Abstract
Nucleosides have been widely used in the treatment of viral diseases, but relatively few have been identified as inhibitors of hepatitis C virus (HCV). The modified ribonucleosides, 2′-C-methyl-adenosine and 2′-O-methyl-cytidine, are potent inhibitors of HCV replication which specifically target the NS5B polymerase. Herein, a more extensive characterization of the effect of these compounds upon HCV replication in subgenomic replicons is reported. A highly selective antireplicative effect induced by the nucleosides in replicon-containing cell lines was maintained during an exponential growth period with potencies which paralleled the reduction of both positive- and negative-strand RNA replication. Moreover, the inhibitory effect closely correlated with the intrinsic metabolic properties of differing replicon clonal lines. Interestingly, while 2′-C-methyl-adenosine elicited similar inhibitory potencies in different cell lines, 2′-O-methyl-cytidine was found to be inactive in one replicon cell line tested, although the corresponding triphosphates comparably inhibited the in vitro activity of replication complexes isolated from these cells and the activity of NS5B polymerase using synthetic templates. The lack of antireplicative effect, attributed to poor intracellular conversion of the 2′-O-methyl-cytidine nucleoside to the active 5′-triphosphate, was reversed using a monophosphate prodrug. Thus, although replicon cells are useful for evaluating the effect of inhibitors upon HCV replication, these findings have important implications for their use in the identification and characterization of nucleosides and other chemotherapeutic agents requiring cellular metabolism.
Hepatitis C virus (HCV) infection affects an estimated 170 million people world wide and is a major cause of hepatocellular carcinoma (39). Currently, the only available treatment is a combination of alpha interferon and ribavirin, which is only effective in 40 to 60% of patients and causes significant side effects. A number of therapeutic approaches has emerged including those that target the viral RNA-dependent polymerase (NS5B) or the serine protease/helicase (NS3), antisense therapies, and therapeutic vaccines (8, 37). However, none has yet yielded a clinically approved drug.
To date, the lack of an infectious cell culture system has hampered the development of HCV antiviral agents. Although full-length cDNAs of HCV are available, viral replication has not been demonstrated. However, the recent advent of stable subgenomic replicon cell lines, capable of RNA replication, has enabled the identification of drug discovery leads (19). Potent inhibitors of the NS3 protease have been identified, one of which, BILN 2061, was found to be effective in phase I clinical trials (1, 18, 26, 28, 36). Several classes of nonnucleoside inhibitors, including benzothiadiazines and disubstituted phenylalanines, have been reported as inhibitors of NS5B polymerase (5, 11, 20, 27, 34, 35, 38). While the clinical usefulness of nucleoside inhibitors (NIs) has been demonstrated for other viruses (7), a limited number of NIs have been identified for HCV, including the 2′-substituted nucleosides which we have previously reported as potent chain terminators of the NS5B polymerase (4, 10, 23, 30, 32, 37).
The development of nucleoside analogs as successful therapeutic agents requires an understanding of the pathways and enzymes needed for activation to the triphosphate form. Nucleoside analogs undergo intracellular phosphorylation by kinases to form the active triphosphate moiety and hence are subject to cell-line-dependent variations in metabolism. The inhibitory activity can be limited by poor uptake, metabolic conversion, and/or competition with endogenous nucleoside triphosphates (NTPs). Further, the degree of phosphorylation can vary depending upon host, tissue, and cell type specificities; regulation by positive and negative feedback loops; phase of the cell cycle; activation state; and viral infection of a cell (31). Additionally, the efficiency of chain termination, the mode of action of known NIs of viral pathogens, depends upon several biochemical parameters including the affinity of the inhibitor NTP for the polymerase, the ratio of the NTP analog to endogenous cellular NTPs, and the relative rates of nucleotide incorporation by the polymerase.
The 2′-substituted nucleoside inhibitors, 2′-C-methyl-adenosine (2′-C-Me-A) and 2′-O-methyl-cytidine (2′-O-Me-C) were previously identified as specific inhibitors of HCV viral replication in a cell-based replicon assay in the absence of cytotoxicity (2, 4, 10). Addition of the nucleosides to cells growing in culture resulted in the intracellular formation of the corresponding triphosphates which are potent, competitive inhibitors of NS5B-catalyzed reactions in vitro. Additionally, the target of this class of nucleosides has been genetically validated by the selection and identification of resistant mutations in replicon cells located in the viral NS5B polymerase (23).
In this study, the specific effect of the nucleosides upon HCV replication was further characterized in two different replicon cell lines derived from parental Huh-7 cells. Although the 2′-C-Me-A analog exhibited selective inhibitory activity in both replicon lines tested, the 2′-O-Me-C analog was inactive in one cell line. This lack of effect upon replication was attributed to poor cell penetration and increased metabolic degradation of the nucleoside by deamination and glycosidic cleavage or demethylation (9, 22). However, the use of a monophosphate prodrug (18a) of the C analog overcame the block in antireplicative effect following its efficient uptake into cells and metabolism to the corresponding triphosphate inhibitor, 2′-O-Me-CTP. Thus, knowledge of the intracellular metabolic pathways of nucleosides combined with mode of action studies is critical to the development of efficacious antiviral agents, the discovery of which can be obscured by differences in clonal properties of replicon cells.
MATERIALS AND METHODS
Materials.
