Abstract
Laribacter hongkongensis, a newly discovered bacterium recently shown to be associated with community-acquired gastroenteritis, is generally resistant to most β-lactams except the carbapenems. We describe the cloning and characterization of a novel chromosomal class C β-lactamase and its regulatory gene in L. hongkongensis. Two genes, ampC and ampR, were cloned by inserting restriction fragments of genomic DNA from L. hongkongensis strain HLHK5 into pBK-CMV to give the recombinant plasmid pBK-LHK-5. The ampR and ampC genes and their promoters were divergently oriented, with the ampR gene immediately upstream of the ampC gene and an intercistronic Lys-R motif, typical of inducible ampC-ampR regulatory systems. The deduced amino acid sequence of the cloned AmpC β-lactamase (pI 8.1) contained consensus motifs characteristic of class C β-lactamases but had identities no greater than 46% to known class C β-lactamases. The kinetic properties of this AmpC were also compatible with those of a class C β-lactamase. PCR of 20 clinical isolates of L. hongkongensis, including HLHK5, showed the presence of both ampC and ampR genes in all isolates. Southern hybridization suggested that the ampC gene of HLHK5 was chromosomally encoded. Subcloning experiments showed that the expression of the ampC gene of HLHK5 was regulated by its ampR gene, which acts as a repressor. The β-lactamase characterized from strain HLHK5 was named LHK-5 (gene, blaLHK-5) and represents the first example of AmpC β-lactamase in the β subdivision of proteobacteria.
Laribacter hongkongensis is a facultative anaerobic, nonsporulating, gram-negative, seagull-shaped or spiral rod first isolated from the blood and empyema thoracis of a patient with alcoholic liver cirrhosis in Hong Kong (31). Subsequently, it was isolated from the stools of six patients with community-acquired diarrhea (29). By use of a newly developed selective medium, cefoperazone MacConkey agar, L. hongkongensis has recently been shown to be associated with community-acquired gastroenteritis (14, 30). The isolation of L. hongkongensis from patients with gastroenteritis correlated with a history of travel and the consumption of fish. Moreover, the bacterium has been found in the intestines of freshwater fish, which may be the source of human infections (30). L. hongkongensis may be a globally emerging pathogen, as the travel histories of patients suggested that it is present in at least four continents, including Asia, Europe, Africa, and Central America (29, 30).
L. hongkongensis is generally resistant to β-lactams, including broad-spectrum penicillins and cephalosporins, but is susceptible to carbapenems, amoxicillin-clavulanate, quinolones, and aminoglycosides (14, 29, 30, 31). Antibiotic treatment is usually not necessary in patients with Laribacter gastroenteritis. However, a quinolone and amoxicillin-clavulanate would be the antibiotic of choice in immunocompromised adults and children, respectively. Since the bacterium displays extensive resistance to β-lactams, cephalosporins, which are sometimes used to treat bacterial gastroenteritis, may not be useful (30). At the moment, the mechanism of β-lactam resistance in L. hongkongensis has not been investigated. Here we report on the cloning and characterization of a novel class C β-lactamase gene from clinical isolates of L. hongkongensis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used for cloning in this study are listed in Table 1. L. hongkongensis strain HKU1 was isolated from the blood culture and empyema pus of a patient with bacteremic empyema thoracis (31). The other 19 isolates of L. hongkongensis (strains HLHK2 to HLHK20) were recovered from the stools of patients with community-acquired gastroenteritis (14, 29, 30). The identities of all L. hongkongensis isolates were confirmed phenotypically by standard conventional biochemical methods and genotypically by 16S rRNA gene sequencing.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] Su− (nonsuppressing) λr (lambda resistant) | Stratagene |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pBK-CMV phagemid | Cloning vector; neomycin and kanamycin resistant | Stratagene |
| pACYC184 | Cloning vector; chloramphenicol and tetracycline resistant | NEB, Beverly, Mass. |
| pBK-LHK-5 | Recombinant plasmid containing a 2.5-kb Sau3A-digested fragment from genomic DNA of L. hongkongensis HLHK5 in pBK-CMV | This report |
| pLHKC | Recombinant plasmid containing ampC gene of L. hongkongensis HLHK5 in pACYC184 | This report |
| pLHKCR | Recombinant plasmid containing ampC and ampR genes of L. hongkongensis HLHK5 in pACYC184 | This report |
Susceptibility testing.
Antibiotic powders with known potencies, including sulbactam (Pfizer Corporation, Hong Kong, China) and clavulanic acid (SmithKline Beecham, Hong Kong, China), were kindly provided by the manufacturers. All other antibiotics were purchased from Sigma (St. Louis, Mo.). The MICs of the β-lactam antibiotics were determined by the broth macrodilution method for nonfastidious, aerobic bacteria, according to NCCLS guidelines (21), with Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.) and an inoculum of 5 × 105 CFU per ml with incubation at 35°C for 20 h. The MICs of ampicillin and cefoperazone were determined alone and in combination with 2 μg of clavulanic acid per ml or 4 μg of sulbactam per ml. Control strains Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 were included with each run.
