Abstract
The contribution of integrons to the dissemination of extended-spectrum β-lactamases (ESBL) was analyzed on all ESBL-producing Escherichia coli isolates from 1988 to 2000 at Ramón y Cajal Hospital. We studied 133 E. coli pulsed-field gel electrophoresis types: (i) 52 ESBL-producing clinical strains (C-ESBL) (16 TEM, 9 SHV, 21 CTX-M-9, 1 CTX-M-14, and 5 CTX-M-10); (ii) 43 non-ESBL blood clinical strains (C-nESBL); and (iii) 38 non-ESBL fecal isolates from healthy volunteers (V-nESBL). Class 1 integrons were more common among C-ESBL (67%) than among C-nESBL (40%) or V-nESBL (26%) (P < 0.001) due to the high number of strains with blaCTX-M-9, which is linked to an In6-like class 1 integron. Without this bias, class 1 integron occurrence would be similar in C-ESBL and C-nESBL groups (47% versus 40%). Occurrence of class 2 integrons was similar among clinical and community isolates (13 to 18%). No isolates contained class 3 integrons. The relatively low rate of class 1 integrons within transferable elements carrying blaTEM (23%) or blaSHV (33%) and the absence of class 2 integrons in all ESBL transconjugants mirror the assembly of translocative pieces containing blaTEM or blaSHV on local available transferable elements lacking integrons. The low diversity of class 1 integrons (seven types recovered in all groups) might indicate a wide dissemination of specific genetic elements in which they are located. In our environment, the spread of genetic elements encoding ESBL has no major impact on the dispersion of integrons, nor do integrons have a major impact on the spread of ESBL, except when blaESBL genes are within an integron platform such as blaCTX-M-9.
The most obvious risk factor for the dissemination of genes encoding extended-spectrum β-lactamases (ESBL) is directional selection of resistant strains following the use of β-lactam antibiotics, particularly expanded-spectrum cephalosporins (5). Nevertheless, the investigation of other factors might be critical for predicting the potential spread and evolution of ESBL-producing strains.
Genes encoding ESBL are usually located on conjugative plasmids (such as blaTEM or blaSHV), although many of the most recently described ESBL genes are frequently found within integron-like structures (such as blaCTX-M, blaGES, or blaVEB-1) (4-6, 17). On the other hand, ESBL-producing isolates are usually resistant to other antibiotics such as aminoglycosides, tetracyclines, chloramphenicol, trimethoprim, sulfonamides, or quinolones, often due to the presence of different resistance genes on transferable elements such as plasmids, transposons, or integrons and/or genetic structures generated by combinatorial evolution of different interactive pieces (2, 17, 27, 29, 39-41). The fact that ESBL genes could be acquired by strains harboring particular integrons may enlarge the possibilities of selection of these isolates by a variety of different antimicrobials. Moreover, ESBL genes can be located on integrons, which may facilitate the spread of such genetic elements (4, 6).
Integrons are natural highly efficient recombination and expression systems able to capture genes as part of genetic elements known as gene cassettes (32). Five integron classes related to antibiotic resistance have been described based on the homology of their integrase genes (1, 3, 16, 32; Gene Bank accession no. AJ277063). Class 1 integrons are the most commonly found in nosocomial and community environments, followed by class 2 integrons, other integron classes being scarcely reported to date (12, 31, 32, 42). An increase in the rate of clinical isolates containing antibiotic resistance integrons (ARI) has been observed in several institutions during the last years, with changes in the gene cassette content of class 1 integron over time (34, 43). The widespread presence of Enterobacteriaceae containing ARI observed among the community-based population indicates the existence of a substantial reservoir potentially feeding multidrug resistance in the nosocomial setting (19, 23). ESBL located on integron-like structures are also being increasingly reported worldwide (4, 6). Although independent association between integrons and particular antimicrobial agents (including ampicillin and/or piperacillin and cefuroxime) has been suggested (20), the association of these genetic elements with antibiotic resistance in ESBL-producing isolates has not been previously explored.
In this study we investigated the occurrence, distribution, and cassette content of integrons among all different ESBL-producing Escherichia coli clones isolated from patients at the Ramón y Cajal University Hospital in Madrid (Spain) during 12 years (1988 to 2000). To evaluate the contribution of class 1, class 2, and class 3 integrons to the dissemination of the different ESBL types (TEM, SHV, and CTX-M) and the antibiotic resistance genes frequently associated with ESBL-producing organisms, the presence of these integrons was also investigated among non-ESBL-producing E. coli isolates from patients attending the same hospital and from healthy volunteers.
MATERIALS AND METHODS
Bacterial strains and epidemiological background.