Radiolabeled nucleotides were purchased from Amersham. Ultrapure nucleoside triphosphates were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.). The 2′-O-Me-C triphosphate was purchased from Trilink (San Diego, Calif.). 2′-O-Me-C and 2′-O-Me-5-iodo-C were purchased from Berry Associates (Dexter, Mich.). The 2′-O-Me-C S-acetyl thioester (SATE) was synthesized according to the method described in reference 29. Compounds 2′-C-Me-A, 2′-C-Me-A triphosphate, and 8-bromo-2′-C-Me-A were synthesized according to the general procedures previously described (4). Radiolabeled compounds [8-3H]2′-C-Me-A, [5-3H]2′-O-Me-C, and [5-3H]2′-O-Me-SATE were prepared by the Tritium Custom Preparation group at Amersham Pharmacia Biotech Ltd (Cardiff, Wales). The specific activity of [5-3H]2′-O-Me-C was 23 Ci/mmole, the specific activity of [8-3H]2′-C-Me-A was 18.5 Ci/mmole, and the specific activity of [5-3H]2′-O-Me-SATE was 3 Ci/mmole.
NS5B enzyme assay on template t500.
Enzyme activity was monitored in reactions catalyzed by NS5B Δ21 (NS5B protein with a C-terminal truncation of 21 amino acids) by determining the incorporation of radiolabeled NTPs into a heteromeric RNA template as described previously (3). The inhibitor concentration at which the enzyme-catalyzed rate is reduced by half (IC50) was determined by fitting the relative rate data to the Hill equation.
In situ RNase protection assay (RPA).
HBI10A and HBIII27 replicon cells were grown and assayed as previously described (4, 25). Compound or dimethyl sulfoxide (DMSO) (1%) was added to cells at 80% confluence (80,000 cells/well) in Cytostar 96-well plates (Amersham Biosciences) grown in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 0.8 mg/ml of G418 and then incubated for 24 h at 37°C/5% CO2. Cells were fixed in 10% formalin-phosphate-buffered saline, permeabilized in 0.25% Triton X-100, and then hybridized with an antisense 33P-labeled RNA probe (1.0 × 104 to 2.0 × 104 cpm/μl) in formamide hybridization buffer (Amersham Biosciences) at 50°C overnight. The RNA probe was generated with T7 runoff transcription and had a sequence complementary to nucleotides 1184 to 1481 of the NS5B gene. Plates were treated with 20 μg/ml of RNase A at room temperature for 30 min, washed with 0.25× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer at room temperature and at 65°C for 20 min for each wash, and then counted in a TopCount plate reader. The 50% effective concentration (EC50) was determined as a percentage of the DMSO control by fitting the data to a 4-parameter equation using Kaleidagraph software (Synergy Software, Reading, Pa.).
Cytotoxicity assay (MTS).
Cytotoxicity was measured by the MTS assay as previously described (4). The 50% cytotoxic concentration (CC50) was determined as a percentage of the DMSO control by fitting the data to a 4-parameter equation as described above.
HCV CRC replicase isolation.
HCV crude replication complexes (CRC) were isolated and fractionated from HBI10A and HBIII27 replicon cells grown by passage at 1:5 in DMEM containing 10% FBS and G418 (0.8 mg/ml) as described previously (23, 33). Cells were scraped into cold phosphate-buffered saline, washed, pelleted, swelled at 5 × 107 cells/ml in ice-cold hypotonic lysis buffer (10 mM Tris-Cl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml of aprotinin) for 10 min, and Dounce homogenized. Nuclei were pelleted, and crude replication complexes were pelleted from the cytoplasmic extract by centrifugation at 30,000 × g for 25 min, resuspended in 10 mM Tris-Cl, pH 8.0, 10 mM NaCl, 15% glycerol at 2.5 ×105 cell equivalents/μl and 1 μg of protein/μl, and stored at −70°C.
HCV CRC replicase assay.
RNA was synthesized in vitro in a reaction mixture containing 20 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 5 mM dithiothreitol, 5 mM KCl, 20 μg/ml of actinomycin D, 20 μCi [32P]CTP, 0.5 mM (each) ATP, GTP, and UTP, 10 μM CTP, 2.5 mM phosphoenol pyruvate, 0.5 μg of pyruvate kinase (2 units), 4 units of RNasin, and 2.5 μg of CRC replicase at 35°C for 60 min. RNA transcripts purified by phenol extraction and ethanol precipitation were electrophoresed on glyoxal denaturing agarose gels and quantified by phosphorimager analysis. Compound or DMSO (5%) was added to the reaction mixture 5 min prior to initiation with NTPs. 2′-C-Me-ATP was tested in the presence of 80 μM ATP. The IC50 was determined as a percentage of the DMSO control by fitting the data to a 4-parameter fit function using Kaleidagraph software (Synergy Software, Reading, Pa).
Northern blot analysis.
The RNA from 5 ×105 Huh-7 or HBI10A cells cultured for 48 h was extracted with Trizol (Gibco-BRL) and purified using RNeasy columns (QIAGEN). RNA was divided and electrophoresed on 1.0% glyoxal denaturing gels, blotted, and probed with 32P-radiolabeled NS5B sense or antisense RNA probes (1 × 106 to 2 × 106 cpm/ml) overnight at 65°C. Blots were washed and quantified on a phosphorimager. The copy number was determined from a known quantity of control transcripts run in parallel as described previously (33).
Intracellular metabolism studies.