Cloning experiments and recombinant plasmids.
A genomic DNA library was constructed as described in our previous publications (26, 28). Briefly, total genomic DNA of an L. hongkongensis strain, strain HLHK5, was extracted from 100 ml of culture broth and was partially digested with Sau3A (Roche, Mannheim, Germany). The partial digests with fragments of 2 to 4 kb were then ligated to the BamHI site of the vector provided by the ZAP Express vector kit (Strategene, La Jolla, Calif.), and a phage expression library was constructed according to the instructions of the manufacturer. The library had at least 1 million independent phage plaques, with more than 95% containing inserts of an average size of 2.5 kb, as checked by restriction enzyme digestion of 100 clones with SalI and XbaI (Roche). The DNA inserts of phage clones were excised with the ExAssist helper phage in XL1-Blue MRF′ cells (Strategene), which yielded the pBK-CMV phagemid vector. E. coli XLOLR cells were then infected with the pBK-CMV phagemid, which yielded the pBK-CMV plasmid with the cloned inserts in E. coli XLOLR cells. Antibiotic-resistant clones were selected on Luria-Bertani (LB) plates containing 32 μg/ml cefoperazone and 50 μg/ml kanamycin. Recombinant plasmid DNA was prepared with a High Pure plasmid isolation kit (Roche) from 1-ml LB broth cultures incubated with 32 μg/ml cefoperazone and 50 μg/ml kanamycin overnight at 37°C. The sizes of the inserted fragments were estimated according to the bacteriophage λ AvaII digest DNA marker (MBI Fermentas).
DNA sequencing, PCR amplification, and sequence analysis.
The primers used in this study are listed in Table 2. Cloned DNA fragments in pBK-CMV were sequenced with vector primers of pBK-CMV (T3 and T7) and synthetic primers (LPW606 and LPW608), designed from the sequencing data of the first round of the sequencing reaction. Bidirectional DNA sequencing was performed with an ABI automatic sequencer (Perkin-Elmer, Norwalk, Conn.), according to the instructions of the manufacturer (26). The DNA sequence was analyzed by a search with the BLAST program on the National Center for Biotechnology Information server at the National Library of Medicine (Bethesda, Md.; http://www.ncbi.nlm.nih.gov). The searches were performed at both the protein and the DNA levels.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Nucleotide positions corresponding to the insert of pBK-LHK-5 shown in Fig. 1 | Purpose |
|---|---|---|---|
| LPW606 | GGCAGGCAGCAACCATTC | 673 to 690 | Sequencing of insert of pBK-LHK-5 |
| LPW608 | ATTGACCTGTCTGCACCG | 1412 to 1429 | Sequencing of insert of pBK-LHK-5 and ampC gene |
| LPW621 | GGCTATCTGTTGCAGCAACC | 19 to 38 | PCR and sequencing of ampR gene |
| LPW622 | CGACCAGTGCAGTAAACGGT | 1385 to 1366 | PCR and sequencing of ampR gene and PCR for preparation of the 298-bp blaLHK-5 probe |
| LPW663 | CGGATTACCCCATTTTCCCG | 1088 to 1107 | PCR and sequencing of ampC gene and PCR for preparation of the 298-bp blaLHK-5 probe |
| LPW664 | CATGTCTGCCCGGAATGTGC | 2405 to 2386 | PCR and sequencing of ampC gene |
| LPW665 | CAATCTAACGAGGGCTGAGT | −118 to −99 | PCR and sequencing of ampR gene |
| LPW679 | TGTTCGACTCGTCGATAGGC | 239 to 220 | PCR and sequencing of ampR gene |
| LPW697 | ATTGCGGTCGGCATTACAGC | 1906 to 1925 | Sequencing of ampC gene |
| LPW1946 | GATCAAAGGCCTGGCTGGC | 878 to 896 | PCR of ampC-ampR gene for construction of recombinant plasmid pLHKCR |
| LPW1947 | GGTCGAACCATCGTGAACC | −2 to 17 | PCR of ampC gene for construction of recombinant plasmid pLHKC |
| LPW1948 | ATGTCTGCCCGGAATGTGC | 2405 to 2387 | PCR of ampC and ampC-ampR genes for construction of recombinant plasmids pLHKC and pLHKCR |
The genomic DNA of 20 clinical isolates of L. hongkongensis (isolates HKU1 and HLHK2 to HLHK20) was extracted as described previously (31). The prevalence of the cloned β-lactamase genes of L. hongkongensis was studied by PCR with laboratory-designed primers specific for the ampC-coding region (primers LPW663 and LPW664) and the ampR-coding region (primers LPW621, LPW622, LPW665, and LPW679). The PCR products of seven L. hongkongensis strains (strains HKU1 and HLHK2 to HLHK7) were also sequenced and analyzed by using additional sequencing primers specific for ampC genes (primers LPW608 and LPW697). The deduced protein sequences of ampC and ampR were compared with known sequences in the GenBank database by multiple-sequence alignment with the CLUSTAL W program (25). The phylogenetic relationships of the ampC and ampR genes of L. hongkongensis to the corresponding related genes were determined with the Clustal X program (version 1.81) (11) and by the neighbor-joining method with GrowTree software (Genetics Computer Group, Inc., San Diego, Calif.).