The 133 E. coli isolates included in this study were divided into three groups on the basis of the source and ESBL production. Only one isolate per person was studied, and all pulsed-field gel electrophoresis-related isolates were excluded. ESBL-producing E. coli clinical isolates (C-ESBL, n = 52) were obtained from clinical specimens of patients located in different wards of our institution (Hospital Ramón y Cajal, Madrid, Spain) between 1988 and 2000 (40.4% in surgical wards, 21.1% in intensive care units, 15.4% in medical wards, and 23.1% outpatients). These isolates were recovered from urine (n = 25), rectal swabs (n = 7), respiratory samples (n = 6), blood (n = 5), wounds (n = 5), and other samples (n = 4). Strains corresponding to a control group of clinical non-ESBL-producing E. coli isolates (C-nESBL, n = 43) were obtained from blood cultures of patients at our hospital during the same period of time in surgical wards (39.5%), medical wards (18.6%), intensive care units (16.3%), and the emergency room (25.6%). Community-type non-ESBL-producing E. coli strains were isolated from feces of healthy volunteers living in Madrid during 2001 (V-nESBL, n = 38) without exposure to antibiotics or hospital environment in the 3 months previous to the sample recovery. For samples from this group, 0.5 g of feces was suspended in 0.5 ml of normal saline and 200 μl was inoculated into MacConkey agar. Only one colony per colonial morphology and resistance phenotype from each person was selected for further analysis. Preliminary susceptibility testing was performed by using the automated PASCO (Difco, Detroit, MI) or WIDER (Fco. Soria Melguizo, Madrid, Spain) system. Bacterial identification was performed using these commercial systems and/or standard biochemical tests.
Clonal and phylogenetic analysis of E. coli isolates.
Chromosomal DNA was prepared as previously described using XbaI as a restriction enzyme (Amersham Life Sciences, Uppsala, Sweden) (18). DNA fragments were separated by electrophoresis in 1.2% agarose gels (pulsed-field agarose certified; Bio-Rad, Hemel Hempstead, United Kingdom) and 0.5× Tris-borate-EDTA buffer by using a contour-clamped homogeneous electric field (CHEF-DRIII system; Bio-Rad) and the following electrophoresis conditions: 12°C at 6 V/cm for 27 h with pulse times ranging from 10 to 40s. Clonal relationships were established following criteria by Tenover et al. (38).
A multiplex PCR assay described by Clermont et al. (8) was performed to assign phylogenetic groups among E. coli isolates. In this analysis, three pairs of primers were used in order to amplify the chuA and yjaA genes and an anonymous DNA fragment, which have been found to be specific phylogenetic group markers (8, 15) (Table 1).
TABLE 1.
Primers sequences and PCR conditions used in this study
| Primer | Oligonucleotide sequence (5′ to 3′) | GenBank accession no. or reference (gene) | Position of amplicon | PCR conditions | Reference(s) |
|---|---|---|---|---|---|
| TEM-F | ATA AAA TTC TTG AAG AC | 36 (blaTEM-1) | 208-228 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C; 1 cycle of 10 min at 72°C | 30 |
| TEM-R | TTA CCA ATG CTT AAT CA | 1075-1055 | |||
| SHV-F | GGG TTA TTC TTA TTT GTC GC | M59181 (blaSHV-1) | 58-77 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 56°C, 1 min at 72°C; 1 cycle of 10 min at 72°C | 30 |
| SHV-R | TTA GCG TTG CCA GTG CTC | 988-971 | |||
| CTX-M-9-F | GTG ACA AAG AGA GTG CAA CGG | AF174129 (blaCTXM-9 in In60) | 6339-6359 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 62°C, 1 min at 72°C; 1 cycle of 10 min at 72°C | 9, 33 |
| CTX-M-9-R | ATG ATT CTC GCC GCT GAA GCC | 7195-7175 | |||
| CTX-M-10-F | CCG CGC TAC ACT TTG TGG C | AY598759 (blaCTXM-10 in pRYC21) | 4924-4942 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C; 1 cycle of 10 min at 72°C | 9, 25 |
| CTX-M-10-R | TTA CAA ACC GTT GGT GAC G | 5885-5867 | |||
| ChuA.1 | GAC GAA CCA ACG GTC AGG AT | 1 cycle of 12 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 59°C, 30 s at 72°C; 1 cycle of 7 min at 72°C | 8 | ||
| ChuA.2 | TGC CGC CAG TAC CAA AGA CA | ||||
| YjaA.1 | TGA AGT GTC AGG AGA CGC TG | 8 | |||
| YjaA.2 | ATG GAG AAT GCG TTC CTC AAC | ||||
| TspE4C2.1 | GAG TAA TGT CGG GGC ATT CA | AF222188 | 8-28 | 8 | |
| TspE4C2.2 | CGC GCC AAC AAA GTA TTA CG | 160-141 | |||