Intracellular metabolism studies with Huh-7, HBI10A, and HBIII27 cells were performed as described previously (4). In brief, Huh-7 cells were plated in complete DMEM containing 10% FBS and HBI10A and HBIII27 cells were plated in the same containing G418 (0.8 mg/ml) at 1.5 × 106 cells/60-mm-diameter dish such that cells were 80% confluent at the time of compound addition. Tritiated compound was incubated at 2 μM in the cell medium for various time periods. Cells were collected, washed with phosphate-buffered saline, and counted. The cells were then extracted in 70% methanol, 20 mM EDTA, 20 mM EGTA and centrifuged. The lysate was dried, and radiolabeled nucleotides were analyzed using ion-pair reverse-phase (C18) high-performance liquid chromatography (HPLC) on a Waters Millenium system connected to an in-line β-RAM scintillation detector (IN/US Systems). The HPLC mobile phases consisted of (i) 10 mM potassium phosphate with 2 mM tetrabutylammonium hydroxide and (ii) 50% methanol containing 10 mM potassium phosphate with 2 mM tetrabutylammonium hydroxide. Peak identification was made by comparison of retention times to standards.
Nucleoside conformation predictions.
Nucleoside conformational preferences were calculated by molecular mechanics using the MMFFs force field and a dielectric constant of 50. For each nucleoside, 1,000 conformers were generated using the JG distance geometry program (S. K. Kearsley, unpublished) written using previously described theory and algorithms (6, 17) and minimized to low gradient using BatchMin (24). To estimate the barrier to interconversion between the Northern and Southern conformers and the steepness of the minima, the conformers generated by distance geometry were also subjected to constrained minimization, holding one of the ribose ring dihedral angles at a random value between −42 and +42 degrees by means of a harmonic force constant of 1000 kJ · mole−1 · rad−2.
RESULTS
Inhibition of HCV RNA replication in subgenomic replicon cells.
Previously, we reported upon the identification of 2′-substituted nucleoside analogs as inhibitors of replication in replicon cells (HBI10A) which harbor and express subgenomic HCV RNA (2, 4). Using an in situ RPA which provides a direct measurement of the potency of inhibitors on positive-strand RNA synthesis in cells, EC50s of 0.25 and 21 μM at 24 h were obtained, respectively, for 2′-C-Me-A and 2′-O-Me-C. A time-dependent effect on HCV RNA replication observed in HBI10A replicon cells demonstrated the high degree of selectivity and potency of this class of nucleosides. As shown in Fig. 1 and Table 1, two- to threefold increases in the potencies of 2′-C-Me-A (EC50 = 0.1 μM) and 2′-O-Me-C (EC50 = 11 μM) were observed at 72 h. The inhibitory activity of both compounds was observed in the absence of cytotoxicity when tested at up to 100 μM by the MTS assay throughout a 48-h period. At 72 h post-compound addition, slight toxicity (CC50 = 100 μM) was elicited by 2′-C-Me-A in HBI10A cells; however, a 1,000-fold selective index for HCV RNA replication was maintained. A cytotoxic effect for 2′-O-Me-C was not observed at the highest concentration tested, 100 μM (selective index >10-fold). By [3H]thymidine uptake, neither nucleoside had an effect when tested at up to 100 μM during a 48-h period; however, at 72 h, a CC50 of 30 μM was obtained for 2′-C-Me-A (data not shown).
FIG. 1.
Time-dependent inhibition of nucleoside analogs in the cell-based HCV replicon assay. The inhibition of HCV replication at the indicated levels and times by 2′-C-Me-A and 2′-O-Me-C was determined in HBI10A replicon cells and quantified by in situ RPA as described in Materials and Methods. The lines represent the best fit of the data to a four-parameter equation.
TABLE 1.
Inhibitory activity and cytotoxic effect of nucleosides in replicon cell-based assay
| Compound | Replicon IC50 (μM)a of cell type at:
|
Toxicity (CC50 [μM]) of cell type at 72 h
|
||||||
|---|---|---|---|---|---|---|---|---|
| 24 h
|
48 h
|
72 h
|
||||||
| HBI10A | HBIII27 | HBI10A | HBIII27 | HBI10A | HBIII27 | HBI10A | HBIII27 | |
| 2′-C-Me-A | 0.3 ± 0.1 | 0.5 ± 0.2 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.1 ± 0.01 | 0.3 ± 0.1 | 100 | 33 |
| 2′-O-Me-C | 21.0 ± 1.8 | >100 | 12.0 ± 0.9 | >100 | 11.0 ± 0.4 | >100 | >100 | >100 |
The IC50 is the average ± standard deviation of the results from at least three determinations.
Interestingly, the nucleosides exhibited differential activities in another replicon line (HBIII27) derived from a different Huh-7 cell clone (25). While 2′-C-Me-A elicited an EC50 of 0.3 μM similar to that obtained in HBI10A replicon cells, 2′-O-Me-C was inactive in the HBIII27 line when tested at concentrations of up to 100 μM for 72 h (Table 1). In this clonal replicon line, 2′-C-Me-A exhibited a CC50 of 33 μM by the MTS assay, indicating a greater sensitivity of these cells to toxic effects, thereby reducing the selectivity of the nucleoside to 100-fold.
Replication of both positive- and negative-strand RNAs in replicon cells was inhibited.