Southern hybridization of β-lactamase gene.
DNA from L. hongkongensis strain HLHK5 was used for Southern hybridization analysis. Plasmid DNA was prepared with a High Pure plasmid isolation kit (Roche) and a Large Construct kit (QIAGEN, Hilden, Germany). The eluent was subjected to agarose gel electrophoresis and pulsed-field gel electrophoresis (PFGE), as described previously, with slight modifications (13). Agarose plugs containing total DNA and total DNA digested with SpeI were also prepared and subjected to PFGE. The restriction enzyme SpeI did not cut within the 298-bp blaLHK-5 probe region prepared from the sequences obtained by PCR with primers LPW663 and LPW622 at an annealing temperature of 55°C targeted within the ampC gene. Southern blot hybridization with Hybond N+ membranes (Amersham International, Little Chalfont, United Kingdom) was modified from our previously published protocol (27, 30). After hybridization with the digoxigenin-labeled probe (the denatured PCR product), the nylon membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl with 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at room temperature for 30 min, followed by washing with 0.5× SSC-0.1% SDS at room temperature for 30 min and 0.1× SSC-0.1% SDS at 55°C for 30 min.
β-Lactamase assays and IEF analysis.
β-Lactamase from cultures of E. coli XLOLR harboring pBK-LHK-5 was extracted by sonication as described in our previous publications (4, 10). Isoelectric focusing (IEF) analysis of the β-lactamase was performed with an ampholine gel (Pharmacia, Hong Kong, China) from pI 3.5 to pI 9.5. Enzyme extracts from strains expressing TEM-1 (pI 5.4), OXA-1 (pI 7.4), and SHV-1 (pI 7.6) were used as controls. The pI value of each enzyme was determined by overlaying the gel with nitrocefin (24). Substrate and inhibition assays were performed as described previously (4). The wavelengths were 235 nm for benzylpenicillin and cefoxitin; 255 nm for cephaloridine, cephalothin, and cefoperazone; 257 nm for ceftazidime and cefepime; and 297 nm for meropenem. The Vmax and Km values were calculated by use of the Lineweaver-Burk plot with the built-in Enzymatic Mechanism software (3). All kinetic studies were performed in triplicate.
Regulation of ampC expression.
Expression of ampC of L. hongkongensis was studied after it was cloned into a plasmid of relatively low copy number (20 to 30 copies), pACYC184. By using the DNA sequence of the cloned fragment of recombinant plasmid pBK-LHK-5 with L. hongkongensis HLHK5 as the template, primers LPW1946 and LPW1948 were used for amplification of the ampC and ampR genes and primers LPW1947 and LPW1948 were use for amplification of the ampC gene. The PCR products obtained were cloned into the BamHI site of pACYC184. The resulting recombinant plasmids, pLHKC and pLHKCR (Table 1), were transformed into E. coli TOP10, and both constructs were resequenced. The MICs for E. coli TOP10 harboring the full-length (pLHKCR) or ampR deletion (pLHKC) constructs with or without imipenem (0.0625 μg/ml) or cefoxitin (8 μg/ml) were determined (6). The disk approximation test was also performed by using disks of cefoxitin (30 μg) or imipenem (10 μg) placed against ampicillin (10 μg), cephaloridine (30 μg), ceftriaxone (30 μg), and cefoperazone (70 μg) disks on Mueller-Hinton agar plates inoculated with the E. coli clones, as described previously (17, 23). The plates were examined after overnight incubation at 37°C.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the blaCLHK gene have been lodged within the GenBank sequence database under accession numbers AY632070 for blaCLHK-1, AY632071 for blaCLHK-2, AY632072 for blaCLHK-3, AY632073 for blaCLHK-4, AY632074 for blaCLHK-5, AY632075 for blaCLHK-6, and AY632076 for blaCLHK-7.
RESULTS
Antibiotic susceptibilities.
The MICs of β-lactams for seven strains of L. hongkongensis (strain HKU1 and HLHK2 to HLHK7) showed that they were resistant to all β-lactams except imipenem and meropenem (Table 3). When LHK5 was expressed in E. coli XLOLR (pBK-LHK5), it conferred resistance to ampicillin, ampicillin-clavulanic acid, ampicillin-sulbactam, cefuroxime, ceftazidime, cefoperazone, and cefoperazone-sulbactam; but less resistance to cefotaxime and cefepime was evident.