| IntI1-F | GGT CAA GGA TCT GGA TTT CG | U49101 (int1) | 786-766 | 1 cycle of 12 min at 94°C; 30 cycles of 30 s at 94°C, 30s at 62°C, 1 min at 72°C; 1 cycle of 8 min at 72°C | 23 |
| IntI1-R | ACA TGC GTG TAA ATC ATC GTC | 303-324 | |||
| 5′CS | GGC ATC CAA GCA GCA AG | M73819 (class 1 integron) | 1190-1206 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 60°C, 2 min at 72°C; 1 cycle of 10 min at 72°C | 21 |
| 3′CS | AAG CAG ACT TGA CCT GA | 1342-1326 | |||
| IntI2-F | CAC GGA TAT GCG ACA AAA AGG T | L10818 (int2) | 219-240 | Same as for int1 | 23 |
| IntI2-R | GTA GCA AAC GAG TGA CGA AAT G | 1007-986 | |||
| attI2-F | GAC GGC ATG CAC GAT TTG TA | AY183453 (class 2 integron) | 1943-1962 | 1 cycle of 12 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 58°C, 3.5 min at 72°C; 1 cycle of 10 min at 72°C | This work |
| orfX-R | GAT GCC ATC GCA AGT ACG AG | 4848-4928 | 42 | ||
| IntI3-F | AGT GGG TGG CGA ATG AGT G | D50438 (int3) | 178-196 | 1 cycle of 12 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 60°C, 1 min at 72°C; 1 cycle of 8 min at 72°C | 12 |
| IntI3-R | TGT TCT TGT ATC GGC AGG TG | 777-758 |
Antimicrobial susceptibility and ESBL identification.
Susceptibility to 13 non-β-lactam antibiotics was determined by the standard disk diffusion method following NCCLS guidelines (24). Disks were purchased from OXOID (Basingstoke, England). All intermediate-susceptible strains were considered nonsusceptible strains. The presence of an ESBL phenotype was presumptively determined by the standard double-disk synergy test. Characterization of ESBL was performed by isoelectric focusing, amplification of bla genes by PCR using primers and conditions showed in Table 1, and further sequencing of PCR products.
Conjugation of ESBL-producing isolates was performed by broth and/or filter mating standard methods using E. coli strain BM21 (nalidixic acid resistant, lactose fermentation positive, and plasmid free) or E. coli strain BM21R (a rifampin-resistant mutant of E. coli BM21) as the recipient (9).
Detection and characterization of class 1, 2, and 3 integrons.
Detection of class 1 and class 2 integrons was performed by PCR using genomic DNA from wild-type strains and the corresponding transconjugants as the template. The primers used for detection and characterization of integrons are shown in Table 1. For class 1 integrons, two primer sets were used: IntI1-F/IntI1-R for amplifying the intI1 gene (23) and 5′CS/3′CS for amplifying the integron variable region-containing gene cassettes (21). For class 2 integrons, the primers used were IntI2-F/IntI2-R for amplifying the intI2 gene (23) and attI2-F/orfX-R (this study; 42) to characterize the gene cassette arrays in class 2 integrons. The primers attI2-F and orfX-R bind, respectively, to attI2 and to orfX, which is situated within Tn7 in a position just downstream of the cassette region. PCR products were separated by electrophoresis on 0.8% (wt/vol) agarose gels and were visualized under UV light after staining with ethidium bromide. The presence of class 3 integrons was determined by dot blot hybridization. Chromosomal DNA was transferred to a Hybond N+ nylon membrane (Amersham Life Science, Arlington Heights, Ill.) and hybridized to a intI3 probe generated by PCR using genomic DNA from Klebsiella pneumoniae FFUL 22K as the template. Labeling and detection were performed with the ECL Random Prime labeling and detection system (Amersham Life Science) following the manufacturer's instructions.
Typing of each class 1 and class 2 integron was performed by restriction fragment length polymorphism (RFLP) analysis. PCR products corresponding to the amplification of the 5′CS-3′CS region of class 1 integrons, and of the attI2-orfX region of class 2 integrons, were purified using a QIAquick PCR purification kit (QIAgen, Hilden, Germany) and further digested with AluI or HaeIII, respectively. The fragments obtained were separated in a 2.5% agarose gel and visualized under UV light after staining with ethidium bromide. Integron types were designed by roman numerals. The subindex indicates the class to which each integron belongs. Amplified DNA fragments corresponding to the variable regions of each distinct class 1 and class 2 RFLP type integrons were sequenced.
Statistic analysis.
Statistic significance for comparison proportions was calculated by the chi-square test (P < 0.05 was considered to be statistically significant).
RESULTS
Epidemiological background of E. coli isolates.