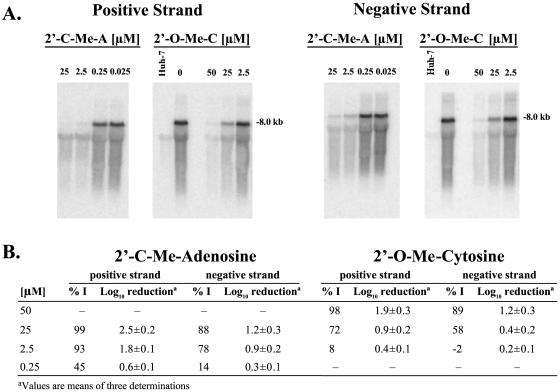
Detection by in situ RPA indicated that the nucleosides inhibited the synthesis of positive-strand HCV RNA in replicon cells. To confirm that the observed inhibitory effect was also attributed to the reduction of negative-strand RNA replication, Northern blot hybridization of RNA isolated from compound-treated cells was performed. The nucleoside analogs inhibited the replication of both positive- and negative-strand HCV RNAs in replicon cells when probed with antisense and sense NS5B probes, respectively (Fig. 2A). As summarized in Fig. 2B, the inhibition of positive-strand RNA synthesis by both nucleosides at 48 h measured by Northern blot analysis was comparable to the potency observed by in situ RPA in replicon cells and the inhibition of negative-strand RNA synthesis was slightly less potent.
FIG. 2.
Effect of nucleosides on positive- and negative-strand replicon RNA synthesis in replicon cells. The inhibition of positive- and negative-strand RNA synthesis was measured by Northern blot analysis of HCV RNA isolated from Huh-7 and HBI10A cells which were cultured for 48 h in the presence of 2′-C-Me-A and 2′-O-Me-C. Total RNA was extracted from cells by phenol extraction and ethanol precipitation. The RNA from 2.5 × 105 cells per lane was electrophoresed on glyoxal denaturing gels and probed with [32P]-radiolabeled NS5B sense or antisense RNA probed as indicated in panel A. Results quantified by phosphorimager analysis are summarized in panel B. %I, percent inhibition.
The effect on HCV RNA copy number was also determined from Northern blot analysis in comparison to known molar amounts of HCV RNA transcripts run in parallel on the same blot. With 25 μM 2′-C-Me-A, a 2.5 log10 reduction was observed in the synthesis of positive-strand RNA and that of negative-strand RNA was reduced by 1.2 log10 (Fig. 2B). The inhibitory potency of the 2′-C-Me-A analog was evident from log10 reductions of 1.8 and 0.9, observed correspondingly for positive- and negative-strand RNAs at 2.5 μM, the EC95 for this compound. Similarly, the 2′-O-Me-C analog elicited log10 reductions of 1.9 and 1.2 for positive- and negative-strand RNAs, respectively, at 50 μM, five times the EC50.
NTP analogs inhibited the activity of HCV replication complexes.
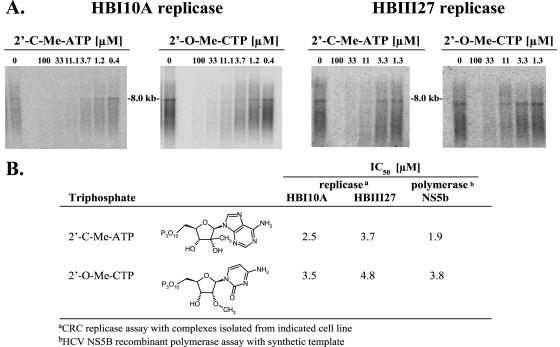
To determine whether the corresponding triphosphates of the nucleoside analogs directly targeted HCV replication, replication complexes which catalyze RNA synthesis were isolated from HBI10A and HBIII27 replicon cells. As shown in Fig. 3A and B, in HBI10A cells, 2′-C-Me-ATP and 2′-O-Me-CTP elicited IC50s of 2.5 and 3.5 μM, respectively, in an in vitro assay which measures the activity of isolated replication complexes. Although 2′-O-Me-C was substantially less active in the HBI10A replicon cells than 2′-C-Me-A, the triphosphate analogs inhibited the activity of replication complexes isolated from the cells with potencies comparable to those observed in the assay of purified HCV NS5B polymerase (Fig. 3B). Both 2′-C-Me-ATP and 2′-O-Me-CTP also inhibited the in vitro activity of replication complexes isolated from HBIII27 replicon cells with potencies similar to the inhibition of replication complexes from HBI10A cells and NS5B polymerase. Hence, the lack of antireplicative effect in HBIII27 cells was not attributed to an inability of replication complexes to be inhibited by 2′-C-Me-ATP or 2′-O-Me-CTP.
FIG. 3.
Inhibition of HCV CRC replicase activity by nucleoside triphosphate analogs. The effect of 2′-C-Me-ATP and 2′-O-Me-CTP were tested in an in vitro reaction with HCV replicase complexes isolated from HBI10A and HBIII27 replicon cells as described in Materials and Methods. RNA products were purified by phenol extraction and ethanol precipitation, electrophoresed on a glyoxal denaturing agarose (0.7%) gel (A), and quantitated by phosphorimager analysis. The IC50s determined for each NTP in the replicase and NS5B polymerase reactions are summarized in panel B.
Lack of inhibition in HBIII27 replicon cells is attributed to metabolic degradation of 2′-O-Me-C.