TABLE 3.
MICs of β-lactams for seven clinical isolates of L. hongkongensis, E. coli XLOLR harboring recombinant plasmid pBK-LHK-5, and E. coli XLOLR
| Bacterial strain | MIC (μg/ml)b
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMP-CA | AMP-SBM | FOX | CXM | CAZ | CTX | CPZ | CPZ-SBM | FEP | ATM | IPM | MPM | |
| L. hongkongensis | |||||||||||||
| HKU1 | 32 | 32 | 16 | 16 | 16 | >256 | 64 | >256 | >256 | 64 | >256 | 0.0625 | 0.0625 |
| HLHK2 | 128 | 128 | 64 | 128 | 256 | >256 | >256 | >256 | >256 | >256 | >256 | 0.0625 | 0.0625 |
| HLHK3 | 256 | 256 | 64 | 64 | 32 | >256 | 128 | >256 | >256 | 32 | >256 | 0.0625 | 0.03125 |
| HLHK4 | 256 | 256 | 128 | 128 | 64 | >256 | 256 | >256 | >256 | >256 | >256 | 0.0625 | 0.0625 |
| HLHK5 | 256 | 64 | 128 | 64 | 64 | >256 | 256 | >256 | >256 | 128 | >256 | 0.0625 | 0.0625 |
| HLHK6 | 256 | 256 | 128 | 64 | 16 | 256 | 128 | >256 | >256 | 128 | >256 | 0.125 | 0.0625 |
| HLHK7 | 64 | 64 | 64 | 64 | 32 | 128 | 128 | >256 | >256 | 2 | >256 | 0.0625 | 0.0625 |
| E. coli | |||||||||||||
| XLOLR(pBK-LHK-5)a | 64 | 64 | 32 | 16 | 128 | 64 | 32 | >256 | >256 | 32 | >256 | 0.125 | 0.0625 |
| XLOLR | 8 | 4 | 4 | 16 | 32 | 4 | 8 | 8 | 8 | 8 | 128 | 0.125 | 0.0625 |
This strain, which harbors the recombinant plasmid pBK-LHK-5, produced the LHK-5 β-lactamase.
AMP, ampicillin; AMP-CA, ampicillin plus clavulanic acid (2 μg/ml); AMP-SBM, ampicillin plus sulbactam (4 μg/ml); FOX, cefoxitin; CXM, cefuroxime; CAZ, ceftazidime; CTX, cefotaxime; CPZ, cefoperazone; CPZ-SBM, cefoperazone plus sulbactam (4 μg/ml); FEP, cefepime; ATM, aztreonam; IPM, imipenem; MPM, meropenem.
Cloning and sequence analysis of blaCLHK-5.
Six identical recombinant E. coli XLOLR clones were obtained. One clone with an insert of 2,529 bp (pBK-LHK-5) was selected for further analysis. The insert contained two open reading frames (ORFs), one of 1,173 bp that encoded a 390-amino-acid sequence with identities of no greater than 46% to known class C β-lactamases and the other of 888 bp that encoded a 295-amino-acid sequence with identities of no greater than 65% to known ampR genes (Fig. 1). The two ORFs are divergently oriented with overlapping −35 and −10 promoter regions. A Lys-R motif (CAAAATCAATTCA) was also found inside the 190-bp ampC-ampR intercistronic region (Fig. 2) (8, 15). The deduced amino acid sequence of the ampC gene contained the consensus sites, SLSK, which is characteristic of serine β-lactamases (20), and YSN and KTG, which are characteristic of class C β-lactamases (12, 18). At amino acid positions 247 to 250, where another structural element characteristic of class C β-lactamases was expected, the amino acid residues DEET were found (Fig. 1) (22). The deduced amino acid sequence of the ampR gene contained a helix-turn-helix domain at the N terminus (Fig. 1) (8, 15).
FIG. 1.
Nucleotide and deduced amino acid sequences of the insert of pBK-LHK-5 containing the ampC- and ampR-coding regions. The deduced amino acid sequences are designated in single-letter code. The putative promoter sequences, represented by −35 and −10 regions, and the Lys-R motif are boxed and indicated by arrows. The start codons are indicated by arrows, and the stop codons are underlined. The shaded areas represent the predicted helix-turn-helix motif of the LysR family. For AmpC, the consensus sites characteristic of serine β-lactamases or class C β-lactamases are in boldface and boxed.
FIG. 2.
