Abstract
Our laboratory recently recovered Chlamydia trachomatis in tissue culture from a urogenital specimen which tested negative by commercial plasmid-based PCR. Immunotyping and omp1 sequencing identified the isolate as a serovar E isolate. Further investigation by PCR and Southern hybridization indicated that this isolate lacks the chlamydial cryptic plasmid.
Chlamydia trachomatis contains 7 to 10 copies of a 7.5-kb cryptic plasmid, pCT, first identified in 1980 (7, 9). The nucleotide sequence of pCT is known for four serovars; the sequences are highly conserved, showing less than 1% nucleotide substitution (1). pCT contains eight open reading frames (ORFs), the products of some of which bear similarity to known proteins: ORF1 contains a dnaB-like gene (12), ORF3 encodes a 28-kDa antigen (2), and ORF8 encodes a hypothetical recombinase-like protein (3). However, the functions of the eight pCT genes have not been characterized (3, 6, 12, 13). Consequently, the role of pCT in the development and pathogenesis of C. trachomatis is unknown. The evolutionary preservation of the plasmid and its nucleotide sequence, however, suggests that it is vital to C. trachomatis.
Only two isolates of C. trachomatis which do not contain pCT have been characterized. The first was a specimen isolated from a homosexual male with proctocolitis, but it is no longer available (11). It was serotyped as L2. The other was a urethral isolate from a male with urethritis. The omp1 nucleotide sequence was most similar to that of serovar B (4). It was propagated in cell culture initially, but subsequently it could not be recovered. We report here an isolate of C. trachomatis which was recovered from the urethra of an asymptomatic male. The isolate grows in cell culture but does not contain pCT or genes borne by pCT. Since pCT is frequently the target of diagnostic tests, we also set out to determine the prevalence of plasmidless strains in a high-risk, pCT PCR-negative population.
Clinical isolate C599 was obtained from a 26-year-old asymptomatic male who was enrolled in a study which compared various molecular techniques for diagnosis of chlamydia. Both urine and urethral swab specimens were obtained. Chlamydia organisms from the urethral swab specimen were expanded in McCoy cell monolayers (5), and inclusions were stained with a genus-specific antilipopolysaccharide antibody (8). Amplification of DNA from urine was done as described by the manufacturer (PCR was done with the Amplicor kit from Roche, and LCR was done with the LCx kit from Abbott Labs). In addition to testing urine specimens by pCT PCR and LCR, pCT amplification was performed on transport media containing the original specimen and on C599-infected McCoy cells to ensure that pCT had not been lost during in vitro expansion of the C599 population.
DNA extractions and PCRs were done as previously described (14). In-house pCT primers were designed based on the published sequences of pCT in GenBank. Both primers hybridize to ORF8 (1): pCT-FOR (5′-ACA GCG GTT GCT CGA AGC AC-3′) was the 5′ primer, and pCT-REV (5′-GCA TTG GAC CGC ATC ACT CA-3′) was the 3′ primer. The resulting product was 720 bp in length. omp1 PCR primers were designed based on published omp1 sequences and amplify the full-length gene (14). omp1 PCR of the swab specimen required a nested reamplification due to the low yield of chlamydial organisms in the specimen. omp1 PCR was carried out with purified DNA from both the original specimen and the expanded population of the isolate. omp1 amplicons were subjected to automated sequencing (Merlin Core Services, Bio 101, Vista, Calif.). Sixty-eight additional isolates from the study population were screened by omp1 PCR. The isolates were from patients who were at high epidemiological risk for sexually transmitted infections (prevalence of chlamydial infections, 15.9%) but who had been diagnosed as uninfected by culture and by commercial pCT amplification.
Southern hybridizations were done with the Genius system (Boehringer Mannheim, Indianapolis, Ind.). Total DNA from C599- and E/UW5-infected McCoy cells was digested with BamHI and probed with full-length pCT randomly primed with digoxigenin (DIG). An omp1 probe also was made by amplifying omp1 from C599 with the incorporation of dUTP-DIG. Visualization of the hybridization signal was done with an anti-DIG–alkaline phosphatase antibody conjugate and the CDP-Star chemiluminescent substrate.