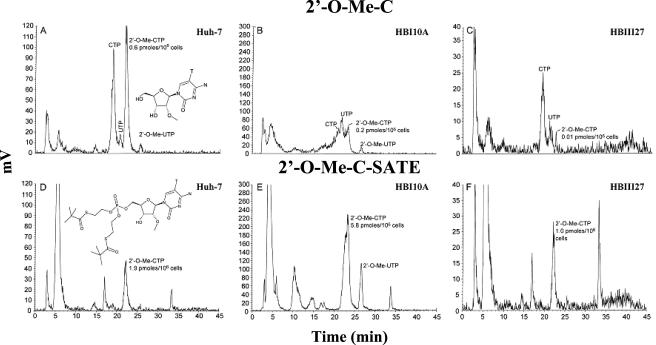
Previously, the intracellular metabolism of 2′-C-Me-A and 2′-O-Me-C was studied in parental Huh-7 cells and clonal HBI10A replicon cells (4). Nucleoside 2′-C-Me-A was efficiently converted to its corresponding triphosphate in both cell lines. In contrast, 2′-O-Me-C exhibited less robust cellular penetration into replicon cells and yielded much less triphosphate with extensive metabolism to UTP and CTP, suggesting that this molecule was subject to intracellular deamination and either demethylation of the 2′-methoxy group or base transfer following ribose deglycosylation. Similarly, the metabolic fate of 2′-O-Me-C was evaluated with HBIII27 cells to determine whether insufficient levels of intracellular NTPs could account for the lack of effect upon replication in HBIII27 cells. As summarized in Table 2, 2′-O-Me-C yielded a 20-fold-lower level of triphosphate in HBIII27 cells than in HBI10A cells, although the total uptake of nucleoside was not changed significantly. Note that the major species detected in Huh-7 parental cells was 2′-O-Me-CTP, whereas UTP and CTP were the predominant species in clonal HBI10A and HBIII27 replicon cells (Fig. 4).
TABLE 2.
Antiviral effect and intracellular metabolite levels
| Compound | Replicon EC50 (μM)a for cell type:
|
pmol of NTP/106 cellsb of type:
|
pmol of N/106 cellsc of type:
|
|||
|---|---|---|---|---|---|---|
| HBI10A | HBIII27 | HBI10A | HBIII27 | HBI10A | HBIII27 | |
| 2′-O-Me-cytidine | 21.2 | >100 | 0.2 | 0.010 | 1.3 | 0.5 |
| 2′-O-Me-cytidine (SATE) | 3.2 | 4.0 | 5.8 | 1.0 | 22.5 | 15.0 |
| 2′-C-Me-cytidine | 4.8 | NDd | 0.8 | ND | 3.0 | ND |
Replicon activity was determined 24 h post-compound addition by in situ RPA.
Total NTP measured at 24 h.
Total radiolabeled metabolites measured at 24 h.
ND, not determined.
FIG. 4.
Intracellular metabolism of 2′-O-Me-C and 2′-O-Me-C SATE detected in replicon cells. Nucleosides 5-[3H]-2′-O-methyl-cytosine and 7-[3H]-SATE were added to Huh-7, HB1, or HBIII27 cells at a 2 μM concentration for 24 h and then extracted and analyzed by HPLC. The HPLC radiochromatograms of metabolites extracted from Huh-7, HB1, and HBIII27 replicon cells treated with 2′-O-Me-C (A to C) and 2′-O-Me-C SATE (D to F) are shown.
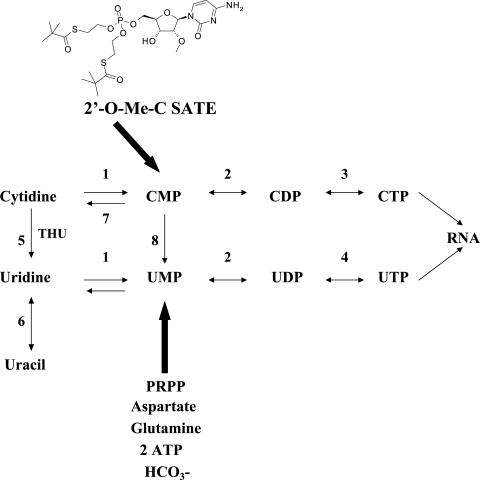
The data above suggested that 2′-O-Me-C was being deaminated to uridine, generating UTP in the replicon cells, as illustrated by the pathway in Fig. 5. In the presence of tetrahydrouridine (THU), a transition state analog inhibitor of cytidine deaminase, a reduction in the amount of UTP and a coincident increase in the amount of CTP were observed; however, the amount of 2′-O-Me-CTP did not increase (data not shown). This again indicated that in addition to deamination, demethylation or base transfer following glycosidic cleavage occurred. In contrast, a 2′-C-Me-C analog was not subject to detectable nucleoside degradation and 2′-C-Me-CTP was the major intracellular species identified. Consequently, 2′-C-Me-C elicited an EC50 of 4.8 μM in HBI10A cells (Table 2).
FIG. 5.
De novo and salvage biosynthesis of ribonucleotides. Enzymes: 1, uridine/cytidine kinase; 2, uridine/cytidine monophosphate kinase; 3, CTP synthetase; 4, uridine/cytidine diphosphate kinase; 5, cytidine deaminase; 6, uridine phosphorylase; 7, 5′-nucleotidase; 8, cytidylate deaminase.
2′-C-Me-C and 2′-O-Me-C analogs are conformationally distinct.
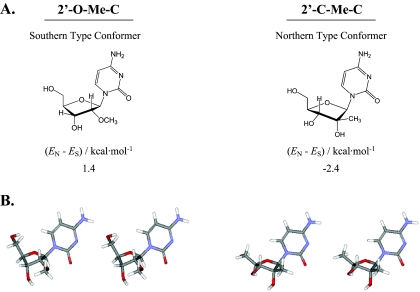
We speculated that the difference in deamination of the 2′-C-Me-C and 2′-O-Me-C analogs might be attributable to the overall conformation of the nucleosides. Therefore, the low-energy conformer of each nucleoside was calculated, revealing that the preferred conformation according to the calculations was Southern for the 2′-O-Me-C analog and Northern for 2′-C-Me-C (Fig. 6). An additional distinction is the position of the 2′-substituted groups. The methoxy substituted group of 2′-O-Me-C is positioned on the alpha face of the nucleoside and is trans relative to the 4′-hydroxymethyl of the ribose, whereas the methyl group of 2′-C-Me-C is positioned on the beta face of the nucleoside and is cis relative to the 4′ ribose position.