Distribution of the studied isolates among the four E. coli phylogenetic groups is shown in Table 2. Groups B2 and D (associated with extraintestinal pathogenic E. coli) were the most represented among strains associated with the hospital setting (66%), and groups A and B1 (more related to animal or human commensal strains) were the most represented among those of healthy volunteers (68%) (P < 0.001) (10, 15). C-ESBL E. coli isolates mainly corresponded to genogroup D (50%; n = 26/52 isolates), whereas most C-nESBL isolates belonged to the genogroup B2 (44%; n = 19/43 isolates) and most V-nESBL belonged to the genogroup A (50%, n = 19/38). Differences in the ecovar composition of C-ESBL and C-nESBL E. coli clinical isolates regarding the occurrence of the B2 and D groups could be due to the bias created by the origin of the isolates. Incidences of phylogenetic groups A and B1 were similar between ESBL and non-ESBL E. coli clinical isolates (21% versus 28% and 10% versus 9%, respectively).
TABLE 2.
Prevalence of class 1 and class 2 integrons among E. coli isolates from patients at the Hospital Universitario Ramón y Cajal and from healthy volunteers
| E. coli isolate group (no. of PFGE types) | Class 1
|
Class 2
|
Classes 1 + 2 | Strains without integrons | Total % of integrons | Phylogenetic group
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| int1+ | Gene cassettes as part of class 1 integrons | int2+ | Gene cassettes as part of class 2 integrons | A | B1 | B2 | D | ||||
| C-ESBL (52) | 67a (35/52)b | 62 (32/52) | 13 (7/52) | 13 (7/52) | 8 (4/52) | 27 (14/52) | 73 (38/52) | 21 (11/52) | 10 (5/52) | 19 (10/52) | 50 (26/52) |
| C-nESBL (43) | 40 (17/43) | 33 (14/43) | 12 (5/43) | 12 (5/43) | 2 (1/43) | 51 (22/43) | 49 (21/43) | 28 (12/43) | 9 (4/43) | 44 (19/43) | 19 (8/43) |
| V-nESBL (38) | 26 (10/38) | 21 (8/38) | 18 (7/38) | 16 (6/38) | 8 (3/38) | 63 (24/38) | 37 (14/38) | 50 (19/38) | 18 (7/38) | 11 (4/38) | 21 (8/38) |
| Total (133) | 47 (62/133) | 41 (54/133) | 14 (19/133) | 14 (18/133) | 6 (8/133) | 45 (60/133) | 55 (73/133) | 32 (42/133) | 12 (16/133) | 25 (33/133) | 32 (42/133) |
Percentage of isolates.
Number of isolates/total.
Four different ESBL groups were identified among the C-ESBL group: (i) TEM type (31%), (ii) SHV type (17%), (iii) CTX-M-9/14 (42%), and (iv) CTX-M-10 (10%). ESBL-encoding genes were transferred by conjugation to suitable recipients by strains producing TEM (81%), SHV (100%), CTX-M-9/14 (86%), and CTX-M-10 (100%).
E. coli isolates and antibiotic resistance.
The percentages of isolates nonsusceptible to sulfonamide were similar among C-ESBL and C-nESBL isolates and higher than that among V-nESBL isolates (75 to 77% versus 45%, respectively). However, differences in the incidence of nonsusceptibility to other antimicrobials were observed for the two groups of clinical isolates studied, C-ESBL and C-nESBL: streptomycin (73% versus 53%), trimethoprim (60% versus 37%), gentamicin (31% versus 7%), kanamycin (35% versus 12%), chloramphenicol (33% versus 21%), ciprofloxacin (48% versus 28%), and nalidixic acid (56% versus 35%). The occurrence of nonsusceptibility among fecal E. coli V-nESBL isolates for the antibiotics mentioned above was not significantly lower than that obtained for the C-nESBL isolates, except for chloramphenicol (5%), ciprofloxacin (11%), and nalidixic acid (24%).
Prevalence of integrons in C-ESBL versus C-nESBL and V-nESBL isolates.
Prevalence of integrons among the E. coli groups studied is shown in Table 2. Class 1 integrons were more frequently found among C-ESBL (67%) than among C-nESBL (40%) or V-nESBL (26%) E. coli strains (P < 0.001). Among C-ESBL isolates, class 1 integrons were mainly associated with isolates harboring blaCTX-M-9/14 (95%) or blaSHV (67%) and less with blaTEM (50%) or blaCTX-M-10 (0%). Conjugative transfer of an integron with the genetic elements carrying ESBL bla genes was observed more frequently for CTX-M-9/14 (95%) than for SHV (33%)-, TEM (23%)-, or CTX-M-10 (0%)-producing E. coli. A number of C-ESBL-producing E. coli isolates carried more than one class 1 integron (10%, n = 5/52).