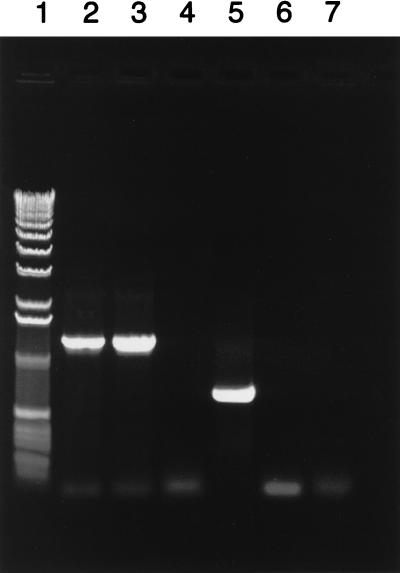
While commercial pCT-based amplification tests gave negative results for both the C599 clinical specimen and expanded population of the isolate, omp1 PCR was positive for both, suggesting that the isolate lacked pCT and did not lose its plasmid after expansion in cell culture. C599 also was negative by the in-house pCT PCR but positive by omp1 PCR (Fig. 1). The nucleotide sequence of the C599 omp1 was identical to that of E/Bour (10), and the E serotype was confirmed by monoclonal antibody staining (8). The 68 pCT PCR-negative, culture-negative patient samples from the study population also were omp1 PCR negative, indicating that plasmidless strains are a relatively rare phenomenon. Ten pCT PCR-positive samples from this population were used as positive controls. The omp1 gene was successfully amplified from all 10 specimens (data not shown).
FIG. 1.
omp1 and pCT PCR of strains C599 and E/UW5. Primers designed for omp1 are located 108 bp upstream of the ATG initiator and ∼80 bp downstream of the TAA terminator, resulting in a PCR product of approximately 1,250 bp. Primers designed for pCT are located in ORF8, producing a PCR product of approximately 720 bp. Lanes: 1, 1-kb DNA size standards (Life Technologies, Gibco BRL); 2, omp1 PCR of C599; 3, omp1 PCR of E/UW5; 4, pCT PCR of C599; 5, pCT PCR of E/UW5; 6, negative omp1 control PCR (no template); 7, negative pCT control PCR (no template). Black-and-white digital images were captured with an Alpha Innotech Alpha-Imager 2000 and annotated by using Adobe Photoshop and Freehand V.8 for Power Macintosh.
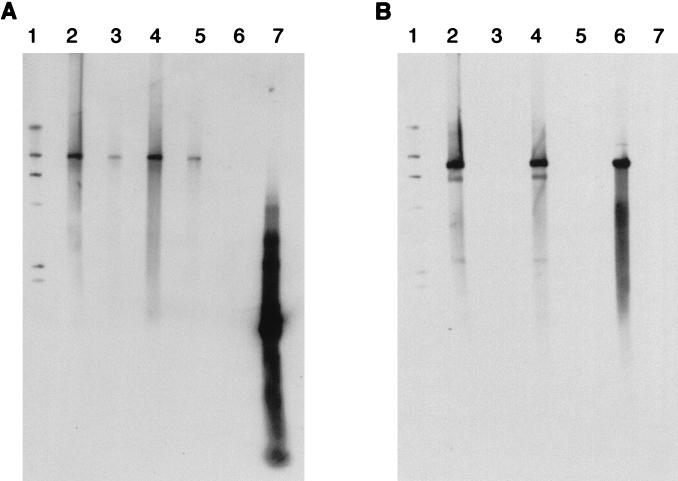
Southern hybridization with an omp1 probe produced a band of approximately 9 kb both in lanes containing C599 DNA and in lanes containing E/UW5 DNA (Fig. 2A). After the blot was stripped and reprobed with the full-length pCT probe, a 7.5-kb band was visible in the E/UW5 DNA-containing lanes but nothing was seen in the C599 DNA-containing lanes (Fig. 2B). This result confirms that the cryptic plasmid is missing from C599 and that its genes are not incorporated into the chlamydial genome.
FIG. 2.