FIG. 6.
Lowest-energy conformations of 2′-O-Me-C and 2′-C-Me-C. Conformations were determined by molecular mechanics calculations and are presented as schematic diagrams with the calculated energy differences obtained between the predicted lowest-energy conformers of the Northern furanose and Southern furanose puckers in panel A. Stereo views are shown as tubes in panel B, colored by element (grey = C, blue = N, red = 0, white = 0).
Antireplicative effect was restored using a monophosphate prodrug.
To ascertain whether the degradative metabolism of 2′-O-Me-C was occurring intracellularly at the nucleoside level, a monophosphate prodrug form, 2′-O-Me-C SATE, was synthesized. When the tritiated SATE compound was added to HBI10A and HBIII27 cells, increases of 25- to 30-fold in total intracellular metabolites and 3- to 10-fold in the concentrations of 2′-O-Me-CTP were detected, respectively, in comparison to the parent nucleoside, 2′-O-Me-C (Fig. 4 and Table 2). Notably, the SATE derivative was converted primarily to 2′-O-Me-CTP and, correspondingly, exhibited an inhibitory effect on HCV replication in HBIII27 cells, similar to that observed in HBI10A cells (Table 2).
DISCUSSION
Previously we identified 2′-substituted nucleosides as inhibitors of HCV replication in replicon-containing cell lines (2, 4, 10, 22). In this report, we further characterized the selective inhibitory effects of this class of nucleosides in replicon cells and have additionally demonstrated that differences in intrinsic potencies attributed to clonal metabolic properties were overcome by intracellular delivery of a monophosphate prodrug form of the nucleoside.
The nucleosides exhibited highly selective inhibition of HCV RNA replication throughout an exponential growth period of 72 h in the replicon cell line, HBI10A. The potencies of the antireplicative effects were 10- and 1,000-fold greater than the cytotoxicity observed for 2′-O-Me-C and 2′-C-Me-A, respectively, and were directly related to the reduction of both positive- and negative-strand RNAs, indicating inhibition of complete cycles of HCV replication. A more pronounced reduction of positive-strand than negative-strand RNA synthesis occurred, as has been previously reported for alpha interferon in replicon cells (12), and is likely attributed to kinetic differences in the replication of positive- and negative-strand RNAs and/or the double-stranded nature of the replicative intermediate (33).
The significant potencies in inhibiting HCV replication were also evident from the effective 1.8 to 1.9 log10 reductions of viral RNA copies at 5 to 10 times the EC50 levels obtained for 2′-O-Me-C and 2′-C-Me-A in HBI10A cells, respectively. It remains to be seen whether the clinical outcome of an anti-HCV agent can be predicted from the effects observed in replicon cell lines, in which persistent replication takes place under selective pressure and the cells are actively growing in contrast to the quiescent state of hepatocytes in vivo. However, recent studies with the protease inhibitor BILN 2061 suggest a reasonable correlation between the in vitro cell-based and enzymatic potencies of a direct antiviral agent and its effectiveness in reducing HCV titer in a clinical setting (18, 28). Similarly, the inhibitory effect of the nucleoside analog NM283, the 3′-valine ester of 2′-C-Me-C, has been demonstrated in primates and correlates well to recently disclosed clinical data in HCV-infected patients (37).
Intriguingly, although the nucleosides were effective in HBI10A and several other replicon cell lines tested (data not shown), in one replicon cell line, HBIII27, the selectivity of the 2′-C-Me-A analog was reduced by 10-fold and the 2′-O-Me-C analog was totally inactive at 100 μM, suggesting significant differences in cellular uptake and/or metabolism. The inhibitory potencies observed in HBI10A and HBIII27 replicon cells correlated with the levels of triphosphates formed intracellularly. A lower amount of intracellular 2′-O-Me-CTP was detected than of 2′-C-Me-ATP, and this difference was reflected by the reduced potency of 2′-O-Me-C in replicon-containing cells. Notably, although 2′-O-Me-C was completely inactive in HBIII27 cells, the inhibition of replication complexes from both cell lines by 2′-O-Me-CTP was comparable. This indicated that the lack of antireplicative activity was not attributed to an inability to inhibit the NS5B target within HBIII27 replication complexes but rather to poor penetration and intracellular conversion of the nucleoside to the active triphosphate in this replicon cell line. Importantly, the efficient phosphorylation of 2′-O-Me-C observed in the heterogeneous parental Huh-7 cells demonstrated that this deficiency was a clonal property of the replicon lines.
The detection of high amounts of radiolabeled intracellular UTP and CTP but little 2′-O-Me-CTP in replicon cells indicated that the 2′-O-Me-C analog was undergoing deamination to uridine and that demethylation of the 2′-methoxy group and/or base transfer following glycosidic bond cleavage by pyrimidine nucleoside phosphorylase was also occurring. In the presence of the cytidine deaminase inhibitor, THU (Fig. 5, 5), a decrease in the formation of the amount of intracellular UTP indicated that deamination of 2′-O-Me-C occurred, although the additional formation of CTP confirmed that demethylation or deglycosylation also took place and, correspondingly, enhanced potency was not observed in THU-treated replicon cells (data not shown). In contrast, the 2′-C-Me-C nucleoside was more efficiently converted to 2′-C-Me-CTP and was not subject to detectable degradation. This difference in metabolic conversion to the triphosphate can be attributed to several factors. Although this study did not address the mechanism responsible for the base changes observed for 2′-O-Me-C, it is likely due to demethylation of the 2′-methoxy group and/or enzymatic cleavage of the glycosidic bond by pyrimidine nucleoside phosphorylases followed by pentosyl transfer of the radiolabeled base (9, 21). Additionally, the threefold-higher concentrations of 2′-C-Me-C in the cell may have contributed to its more efficient phosphorylation compared to a lower level of 2′-O-Me-C, which is more likely to be deaminated to uridine, consistent with the rate-limiting activity of cytidine kinase relative to the abundance of cytidine deaminase in cells (22).
