-174. [PubMed] [Google Scholar]
- 9.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 11.Marcelin, A. G., C. Lamotte, C. Delaugerre, N. Ktorza, H. Ait Mohand, R. Cacace, M. Bonmarchand, M. Wirden, A. Simon, P. Bossi, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, and V. Calvez. 2003. Genotypic inhibitory quotient as predictor of virological response to ritonavir-amprenavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob Agents Chemother. 47:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh, K. C., E. Eiden, and E. McDonald. 1997. Determination of ritonavir, a new HIV protease inhibitor, in biological samples using reversed-phase high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 704:307-313. [DOI] [PubMed] [Google Scholar]
- 13.Masquelier, B., D. Breilh, D. Neau, S. Lawson-Ayayi, V. Lavignolle, J. M. Ragnaud, M. Dupon, P. Morlat, F. Dabis, and H. Fleury. 2002. Human immunodeficiency virus type 1 genotypic and pharmacokinetic determinants of the virological response to lopinavir-ritonavir-containing therapy in protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 46:2926-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, R. L., S. Brun, C. Hicks, J. J. Eron, R. Gulick, M. King, A. C. White, Jr., C. Benson, M. Thompson, H. A. Kessler, S. Hammer, R. Bertz, A. Hsu, A. Japour, and E. Sun. 2001. ABT-378/ritonavir plus stavudine and lamivudine for the treatment of antiretroviral-naive adults with HIV-1 infection: 48-week results. AIDS 15:F1-9. [DOI] [PubMed] [Google Scholar]
- 15.Nijhuis, M., C. A. Boucher, P. Schipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95:14441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkin, N. T., C. Chappey, and C. J. Petropoulos. 2003. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS 17:955-961. [DOI] [PubMed] [Google Scholar]
- 17.Shulman, N., A. Zolopa, D. Havlir, A. Hsu, C. Renz, S. Boller, P. Jiang, R. Rode, J. Gallant, E. Race, D. Kempf, and E. Sun. 2002. Virtual inhibitory quotient predicts response to ritonavir boosting of indinavir-based therapy in HIV-infected patients with ongoing viremia. Antimicrob. Agents Chemother. 46:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torti, C., E. Quiros-Roldan, W. Keulen, L. Scudeller, S. Lo Caputo, C. Boucher, F. Castelli, F. Mazzotta, P. Pierotti, A. M. Been-Tiktak, G. Buccoliero, M. De Gennaro, G. Carosi, and C. Tinelli. 2003. Comparison between rules-based human immunodeficiency virus type 1 genotype interpretations and real or virtual phenotype: concordance analysis and correlation with clinical outcome in heavily treated patients. J. Infect. Dis. 188:194-201. [DOI] [PubMed] [Google Scholar]
- 19.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]