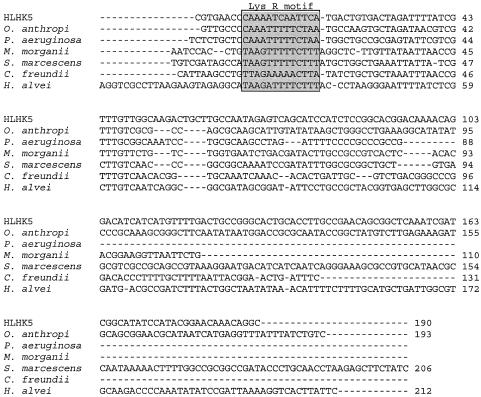
Multiple alignment of the DNA sequences of the ampC-ampR intercistronic regions of the β-lactamases of L. hongkongensis HLHK5, O. anthropi SLO74, P. aeruginosa PAO01, M. morganii GUI-1, Serratia marcescens, C. freundii OS60, and H. alvei HA-1. The Lys-R motif is boxed.
Diversity and phylogenetic analysis of blaCLHK.
The deduced amino acid sequence of blaCLHK-5 had 46% amino acid identities with the AmpC sequences of Sinorhizobium meliloti (GenBank accession no. CAC49440) and Ralstonia metallidurans (GenBank accession no. ZP_00023977) and the β-lactamase of Pseudomonas aeruginosa (GenBank accession no. AAM08945) (Fig. 3). The deduced amino acid sequence of the ampR gene of HLHK5 had 65% amino acid identities with those of Ochrobactrum anthropi (GenBank accession no. CAC04521) and P. aeruginosa PAO1 (GenBank accession no. AAG07496) and 63% amino acid identity with that of Pseudomonas fluorescens PfO-1 (GenBank accession no. ZP_00087575) (Fig. 4). PCR of all 20 isolates of L. hongkongensis showed bands at about 1,300 bp for the ampC gene and bands at about 1,350 bp for the ampR gene. Sequencing of the PCR products of HLHK5 showed that both genes were identical to those obtained from the E. coli XLOLR(pBK-LHK-5) clone. Sequencing of the PCR products of the other six isolates (isolates HKU1, HLHK2 to HLHK4, HLHK6, and HLHK7) showed 95% to 97% amino acid identities of their AmpC sequences to that of HLHK5 (Fig. 3) and 98% to 99% amino acid identities of their AmpR sequences to that of HLHK5 (Fig. 4). The deduced amino acid sequences of the ampC genes were different among all seven isolates. The ampC genes of strains HKU1 and HLHK2 had six and three nucleotide deletions, respectively, resulting in two and one amino acid deletions at positions 38 to 39 and position 283, respectively. The deduced amino acid sequences of the ampR genes of strains HLHK2 and HLHK6 were identical.
FIG. 3.
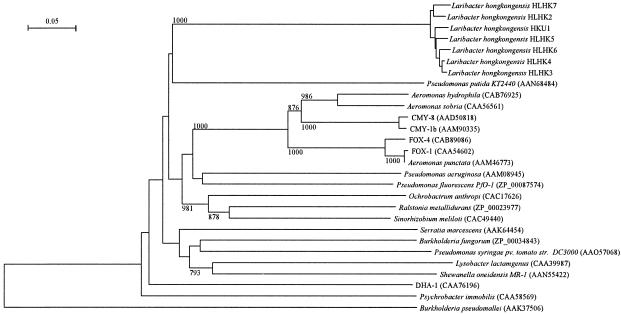
Phylogenetic tree showing the relationships of the AmpC of L. hongkongensis to related chromosomally encoded or plasmid-encoded class C β-lactamases. The tree was constructed by using the neighbor-joining method and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 100 amino acids by use of the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
FIG. 4.
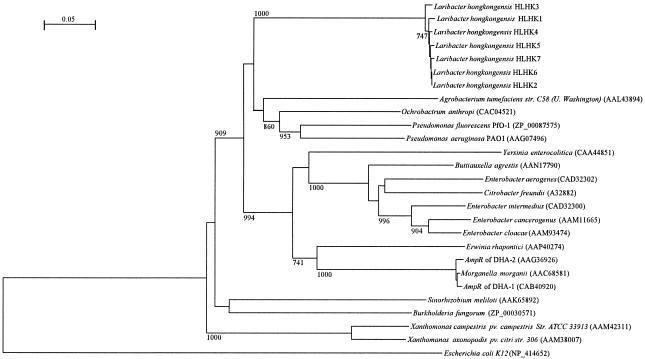
Phylogenetic tree showing the relationships of the AmpR of L. hongkongensis to related AmpR proteins. The tree was constructed by using the neighbor-joining method and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of per 100 amino acids by use of the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
Southern hybridization of β-lactamase gene.
Agarose gel electrophoresis and PFGE did not reveal the presence of a plasmid of ≤250 kb in L. hongkongenesis strain HLHK5 after plasmid extraction with the High Pure plasmid isolation kit (for plasmids of up to 50 kb) and the Large Construct kit (for plasmids up to 250 kb). However, PFGE of total DNA showed genomic DNA and an additional band at about 310 kb which may correspond to a potential plasmid. Genomic DNA, but not the additional band, hybridized with the blaLHK-5-specific probe. A single band of about 150 kb from SpeI-digested total DNA also showed positive hybridization with the blaLHK-5-specific probe.