One plausible explanation for the extensive deamination of 2′-O-Me-C is that it may be a preferred substrate for cytidine deaminase. Structural and enzymatic studies of cytidine deaminase indicate that its preferred substrates are compounds with furanose rings having a Southern type conformation, in which the 3′ carbon is in the exo position below the plane of the ring. Consequently, the 3′ OH in the pseudo axial configuration can interact tightly with the enzyme, which is essential for activity (16, 40). Additionally, nucleosides having 2′-ribose substitutions on the alpha face of the nucleoside which are positioned trans relative to the 4′-hydroxymethyl of the ribose are better cytidine deaminase substrates than nucleosides having beta face substituents positioned cis relative to the 4′-ribose position (15, 16). Consistent with this, the lowest energy conformation predicted for the sugar of the 2′-O-Me-C analog, is of the Southern type, and its 2′-substituted methoxy group is trans relative to the 4′-hydroxymethyl of the ribose (Fig. 6). In contrast, 2′-C-Me-C, which prefers a Northern conformation and has a 2′-methyl group in the cis position relative to the 4′-hydroxymethyl of the ribose position, exhibits less deamination.
The observed lack of effect upon HCV replication in HBIII27 replicon cells was remedied by the use of a SATE (18a) monophosphate prodrug of 2′-O-Me-C. In contrast to the 2′-O-Me-C parent nucleoside, the SATE derivative was efficiently converted to 2′-O-Me-CTP in both replicon cell lines, averting the degradative effects of the replicon clones. Although cytidine monophosphates can also be deaminated by cytidylate deaminase (22), little by-product UTP or CTP was detected, suggesting that deamination predominantly occurred at the nucleoside level. This may be attributed to differences in levels of the cytidine versus cytidylate deaminases and/or substrate affinities of these enzymes. Additionally, the prodrug appears to bypass the restriction in nucleoside phosphorylation which occurs at the monophosphorylation step catalyzed by nucleoside kinase (Fig. 5). Following intracellular delivery of the prodrug, the released monophosphate can be efficiently phosphorylated by mono- and diphosphate kinases which are less specific and generally present at higher intracellular concentrations than the nucleoside kinase (22, 31). This, combined with better penetration of the SATE derivative into cells than of the 2′-O-Me-C parent, results in higher intracellular concentrations of 2′-O-Me-CTP.
Given that several intracellular metabolic parameters can influence the ability of nucleoside analogs to become effective and safe therapeutic agents, studies of intracellular metabolism and toxicity were critical to the development of this compound class. The differential antiviral and metabolic effects cited in this study demonstrate the importance of clonal variability among individual replicon lines in assessing cellular processes, a relevant concern in screening libraries of nucleosides for new lead structures and evaluating the potency of anti-HCV agents requiring metabolism. As such, a combined investigation of metabolic, antiviral, and biochemical properties was key to the successful identification of 2′-substituted nucleosides as selective HCV antiviral agents.
REFERENCES
- 1.Andrews, D. M., H. M. Chaignot, B. A. Coomber, M. D. Dowle, S. L. Hind, M. R. Johnson, P. S. Jones, G. Mills, A. Patikis, T. J. Pateman, J. E. Robinson, M. J. Slater, and N. Trivedi. 2003. The design of potent, non-peptidic inhibitors of hepatitis C protease. Eur. J. Med. Chem. 38:339-343. [DOI] [PubMed] [Google Scholar]
- 2.Carroll, S. S., D. B. Olsen, P. L. Durette, B. Bhat, P. Dande, and A. B. Eldrup. December 2003. Preparation of nucleoside derivatives as inhibitors of RNA-dependent RNA viral polymerase. WO patent 2,004,000,858.
- 3.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 5.Chan, L., T. J. Reddy, M. Proulx, S. K. Das, O. Pereira, W. Wang, A. Siddiqui, C. G. Yannopoulos, C. Poisson, N. Turcotte, A. Drouin, M. H. Alaoui-Ismaili, R. Bethell, M. Hamel, L. L'Heureux, D. Bilimoria, and N. Nguyen-Ba. 2003. Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem. 46:1283-1285. [DOI] [PubMed] [Google Scholar]
- 6.Crippen, G. M., and T. F. Havel. 1988. Distance geometry and molecular conformation. Research Studies Press, Wiley, New York, N.Y.