The occurrence of class 2 integrons was similar among clinical C-ESBL or C-nESBL and commensal V-nESBL (13%, 12%, and 18%, respectively) E. coli isolates. Within the C-ESBL group, these genetic elements were more common among isolates containing blaTEM (19%) than blaCTX-M-9/14 (14%) or blaSHV (11%), whereas they were absent among isolates containing blaCTX-M-10 (0%). Class 2 integrons were not cotransferred with ESBL bla genes in any case. Simultaneous presence of class 1 and class 2 integrons was detected in all E. coli groups (2 to 8%). Gene cassettes were not detected for a number of isolates harboring intI1 (13%, n = 8/62) or intI2 (5%, n = 1/19).
Class 1 integrons were found among all four E. coli phylogenetic groups at similar rates (40 to 56%). Class 2 integrons were more common among E. coli ecovar D (26%) than among strains of the B1 (13%), A (12%), or B2 (3%) groups (Table 3). Class 3 integrons were not detected in any of the isolates studied.
TABLE 3.
Distribution of class 1 and class 2 integrons among different E. coli phylogenetic groups
| Phylogenetic group | Presence of integrons
|
|||
|---|---|---|---|---|
| Class 1 (n = 62) | Class 2 (n = 19) | Classes 1 + 2 (n = 8) | No integrons (n = 60) | |
| A (n = 42) | 40a (17/42)b | 12 (5/42) | 2 (1/42) | 50 (21/42) |
| B1 (n = 16) | 56 (9/16) | 13 (2/16) | 13 (2/16) | 44 (7/16) |
| B2 (n = 33) | 42 (14/33) | 3 (1/33) | 3 (1/33) | 58 (19/33) |
| D (n = 42) | 52 (22/42) | 26 (11/42) | 10 (4/42) | 31 (13/42) |
Percentage of isolates.
Number of isolates/total.
The prevalence of class 1 integrons in C-ESBL isolates dramatically increased along the studied period from 30% during the 1988 to 1995 period to 87% during 1996 to 1998 and reached 70% in 1999 to 2000. Among C-nESBL isolates, these elements increased from 36% (1996 to 1998) to 41% (1999 to 2000). Class 2 integrons also increased over time.
Diversity of E. coli integrons.
Seven different class 1 integrons were identified (Table 4). Types I1, II1, and IV1 were the most prevalent groups. Type IV1, which included a dfrA16 and an aadA2 gene cassette, was the most frequently found among C-ESBL clones, due to the high prevalence of blaCTX-M-9, which is located on In60, an unusual integron that contains the first 5′CS-3′CS region corresponding to type IV1 (33; T. M. Coque, M. C. Varela, A. Oliver, E. Machado, J. C. Galán, F. Baquero, and R. Cantón, abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstract C2-646, 2002). The gene cassettes most commonly found in our collection were those coding for aminoglycoside and/or trimethoprim resistances. Three class 2 integrons were detected, and their sequences were identical to those described for Tn1826 (Type I2), Tn7 (Type II2), and Tn1825 (Type III2) (Table 4 and Fig. 1) (3). Type II2 was the most frequent and widely distributed. Type I2 and type III2 were only found among isolates of the V-nESBL and C-nESBL groups, respectively.
TABLE 4.
Class 1 and class 2 integrons found among E. coli isolates from clinical specimens (ESBL and non-ESBL) and from healthy volunteers
| RFLP type | Length of variable region (bp) | Gene cassettes and order | Resistance phenotypea | No. of isolates | C-ESBL (n = 52)
|
C-nESBL (n = 43) | V-nESBL (n = 38) | Isolation date | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM | SHV | CT-X-M-9/14 | CTX-M-10 | ||||||||
| Class 1 integrons | |||||||||||
| I1 | 1,000 | aadA1 | Sm Sp | 15 | 0 | 4 | 3 | 0 | 6 | 2 | 1988-2001 |
| II1 | 1,500 | dhfrA1-aadA1 | Tmp Sm Sp | 12 | 1 | 2 | 2 | 0 | 5 | 2 | 1997-2001 |
| III1 | 1,800 | dhfrA12-orfF-aadA2 | Tmp Sm Sp | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1997 |
| IV1 | 1,500 | dhfrA16-aadA2 | Tp Sm Sp | 13 | 0 | 0 | 13 | 0 | 0 | 0 | 1996-2000 |
| V1 | 950 | aadA2 | Sm Sp | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1998 |
| VI1 | 1,500 | dhfrA17-aadA5 | Tp Sm Spc | 5 | 1 | 0 | 0 | 0 | 3 | 1 | 1997-2001 |
| VII1 | 800 | aac(6′)-Ib" | Km Ak Tb Nt Gm | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 2000-2001 |
| Class 2 integrons | |||||||||||
| I2 | 1,600 | sat-aadA1-orfX | Str Sm Sp | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 2001 |
| II2 | 1,900 | dhfrA1-sat-aadA1-orfX | Tp Str Sm Sp | 15 | 3 | 0 | 3 | 0 | 5 | 4 | 1991-2001 |
| III2 | 2,300 | orf-sat-aadA1-orfX | Str Sm Sp | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2000 |
Sm, streptomycin; Sp, spectinomycin; Tp, trimethoprim; Km, kanamycin; Ak, amikacin; Tb, tobramycin; Nt, netilmicin; Gm, gentamicin; Str, streptotricin.