Southern hybridization of BamHI-digested C599 and E/UW5 total DNA probed with DIG-labeled omp1 PCR product (A) and DIG-labeled pCT (B). Lanes: 1, DIG II marker bands at (from top to bottom) 23, 9, 6, 4, 2.3, and 2 kb; 2, 1 μg of E/UW5 BamHI-digested DNA; 3, 1 μg of C599 BamHI-digested DNA; 4, 0.5 μg of E/UW5 BamHI-digested DNA; 5, 0.5 μg of C599 BamHI-digested DNA; 6, BamHI-linearized pCT (7.5 kb); 7, omp1 PCR product (1,250 bp). Southern blot X-ray images were scanned at 400 dots per inch with a UMax Powerbook scanner. The images were then combined into one photo by using Adobe Photoshop and Freehand V.8 for Power Macintosh.
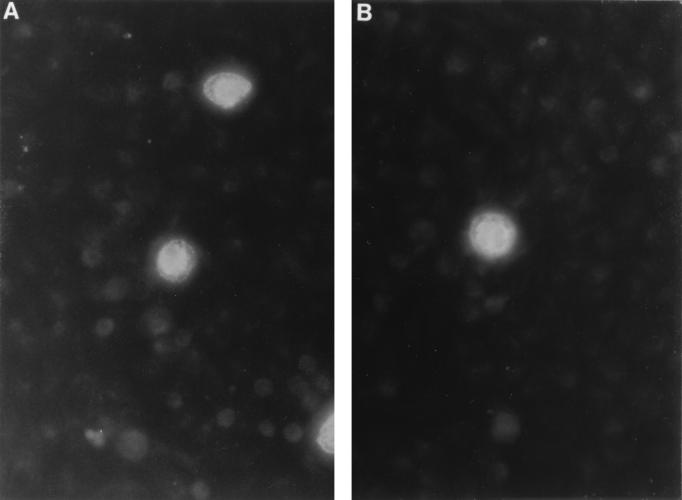
C599 was compared to laboratory strain E/UW5 for inclusion morphology and growth characteristics. Fluorescent microscopic examination of C599 inclusions stained with a serovar E-specific, fluorescein-labeled anti-major outer membrane protein monoclonal antibody did not reveal any significant differences among inclusions of laboratory strain E/UW5 (Fig. 3A) and C599 (Fig. 3B).
FIG. 3.
Fluorescent micrograph of chlamydial inclusion bodies stained with a fluorescein-labeled serovar E-specific anti-major outer membrane protein monoclonal antibody. McCoy cell monolayers were infected with laboratory strain E/UW5 (A) and clinical isolate C599 (B). Infected monolayers were then fixed and stained as previously described (8). Magnification, ×100.
C599 populations can be expanded and the strain can be passaged repeatedly in shell vials when the inoculum contains large numbers of inclusion-forming units (IFU). However, when C599 is inoculated into shell vials at low numbers of IFU, it does not grow as well as the laboratory-adapted E/UW5 strain. This was demonstrated by inoculating McCoy cell monolayers on two separate occasions with either C599 or E/UW5 adjusted to yield less than 250 IFU per vial. The number of IFU detected by fluorescent-antibody staining diminished between the primary and secondary passages of C599, while the E/UW5 population expanded without difficulty (Table 1).
TABLE 1.
Growth rates at two independent times for clinical isolate C599 and laboratory strain E/UW5
| Trial | No. of IFU/100 μl
|
|||
|---|---|---|---|---|
| E/UW5
|
C599
|
|||
| Primary passage | Secondary passage | Primary passage | Secondary passage | |
| 1 | 237 | 861 | 146 | 25 |
| 2 | 218 | 1,757 | 119 | 1 |
Strain C599 was isolated in 1996. In 1993 the patient from whom C599 was isolated also had C. trachomatis isolated from his urethra. The 1993 isolate was recovered from our freezer, and commercial pCT PCR for this isolate was positive. Monoclonal antibody staining established it as a serotype I isolate. Therefore, it appears that the two infections were independent and that this person was not persistently infected from 1993 to 1996 with a strain that lost its plasmid over time. In addition, the patient had negative urethral cultures for C. trachomatis five times between the 1993 and 1996 infections. Subsequent to the isolation of C599 and the patient’s treatment for that infection with a 7-day course of doxycycline, the patient had two negative urethral cultures for C. trachomatis. Therefore, it does not appear that loss of pCT affected this patient’s response to antibiotic therapy or produced a chronic chlamydial infection in this patient.