- 7.De Clercq, E. 2001. Antiviral drugs: current state of the art. J. Clin. Virol. 22:73-89. [DOI] [PubMed] [Google Scholar]
- 8.De Francesco, R., and C. M. Rice. 2003. New therapies on the horizon for hepatitis C: are we close? Clin. Liver Dis. 7:211-242. [DOI] [PubMed] [Google Scholar]
- 9.Desgranges, C., E. De Clercq, G. Razaka, F. Drouillet, I. Belloc, and H. Bricaud. 1986. Deoxyribosyl exchange reactions leading to the in vivo generation and regeneration of the antiviral agents (E)-5-(2-bromovinyl)-2′-deoxyuridine, 5-ethyl-2′-deoxyuridine and 5-(2-chloroethyl)-2′-deoxyuridine. Biochem. Pharmacol. 35:1647-1653. [DOI] [PubMed] [Google Scholar]
- 10.Eldrup, A. B., C. R. Allerson, C. F. Bennett, S. Bera, B. Bhat, M. R. Bosserman, J. Brooks, C. Burlein, S. S. Carroll, P. D. Cook, K. L. Getty, M. MacCoss, D. R. MacMasters, D. B. Olsen, T. Prakash, M. Prhavc, Q. Song, J. E. Tomassini, and J. Xia. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 47:2283-2295. [DOI] [PubMed] [Google Scholar]
- 11.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 12.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Reference deleted.
- 15.Krajewska, E., and D. Shugar. 1975. Alkylated cytosine nucleosides: substrate and inhibitor properties in enzymatic deamination. Acta Biochim. Pol. 22:185-194. [PubMed] [Google Scholar]
- 16.Kries, W., K. A. Watanabe, and J. J. Fox. 1978. Structural requirements for the enzymatic deamination of cytosine nucleosides. Helv. Chim. Acta 61:1011-1016. [Google Scholar]
- 17.Kuszewski, J., M. Nilges, and A. T. Brunger. 1992. Sampling and efficiency of metric matrix distance geometry: a novel partial metrization algorithm. J. Biomol. NMR 2:33-56. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 18a.Lefebvre, I., C. Périgaud, A. Pompon, A.-M. Aubertin, J.-L. Girardet, A. Kirn, G. Gosselin, and J.-L. Imbach. 1995. Mononucleoside phosphotriester derivatives with S-aceyl-2-thioethyl bioreversible phosphate-protecting groups: intracellular delivery of 3′-azido-2′,3′-dideoxythymidine 5′-monophosphate. J. Med. Chem. 38:3941-3950. [DOI] [PubMed] [Google Scholar]
- 19.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 20.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maehara, Y., H. Nakamura, Y. Nakane, K. Kawai, M. Okamoto, S. Nagayama, T. Shirasaka, and S. Fujii. 1982. Activities of various enzymes of pyrimidine nucleotide and DNA syntheses in normal and neoplastic human tissues. Gann 73:289-298. [PubMed] [Google Scholar]
- 22.Marquez, V. E. 1989. Inhibition of cytidine deaminase: mechanism and effect on the metabolism of antitumor agents, p. 91-114. In R. I. Glazer (ed.), Developments in cancer chemotherapy. CRC Press, Boca Raton, Fla.
- 23.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain terminating ribonucleoside analogs which inhibit HCV replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 24.Mohamadi, F., N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson, and W. C. Still. 1990. MacroModel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 11:4. [Google Scholar]
- 25.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 26.Narjes, F., U. Koch, C. Steinkuhler. 2003. Recent developments in the discovery of hepatitis C virus serine protease inhibitors—towards a new class of antiviral agents? Expert Opin. Investig. Drugs 12:153-163. [DOI] [PubMed] [Google Scholar]
- 27.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 28.Pause, A., G. Kukolj, M. Bailey, M. Brault, F. Do, T. Halmos, L. Lagace, R. Maurice, M. Marquis, G. McKercher, C. Pellerin, L. Pilote, D. Thibeault, and D. Lamarre. 2003. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem. 278:20374-20380. [DOI] [PubMed] [Google Scholar]
- 29.Prakash, T. P., S. S. Carroll, B. Bhat, M. Prhavc, D. B. Olsen, A. B. Eldrup, M. MacCoss, J. E. Tomassini, S. M. Galloway, C. H. Hughes, and P. D. Cook. 2005. Synthesis and evaluation of S-acyl-2-thioethyl esters of modified nucleoside 5′-monophosphates as inhibitors of hepatitis C virus RNA replication. J. Med. Chem. 48:1199-1210. [DOI] [PubMed] [Google Scholar]
- 30.Shim, J. H., G. Larson, V. Lai, S. Naim, and J. Z. Wu. 2003. Canonical 3′-dexoyribonucleotides as a chain terminator for HCV RNA-dependent RNA polymerase. Antiviral Res. 58:243-251. [DOI] [PubMed] [Google Scholar]
- 31.Stein, D. S., and K. H. P. Moore. 2001. Phosphorylation of nucleoside analog antiretrovirals: a review for clinicians. Pharmacotherapy 21:11-34. [DOI] [PubMed] [Google Scholar]
- 32.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, A. E. A. Hassan, C. K. Chu, K. W. Pankiewicz, K. A. Watanabe, R. F. Schinazi, and M. J. Otto. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomassini, J. E., E. Boots, L. Gan, P. Graham, V. Munshi, B. Wolanski, J. F. Fay, K. Getty, and R. LaFemina. 2003. An in vitro Flaviviridae replicase system capable of authentic RNA replication. Virology 313:274-285. [DOI] [PubMed] [Google Scholar]
- 34.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, M. P., N. Yao, and Z. Hong. 2003. Promising candidates for the treatment of chronic hepatitis C. Expert Opin. Investig. Drugs 12:1269-1280. [DOI] [PubMed] [Google Scholar]
- 38.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 39.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Xiang, S., S. A. Short, R. Wolfenden, and C. W. Carter, Jr. 1996. Cytidine deaminase complexed to 3-deazacytidine: a “valence buffer” in zinc enzyme catalysis. Biochemistry 35:1335-1341. [DOI] [PubMed] [Google Scholar]