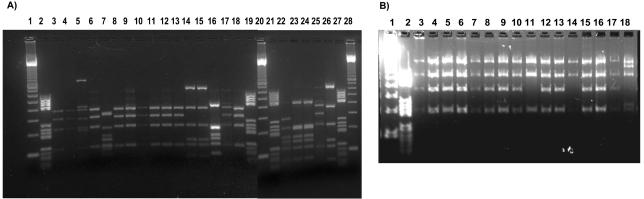
FIG. 1.
RFLP analysis of class 1 and class 2 integrons. (A) AluI digest of class 1 integron DNA from E. coli isolates: lanes 1, 20, and 28, DNA molecular weight marker λ-EcoT14I/BglII digest (Takara Bio Inc.); lanes 2, 19, 21, and 27, DNA molecular weight marker III (Roche Diagnostics); anes 3, 4, 6, 8 to 13, 17, 18, and 25, type I1 (aadA1); lane 7, type III1 (dhfrA12-orfF-aadA2); lanes 14, 15, and 26, type II1 (dhfrA1-aadA1); lanes 16, 23, and 24, type IV1 (dhfrA16-aadA2); lane 22, type V1 (aadA2). (B) HaeII digest of class 2 integron DNA from E. coli isolates: lane 1, DNA molecular weight marker λ-EcoT14I/BglII digest (Takara Bio Inc.); lane 2, DNA molecular weight marker III (Roche Diagnostics); lanes 3 to 10, 12, 13, and 15 to 17, type II2 (dhfrA1-sat-aadA1-orfX); lanes 11 and 14, type I2 (sat-aadA1-orfX); lane 18, type III2 (orf-sat-aadA1-orfX).
The association between antibiotic resistance phenotypes and class 1 or class 2 integrons is shown in Table 5. Integrons were frequently found in resistant strains, although the presence of gene cassettes coding for a particular antibiotic resistance was only demonstrated for trimethoprim, sulfonamide, streptomycin, spectinomycin, amikacin, gentamicin, or kanamycin. Nevertheless, genes coding for resistance to the above antibiotics were not always linked to class 1 integrons. Strains susceptible to trimethoprim, sulfonamide, streptomycin, gentamicin, kanamycin, chloramphenicol, ciprofloxacin, and nalidixic acid did not carry class 1 or 2 integrons.
TABLE 5.
Association between antibiotic resistance and presence of class 1 or class 2 integrons among E. coli isolates from clinical and community environments
| Antibiotica | No. of resistant strains | No. of strains with integrons | Class 1
|
Class 2
|
No. of strains without integrons | ||
|---|---|---|---|---|---|---|---|
| intI1+b | Resistance linked to class 1 integronsc | intI2+b | Resistance linked to class 2 integronsc | ||||
| Tp | 59 | 58 | 51 | 29 | 15 | 15 | 1 |
| Su | 89 | 67 | 61 | 61 | 14 | 0 | 22 |
| Sm | 82 | 60 | 50 | 34 | 18 | 18 | 22 |
| Gm | 21 | 15 | 13 | 2 | 6 | 0 | 6 |
| Km | 26 | 21 | 19 | 1 | 5 | 0 | 5 |
| Cm | 28 | 23 | 23 | 0 | 2 | 0 | 5 |
| Cp | 41 | 33 | 29 | 0 | 9 | 0 | 8 |
| Na | 53 | 39 | 34 | 0 | 11 | 0 | 14 |
Tp, trimethoprim; Su, sulfonamides; Sm, streptomycin; Gm, gentamicin; Km, kanamycin; Cm, chloramphenicol; Cp, ciprofloxacin; Na, nalidixic acid.
Number of isolates containing the intI1 or intI2 gene.
Number of isolates with gene cassettes encoding resistance to the corresponding antibiotic.
DISCUSSION
In this study, comparative information is provided about the evolution of integron content of ESBL and non-ESBL-producing E. coli isolates over more than a decade in a single hospital. The previously described wide dissemination of integrons among both clinical and commensal E. coli human strains (11, 19, 23) and the increase of the class 1 integrons rate among nosocomial isolates during the last decade (34, 43) was confirmed.