Biochemical properties and IEF analysis of LHK-5.
IEF analysis of the E. coli XLOLR(pBK-LHK-5) clone revealed a β-lactamase with an isoelectric point of 8.1. The kinetic parameters of the β-lactamase enzyme are shown in Table 4.
TABLE 4.
Kinetic parameters of LHK-5 β-lactamase in E. coli XLOLR(pBK-LHK-5)a
| Substrate or inhibitor | Relative Vmaxb | Km (μM) | Relative Vmax/Kmb | IC50 (μM)c |
|---|---|---|---|---|
| Substrate | ||||
| Benzylpenicillin | 2.8 ± 0.35d | 356 ± 13 | 0.7 | NAe |
| Cephaloridine | 100 ± 9 | 88 ± 10 | 100 | NA |
| Cephalothin | 101 ± 2 | 56 ± 2 | 159 | NA |
| Cefoxitin | <1 | — | — | NA |
| Ceftazidime | <1 | — | — | NA |
| Cefepime | 1.05 ± 0.09 | — | — | NA |
| Cefoperazone | <1 | — | — | NA |
| Meropenem | <1 | — | — | NA |
| Inhibitor | ||||
| Cloxacillin | NA | NA | NA | 0.0035 ± 0.0 |
| Clavulanate | NA | NA | NA | 138 ± 2.1 |
| Sulbactam | NA | NA | NA | 57 ± 1.6 |
E. coli XLOLR haboring the recombinant plasmid pBK-LHK-5.
Relative to that of cephaloridine, which was taken as 100%.
IC50, concentration of β-lactamase inhibitor that inhibits 50% of the activity after 10 min of incubation at 37°C. Cephaloridine (50% inhibitory concentration, 150 μM) was used as the substrate.
Data shown are the means ± standard deviations of three measurements.
NA, not applicable.
—, in these cases, the hydrolysis parameters could not be determined (Vmax was too low or Km was too high).
Regulation of L. hongkongensis ampC expression.
E. coli harboring the ampR deletion construct (pLHKC) showed higher MICs to ampicillin, cephaloridine, and cefoperzone (128 μg/ml, >256 μg/ml and > 256 μg/ml, respectively) than E. coli harboring the full-length ampR-ampC (pLHKCR) construct (16 μg/ml, 32 μg/ml, and 1 μg/ml, respectively), indicating that the expression of the L. hongkongensis ampC gene is negatively regulated by ampR. However, no increase in the MICs to ampicillin or cefoperazone was observed in the presence of cefoxitin or imipenem. Similarly, no flattening of the ampicillin, cephaloridine, ceftriaxone, and cefoperazone zones of inhibition was observed on a lawn of E. coli expressing pLHKCR.
DISCUSSION
In this study, we identified a β-lactamase from a clinical isolate of L. hongkongensis and showed that the gene was also present in all other strains tested. Unlike most members of the family Enterobacteriaceae with an inducible chromosomal ampC gene, which are usually not resistant to ceftazidime or cefotaxime, unless the ampC gene is expressed at very high levels, L. hongkongensis is generally resistant to most β-lactams except the carbapenems, as in the case in O. anthropi (20). β-Lactamase inhibitors, clavulanic acid and sulbactam, did not restore its susceptibility to β-lactams. Expression of the β-lactamase of L. hongkongensis, LHK-5, in E. coli showed that the recombinant clone also displayed resistance to all β-lactams except cefoxitin and carbapenems. Therefore, the present β-lactamase, which is only distantly related to other class C β-lactamases, behaves differently if it is responsible for all the β-lactam resistance profiles in L. hongkongensis. However, whether the unusual aztreonam resistance in L. hongkongensis can be explained by the presence of this β-lactamase remains to be determined, as the E. coli XLOLR strain used in the present study is also intrinsically resistant to aztreonam. Moreover, the variability of the cefepime MICs of the different strains of L. hongkongensis suggests that the present AmpC may not be responsible for the cefepime resistance observed in some strains of L. hongkongensis. Further investigations should be carried out to determine if additional resistance mechanisms are responsible for the β-lactam resistance in L. hongkongensis.