We report here a plasmid-free urethral isolate of C. trachomatis, serovar E, the omp1 sequence of which is identical to that of E/Bour (10). It is similar to the two other reported plasmidless isolates (4, 11) in that it was obtained from a male patient. However, unlike the other two isolates, C599 was obtained from an asymptomatic person. Therefore, the role of pCT in pathogenesis cannot be defined from these examples. C599 was responsive to antibiotic therapy in vivo, although in vitro susceptibility testing has not been done. Our report is the first to describe a plasmidless strain of one of the common urogenital serovars. The existence of such plasmid-free strains of C. trachomatis argues that pCT does not play a critical role in the chlamydial developmental cycle. Since C599 grows reasonably well in culture, it may prove to be a useful tool in defining the functions of the plasmid-based genes of C. trachomatis.
Acknowledgments
This work was supported by grant AI-31494 to R.B.J. from the National Institute for Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the chlamydial common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–154. doi: 10.1016/0147-619x(90)90034-a. [DOI] [PubMed] [Google Scholar]
- 2.Comanducci M, Cevenini R, Moroni A, Giuliani M M, Ricci S, Scarlato V, Ratti G. Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28kDa antigen. J Gen Microbiol. 1993;139:1083–1092. doi: 10.1099/00221287-139-5-1083. [DOI] [PubMed] [Google Scholar]
- 3.Fahr M J, Sriprakash K S, Hatch T P. Convergent and overlapping transcripts of the Chlamydia trachomatis 7.5 kb plasmid. Plasmid. 1992;28:247–257. doi: 10.1016/0147-619x(92)90056-g. [DOI] [PubMed] [Google Scholar]
- 4.Farencena A, Comanducci M, Donati M, Ratti G, Cevenini R. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun. 1997;65:2965–2969. doi: 10.1128/iai.65.7.2965-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones R B, Van Der Pol B, Katz B P. Effect of differences in specimen processing and passage technique on recovery of Chlamydia trachomatis. J Clin Microbiol. 1989;27:894–898. doi: 10.1128/jcm.27.5.894-898.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahane S, Sarov I. Cloning of a chlamydial plasmid: its use as a probe and in vitro analysis of encoded polypeptides. Curr Microbiol. 1987;14:255–258. [Google Scholar]
- 7.Lovett M, Kuo C C, Holmes K, Falkow S. Plasmids of the genus Chlamydia. In: Nelson J D, Grassi C, editors. Current chemotherapy and infectious disease. Proceedings of the 11th International Congress of Chemotherapy and the 19th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1980. pp. 1250–1252. [Google Scholar]
- 8.Newhall W J, Terho P, Wilde III C E, Batteiger B E, Jones R B. Serovar determination of Chlamydia trachomatis isolates by using type-specific monoclonal antibodies. J Clin Microbiol. 1986;23:333–338. doi: 10.1128/jcm.23.2.333-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986;16:52–62. doi: 10.1016/0147-619x(86)90079-x. [DOI] [PubMed] [Google Scholar]
- 10.Peterson E M, Markoff B A, de la Maza L M. The major outer membrane protein nucleotide sequence of Chlamydia trachomatis, serovar E. Nucleic Acids Res. 1990;18:3414. doi: 10.1093/nar/18.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson E M, Markoff B A, Schachter J, de la Maza L M. The 7.5 kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid. 1990;23:144–148. doi: 10.1016/0147-619x(90)90033-9. [DOI] [PubMed] [Google Scholar]
- 12.Sriprakash K S, MacAvoy E S. Characterization and sequence of a plasmid from the trachoma biovar of Chlamydia trachomatis. Plasmid. 1987;18:205–214. doi: 10.1016/0147-619x(87)90063-1. [DOI] [PubMed] [Google Scholar]
- 13.Sriprakash K S, Pearce B J. Mapping of transcripts encoded by the plasmid in Chlamydia trachomatis. FEMS Microbiol Lett. 1990;71:299–304. doi: 10.1016/0378-1097(90)90237-k. [DOI] [PubMed] [Google Scholar]
- 14.Stothard D R, Boguslawski G, Jones R B. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]