Class 1 integrons were more frequently found among E. coli isolates associated with the clinical setting than among fecal E. coli isolates from healthy human volunteers, suggesting a linkage with particular strains and/or transferable genetic elements in the hospital. In fact, class 1 integrons are frequent in our series among ESBL-producing clinical isolates because of the high proportion of strains containing blaCTX-M-9, a gene linked to In60, an In6-like class 1 integron (28, 33). CTX-M-9 is one of the most common ESBLs found in Spain, and it has been increasingly recovered from E. coli since its first description in 1996 (14, 33). Without this bias, the distribution of class 1 integrons could be considered similar among C-ESBL and C-nESBL isolates (47% versus 40%).
Previous studies have shown the association of ESBL genes with plasmids from bacteria responsible for nosocomial outbreaks and associated with class 1 integrons (39). However, we found a relatively low occurrence of class 1 integrons within different plasmids carrying blaTEM or blaSHV genes (23% and 33%, respectively) and the lack of any integron element on blaCTX-M-10-producing isolates. Conversely, the simultaneous presence of the same class 1 integron types among C-ESBL and C-nESBL isolates recovered from the same hospital wards might indicate a wide dissemination of specific structures in which integrons are located. Indeed, there is a certain specificity of integrons for particular dispersive units (28, 29, 35, 37, 39). If class 1 integrons are more frequently found among hospital clinical isolates, there is a similar occurrence of class 2 integrons among clinical isolates and fecal human isolates (13 versus 18%) from healthy people, suggesting the current absence of privileged selection for strains carrying class 2 integrons in the nosocomial setting.
A great diversity of ARI has been reported in different environments, and changes in the gene cassette content as a result of different genetic events have been described under high antibiotic selective pressure (26, 37, 43). However, our results indicate a low diversity and stability of class 1 integrons, in agreement with other European studies (22). The most common class 1 integrons were types I1, II1, and IV1. Type I1, related to that of Tn21, and type II1 were also the most common integrons found among isolates from different continents (21-23, 28, 42). Type IV1 corresponds to the first 5′CS/3′CS part of In60, in which blaCTX-M-9 is located, and also to the first 5′CS/3′CS part of In36, containing the qnr gene widely disseminated in China (33, 41). Regarding class 2 integrons, we found three out of the five known class 2 integrons, which indicates the occurrence of types other than the widely disseminated Tn7, such as Tn1825 or Tn1826 (3, 42).
The presence of integrons was independently associated with resistance to trimethoprim, sulfonamide, or streptomycin, in agreement with previous studies (20, 22). Resistance to trimethoprim and sulfonamides is usually determined in integrons (11, 32), but in our series we could not detect integrons in a small proportion of trimethoprim-resistant strains or in 25% of sulfonamide-resistant isolates. We cannot discard the presence of unusual class 1 integron structures escaping classical amplification procedures, or of sul2 or sul3 genes, not located in class 1 integrons (13). The finding of a similar rate of nonsusceptibility to sulfonamides among both groups of clinical E. coli isolates was surprising, since the occurrence of class 1 integrons in C-ESBL isolates was much higher than among C-nESBL isolates. Again, that could be explained by a higher frequency of sul2 and sul3 genes in strains belonging to ecovar B2 (13), predominant in the C-nESBL group. The aad genes were also extremely common in our series and were never located at the first position in the integron platform when other gene cassettes were present, suggesting its earlier recruitment by the element (26). Apart from sulfonamides, trimethoprim, or streptomycin-spectinomycin resistance, we only found gene cassettes coding for kanamycin and gentamicin resistance.
As in other recent studies, resistance to quinolones was more common among integron-containing strains (20). This association could be explained in part by the putative presence of the qnr gene that may be located on an In6-like class 1 integron (41). However, the absence of ORF513 in such strains precludes the presence of this gene as part of an In6-like structure (unpublished results). Finally, most chloramphenicol-resistant strains contained a class 1 integron (82%). Whether genes coding for resistance to this antibiotic are part of an unusual class 1 integron or part of individual cassettes integrated in the chromosome as previously described in old studies deserves further research.
Our results indicate that the selection and spread of genetic elements encoding ESBL has not a major role for integron dispersal, except when blaESBL genes are within an integron platform, such as blaCTX-M-9. In addition, the contribution of integrons to ESBL dissemination seems to be of small significance, as the encoded resistances currently correspond to old antibiotics without significant intensity of selective pressure in the current clinical setting. This situation might eventually change, following possible events of selection of specific broad-spectrum plasmids able to capture integrons (7, 35), or by integron capture of determinants encoding resistances to antibiotics frequently used in the hospital environment (41). Surveillance of the integron content of nosocomial E. coli populations may be critical to predict and prevent the spread of particular antibiotic resistance determinants.
Acknowledgments
Elisabete Machado was supported by a fellowship from Fundação para a Ciência e Tecnologia de Portugal (SFRH/BD/11304/2002). This work was partially supported by research grants from the Fondo de Investigaciones Sanitarias, Ministerio de Sanidad of Spain (FIS 01/412), Ministerio de Ciencia y Tecnología of Spain (SAF 2003-09285), and the European Commission (grant SLMM-CT-2003-503335).