Sequence analysis showed that the insert of the recombinant plasmid contains an ampC gene and an ampR gene. The two genes and their corresponding putative promoters were in opposite orientations, with the ampR gene immediately upstream of the ampC gene and an intercistronic region containing a Lys-R motif. Such an arrangement is typical of inducible ampC-ampR regulatory systems found in many other gram-negative bacteria, including Morganella and Citrobacter (15, 22). The sequences of both genes from the E. coli XLOLR(pBK-LHK-5) clone were identical to those obtained from the parental strain, HLHK5. Both genes were present in all the 20 strains tested. Sequence analysis of the ampC genes of seven strains of L. hongkongensis showed 3% to 5% amino acid differences among their ampC gene sequences. Hybridization experiments suggested that the ampC gene in L. hongkongensis was probably chromosomally encoded and was present in the 150-kb fragment of SpeI-digested total DNA. The result is consistent with the presence of ampC genes in all 20 strains of L. hongkongensis as determined by PCR, despite the absence of plasmids in some strains (unpublished data). The intercistronic region is long (190 bp), and the DNA sequence of this Lys-R motif in L. hongkongensis is more similar to those of O. anthropi (20) and P. aeruginosa (16) than to those of members of the family Enterobacteriaceae (Fig. 2). The 38-bp specific AmpR binding sites which have been outlined for the AmpR protein of Citrobacter freundii was not found, probably because the AmpR of L. hongkongensis is only distantly related to that of C. freundii (15) (Fig. 4).
The AmpC of L. hongkongensis represents a new class C β-lactamase, with less than 50% amino acid sequence homology to known class C β-lactamases. From phylogenetic analysis, it is only distantly related to other chromosomally encoded or plasmid-encoded class C β-lactamases (Fig. 3), with the highest amino acid sequence similarity (46%) to the putative β-lactamase in the 1,683-kb pSymB megaplasmid of the symbiotic bacterium S. meliloti (5). Nevertheless, it possessed properties characteristic of class C β-lactamases. The present ampC gene contained amino acid residues characteristic of serine β-lactamases (SXXK) and class C β-lactamases (YXN and KXG) (12, 18, 20). The kinetic parameters of LHK-5 revealed that the enzyme had strong activities against cephalothin and cephaloridine. Similar to other class C β-lactamases, such as those from O. anthropi (20) and Morganella morganii (22), the present enzyme only poorly hydrolyzed ceftazidime, cefoperazone, and cefoxitin. However, unlike LHK-5, other class C enzymes, when they are cloned into E. coli, usually confer cefoxitin resistance. Therefore, LHK-5 possesses properties more similar to those of the class C enzyme of Hafnia alvei, which neither hydrolyzes nor provides resistance to cefoxitin (6). The hydrolytic activity of LHK-5 was strongly inhibited by low concentrations of cloxacillin but was poorly inhibited by clavulanic acid and sulbactam.
The ampR of L. hongkongensis regulates the expression of ampC by acting as a repressor. However, induction of ampC by cefoxitin or imipenem was not observed in the present study. The reason for the lack of induction in L. hongkongensis remains to be determined. Although the inducible effects of cefoxitin or imipenem have been observed in the ampC-ampR systems of many gram-negative bacteria, a previous study with O. anthropi also failed to demonstrate such effects, and the authors attributed this to the high level of resistance to extended-spectrum cephalosporins conferred by the E. coli clone (20). It has been shown that the inducibility of ampC-ampR systems may depend on the availability of other elements, such as AmpD, AmpE, and AmpG (2). Further studies are required to determine whether the ampC-ampR of L. hongkongensis exhibits a different regulatory profile or whether its induction effect can be demonstrated in other host systems with compatible accessory elements.
The present study represents the first report of an ampC gene and the ampC-ampR system in the β subdivision of the proteobacteria. While ampC-ampR systems have been almost exclusively reported in the γ subdivision of proteobacteria, it has also been recently reported in the α subdivision (9, 20). L. hongkongensis is a member of the family Neisseriaceae under the β subdivision of proteobacteria, in which the ampC gene has not previously been found (MEDLINE search up to June 2004). The presence of ampC genes in three subdivisions of proteobacteria suggests that ampC genes are ancient genes that may have been present before the divergence of these subdivisions or horizontally transferred between bacteria from different subdivisions. Nevertheless, its presence in a huge diversity of bacteria, including both animal pathogens and environmental bacteria, implies that the gene has been maintained even in the absence of selective pressure from clinical antibiotic usage (1, 9). Although it has been proposed that the gene may be involved in peptidoglycan metabolism (7, 19), the exact function of ampC remains to be determined.
Acknowledgments
This work was partly supported by the University Development Fund, University Research Grant Council, The University of Hong Kong, and the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government.
REFERENCES
- 1.Barlow, M., and B. G. Hall. 2002. Origin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, P. M., and I. Chopra. 1993. Molecular basis of β-lactamase induction in bacteria. Antimicrob. Agents Chemother. 37:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., and R. B. Sykes. 1986. Methodology for the study of β-lactamases. Antimicrob. Agents Chemother. 30:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, T. K., P. L. Ho, P. C. Woo, K. Y. Yuen, and P. Y. Chau. 2002. Cloning and expression of class A β-lactamase gene blaA (BPS) in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 46:1132-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girlich, D., T. Naas, S. Bellais, L. Poirel, A. Karim, and P. Nordmann. 2000. Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob. Agents Chemother. 44:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson, T. A., K. D. Young, S. A. Denome, and P. K. Elf. 1997. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179:6112-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins, C. S., M. B. Avison, L. Jamieson, A. M. Simm, P. M. Bennett, and T. R. Walsh. 2001. Characterization, cloning and sequence analysis of the inducible Ochrobactrum anthropi AmpC β-lactamase. J. Antimicrob. Chemother. 47:745-754. [DOI] [PubMed] [Google Scholar]
- 10.Ho, P. L., T. K. Cheung, W. C. Yam, and K. Y. Yuen. 2002. Characterization of a laboratory-generated variant of BPS β-lactamase from Burkholderia pseudomallei that hydrolyses ceftazidime. J. Antimicrob. Chemother. 50:723-726. [DOI] [PubMed] [Google Scholar]
- 11.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 10:403-405. [DOI] [PubMed] [Google Scholar]
- 12.Joris, B., J. M. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J. M. Frere, J. A. Kelly, J. C. Boyington, P. C. Moews, and J. R. Knox. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Eur. J. Biochem. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau, S. K. P., P. C. Y. Woo, H. Tse, K. W. Leung, S. S. Y. Wong, and K. Y. Yuen. 2003. Invasive Streptococcus iniae infections outside North America. J. Clin. Microbiol. 41:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, S. K. P., P. C. Y. Woo, W. T. Hui, M. W. S. Li, J. L. L. Teng, T. L. Que, W. K. Luk, R. W. M. Lai, R. W. H. Yung, and K. Y. Yuen. 2003. Cefoperazone MacConkey agar for selective isolation of Laribacter hongkongensis. J. Clin. Microbiol. 41:4839-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg, F., L. Westman, and S. Normark. 1987. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc. Natl. Acad. Sci. USA 82:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodge, J. M., S. D. Minchin, L. J. Piddock, and J. W. Busby. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem. J. 272:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahlen, S. D., S. S. Morrow, B. Abdalhamid, and N. D. Hanson. 2003. Analyses of ampC gene expression in Serratia marcescens reveal new regulatory properties. J. Antimicrob. Chemother. 51:791-802. [DOI] [PubMed] [Google Scholar]
- 18.Matsumura, N., S. Minami, and S. Mitsuhashi. 1998. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42:176-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morosini, M. I., J. A. Ayala, F. Baquero, J. L. Martinez, and J. Blazquez. 2000. Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadjar, D., R. Labia, C. Cerceau, C. Bizet, A. Philippon, and G. Arlet. 2001. Molecular characterization of chromosomal class C β-lactamase and its regulatory gene in Ochrobactrum anthropi. Antimicrob. Agents Chemother. 45:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Poirel, L., M. Guibert, D. Girlich, T. Naas, and P. Nordmann. 1999. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob. Agents Chemother. 43:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders, C. C., and W. E. Sanders, Jr. 1979. Emergence of resistance to cefamandole: possible role of cefoxitin-inducible β-lactamases. Antimicrob. Agents Chemother. 15:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siu, L. K., J. Y. Lo, K. Y. Yuen, P. Y. Chau, M. H. Ng, and P. L. Ho. 2000. β-Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo, P. C., P. K. Leung, H. W. Tsoi, and K. Y. Yuen. 2001. Cloning and characterisation of malE in Burkholderia pseudomallei. J. Med. Microbiol. 50:330-338. [DOI] [PubMed] [Google Scholar]
- 27.Woo, P. C., P. K. Leung, H. W. Tsoi, B. Y. Chan, T. L. Que, and K. Y. Yuen. 2002. Characterization of a novel insertion sequence, ISBp1, in Burkholderia pseudomallei. Arch. Microbiol. 177:267-273. [DOI] [PubMed] [Google Scholar]
- 28.Woo, P. C., P. K. Leung, S. S. Wong, P. L. Ho, and K. Y. Yuen. 2001. groEL encodes a highly antigenic protein in Burkholderia pseudomallei. Clin. Diagn. Lab. Immunol. 8:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo, P. C. Y., P. Kuhnert, A. P. Burnens, J. L. L. Teng, S. K. P. Lau, T. L. Que, H. H. Yau, and K. Y. Yuen. 2003. Laribacter hongkongensis: a potential cause of infectious diarrhea. Diagn. Microbiol. Infect. Dis. 47:551-556. [DOI] [PubMed] [Google Scholar]
- 30.Woo, P. C. Y., S. K. P. Lau, J. L. L. Teng, T. L. Que, R. W. H. Yung, W. K. Luk, R. W. M. Lai, W. T. Hui, S. S. Y. Wong, H. H. Yau, and K. Y. Yuen. 2004. Association of Laribacter hongkongensis in community-acquired human gastroenteritis with travel and with eating fish: a multicentre case-control study. Lancet 363:1941-1947. [DOI] [PubMed] [Google Scholar]
- 31.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]