We thank Aida Duarte (University of Lisbon, Lisbon, Portugal), John Maurer (University of Georgia, Athens), Carmen Mendoza (University of Oviedo, Oviedo, Spain), and Hatch Stokes (Macquarie University, Sydney, Australia) for kindly providing control strains for different class 1, 2, and 3 integrons. We also thank Mary Harper for assistance with the English corrections of the manuscript.
Teresa M. Coque and Luisa Peixe are coadvisors of E.M.'s Ph.D. thesis.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallobetalactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquero, F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510-518. [DOI] [PubMed] [Google Scholar]
- 3.Birski, L., and D. Mazel. 2003. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 47:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantón, R., T. M. Coque, and F. Baquero. 2003. Multi-resistant Gram-negative bacilli: from epidemics to endemics. Curr. Opin. Infect. Dis. 16:315-325. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli, A., L. Villa, C. Pezzella, E. Bordi, and P. Visca. 2001. Expanding drug resistance through integron acquisition by IncFI plasmids of Salmonella enterica Typhimurium. Emerg. Infect. Dis. 7:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque, T. M., A. Oliver, J. C. Perez-Diaz, F. Baquero, and R. Canton. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventré, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 11.Fluitz, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein, C., M. S. Lee, S. Sanchez, et al. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grape, M., L. Sundstrom, and G. Kronvall. 2003. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 52:1022-1024. [DOI] [PubMed] [Google Scholar]
- 14.Hernández, J. R., L. Martínez-Martínez, R. Cantón, T. M. Coque, A. Pascual, and the Spanish Group for Nosocomial Infections (GEIH). 2005. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Antimicrob. Agents Chemother. 49:••••-••••. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochhut, B., Y. Lofti, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, G. A., and L. Sutton. 1991. Properties of plasmids responsible for production of extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis, p. 17-31. In N. Woodford and A. P. Johnson (ed.), Methods in molecular medicine, vol 15. Molecular bacteriology: protocols and clinical applications. Humana Press Inc., Totowa, N.J. [Google Scholar]
- 19.Leverstein-van Hall, M. A., A. Paauw, A. T. A. Box, H. E. M. Blok, J. Verhoef, and A. C. Fluit. 2002. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leverstein-van Hall, M. A., H. E. M. Blok, R. T. Donders, A. Paauw, A. C. Fluit, and J. Verhoef. 2003. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187:251-259. [DOI] [PubMed] [Google Scholar]
- 21.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Freijo, P., A. C. Fluit, F.-J. Schmitz, J. Verhoef, and M. E. Jones. 1999. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2000. Methods for diffusion disk antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Oliver, A., T. M. Coque, D. Alonso, A. Valverde, F. Baquero, and R. Cantón. 2005. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49:1567-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partridge, S. R., C. Collis, and R. M. Hall. 2002. Class 1 integrons containing a new cassette, aadA10, with Tn1404 from R151. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston, K. E., E. M. Graffunder, A. M. Evans, and R. A. Venezia. 2003. Survey of plasmid-associated genetic markers in Enterobacteriaceae with reduced susceptibilities to cephalosporins. Antimicrob. Agents Chemother. 47:2179-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosser, S. J., and H.-K. Young. 1999. Identification and characterization of class 1 integrons in bacteria from the aquatic environment. J. Antimicrob. Chemother. 44:11-18. [DOI] [PubMed] [Google Scholar]
- 32.Rowe-Magnus, D. A., and D. Mazel. 2002. The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 292:115-125. [DOI] [PubMed] [Google Scholar]
- 33.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz, F. J., D. Hafner, R. Geisel, P. Follmann, K. Kirchke, K. Köhrer, J. Verhoef, and A. C. Fluit. 2001. Increased prevalence of class 1 integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J. Clin. Microbiol. 39:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorum, H., T. M. Lábee-Lund, A. Solberg, and A. Wold. 2003. Integron-containing IncU plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennstedt, T., R. Szczepanowski, S. Braun, A. Pühler, and A. Sclüter. 2003. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45:239-252. [DOI] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villa, L., C. Pezzella, F. Tosini, P. Visca, A. Petrucca, and A. Carattoli. 2000. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum beta-lactamase gene and a class 1 integron. Antimicrob. Agents Chemother. 44:2911-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, H. S., J. C. Lee, H. Y. Kang, D. W. Ro, J. Y. Chung, Y. S. Jeong, S. H. Tae, C. H. Choi, E. Y. Lee, S. Y. Seol, Y. C. Lee, and D. T. Cho. 2003. Changes in the gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J. Clin. Microbiol. 41:5429-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]